A Survey of Transposon Landscapes in the Putative Ancient Asexual Ostracod Darwinula stevensoni
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction, Screening and Sequencing of a Genomic Fosmid Library
2.2. Illumina Draft Genome of D. stevensoni
2.3. De Novo Identificiation of Ostracod TEs
2.4. Assessing Insertion Sites of TEs from Fosmid Data
2.5. Estimating Single-Copy Gene Content in Fosmid Sequence Data
2.6. Search for Remote Homologies
3. Results
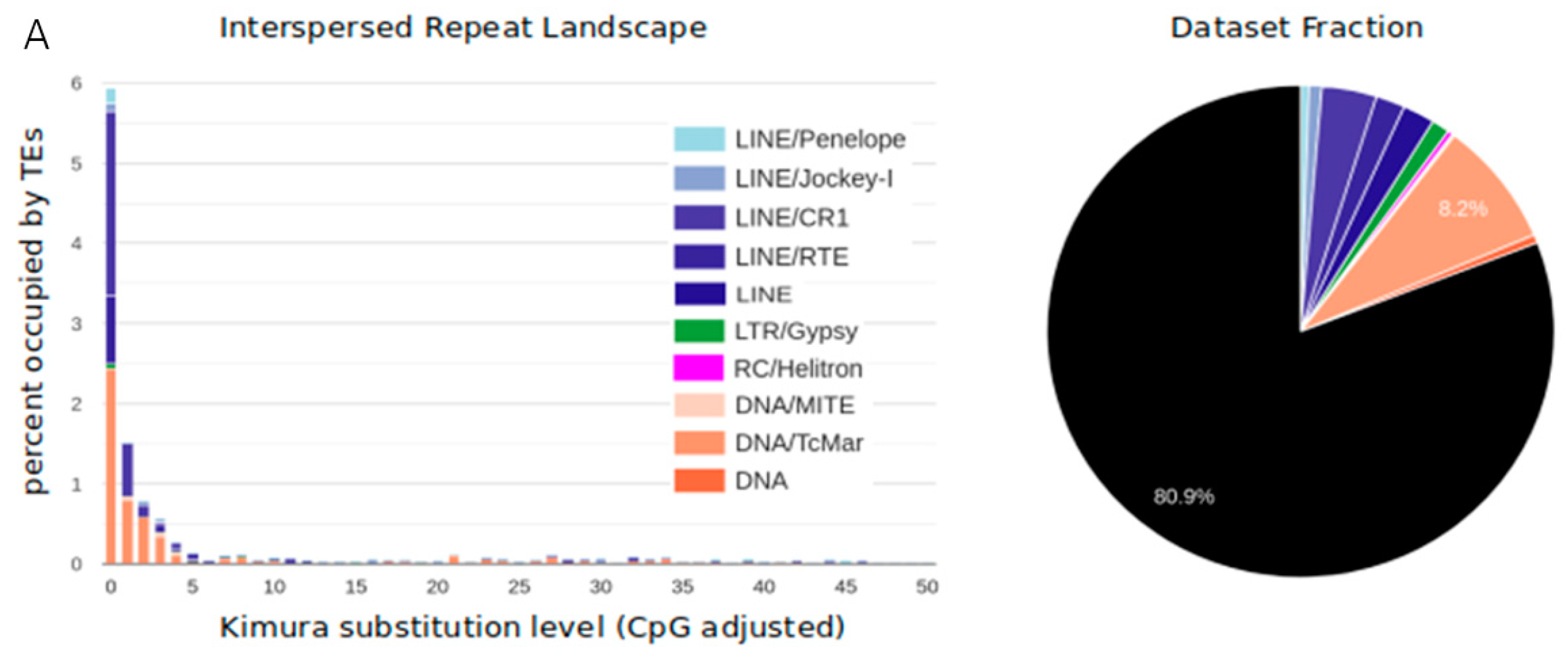
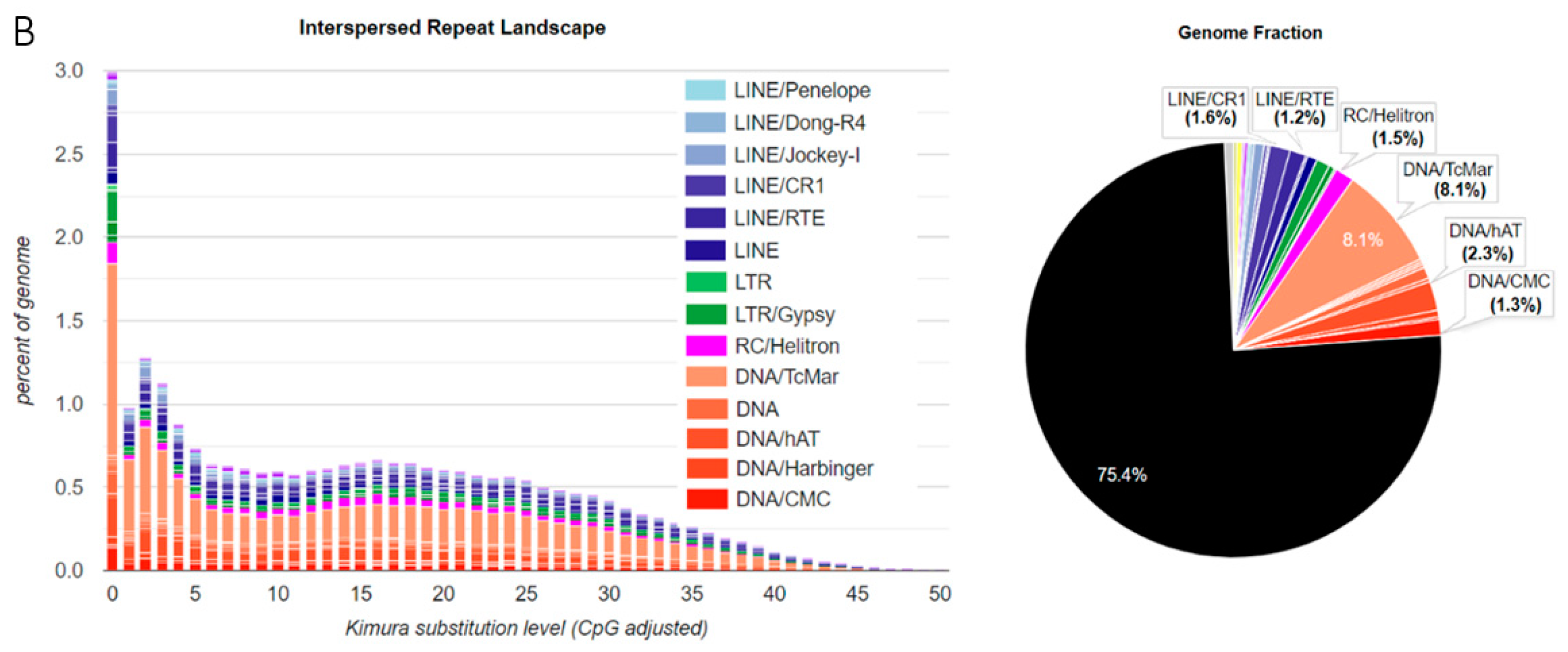
3.1. TE Diversity, Substitution Levels and Abundance
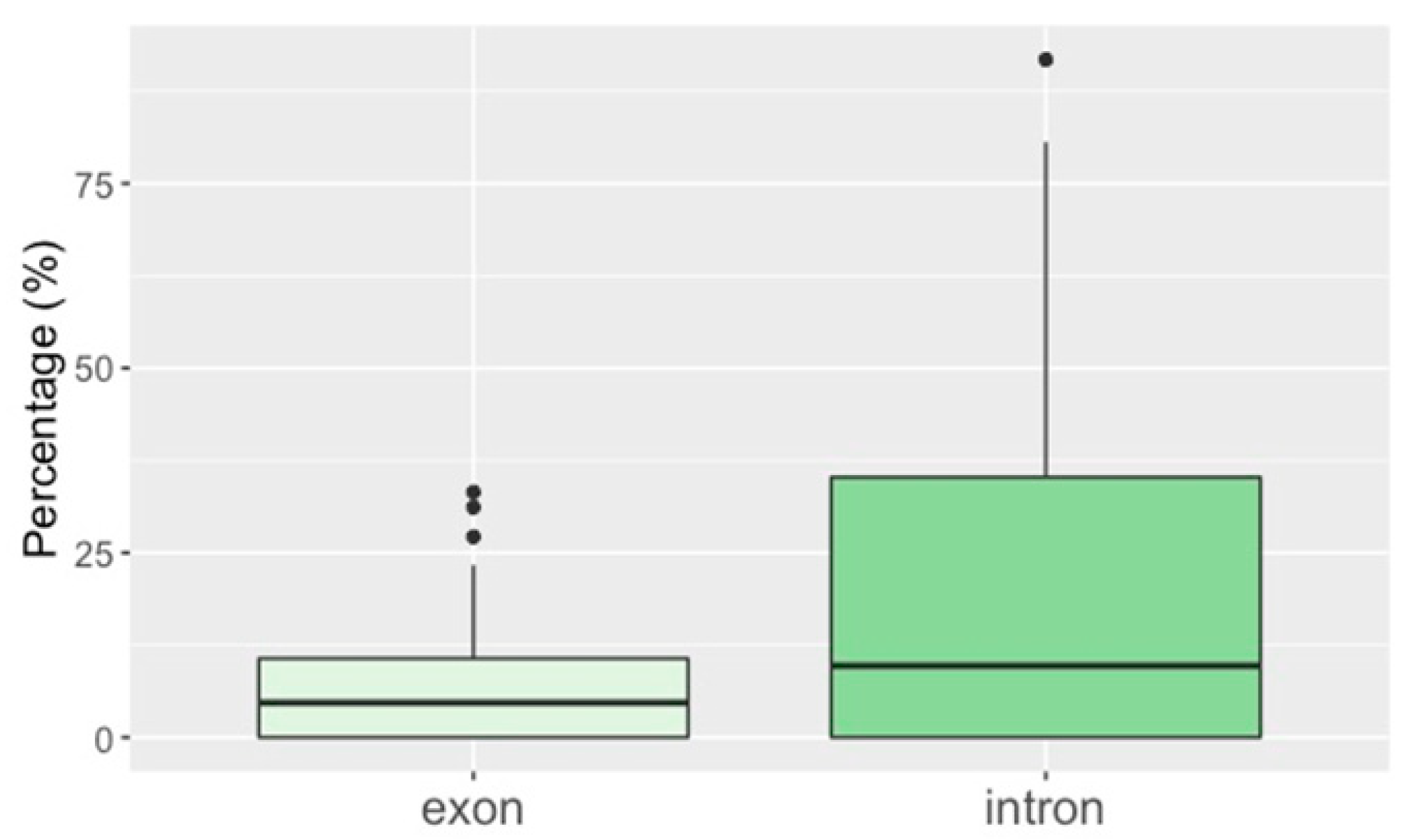
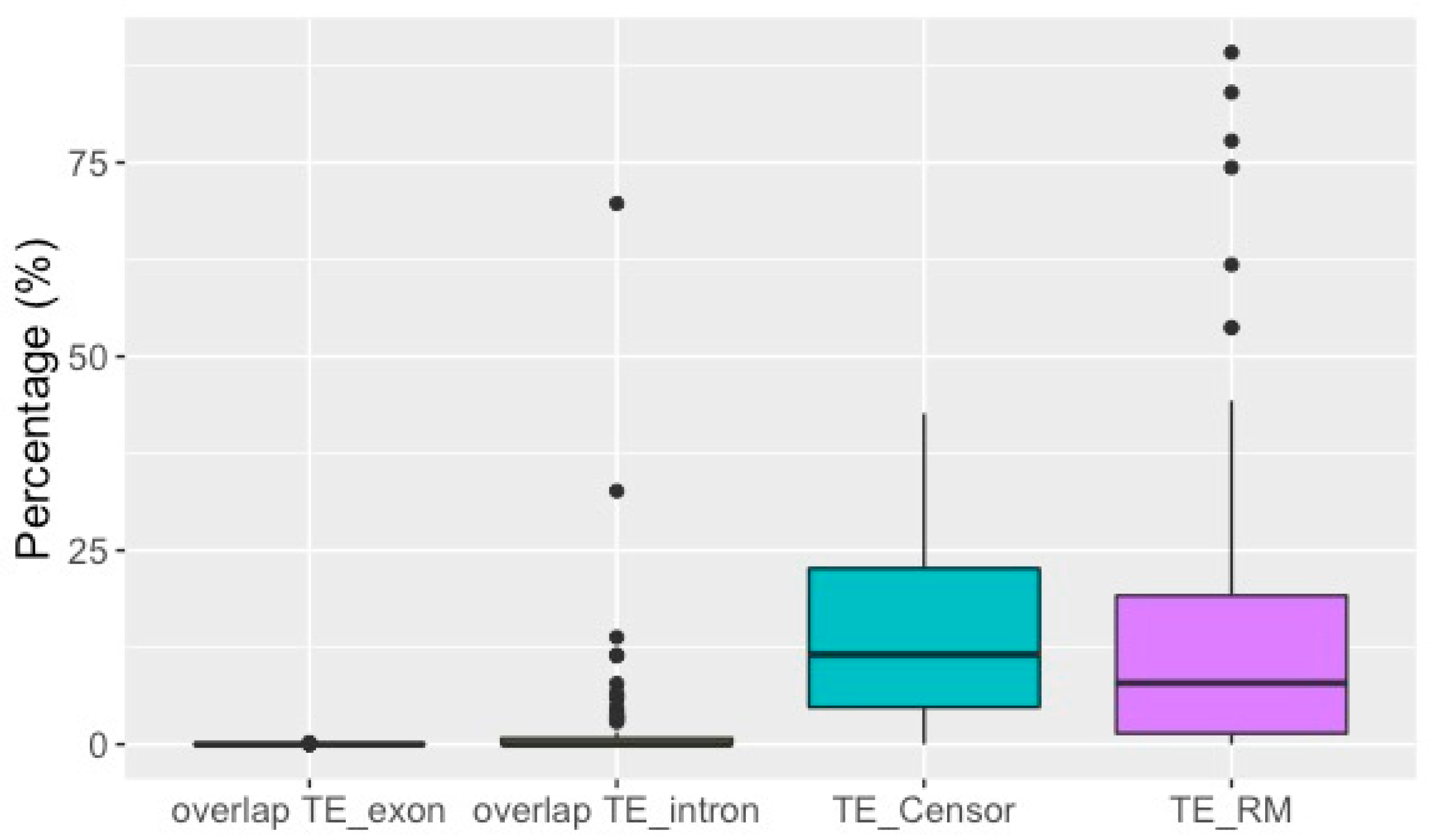
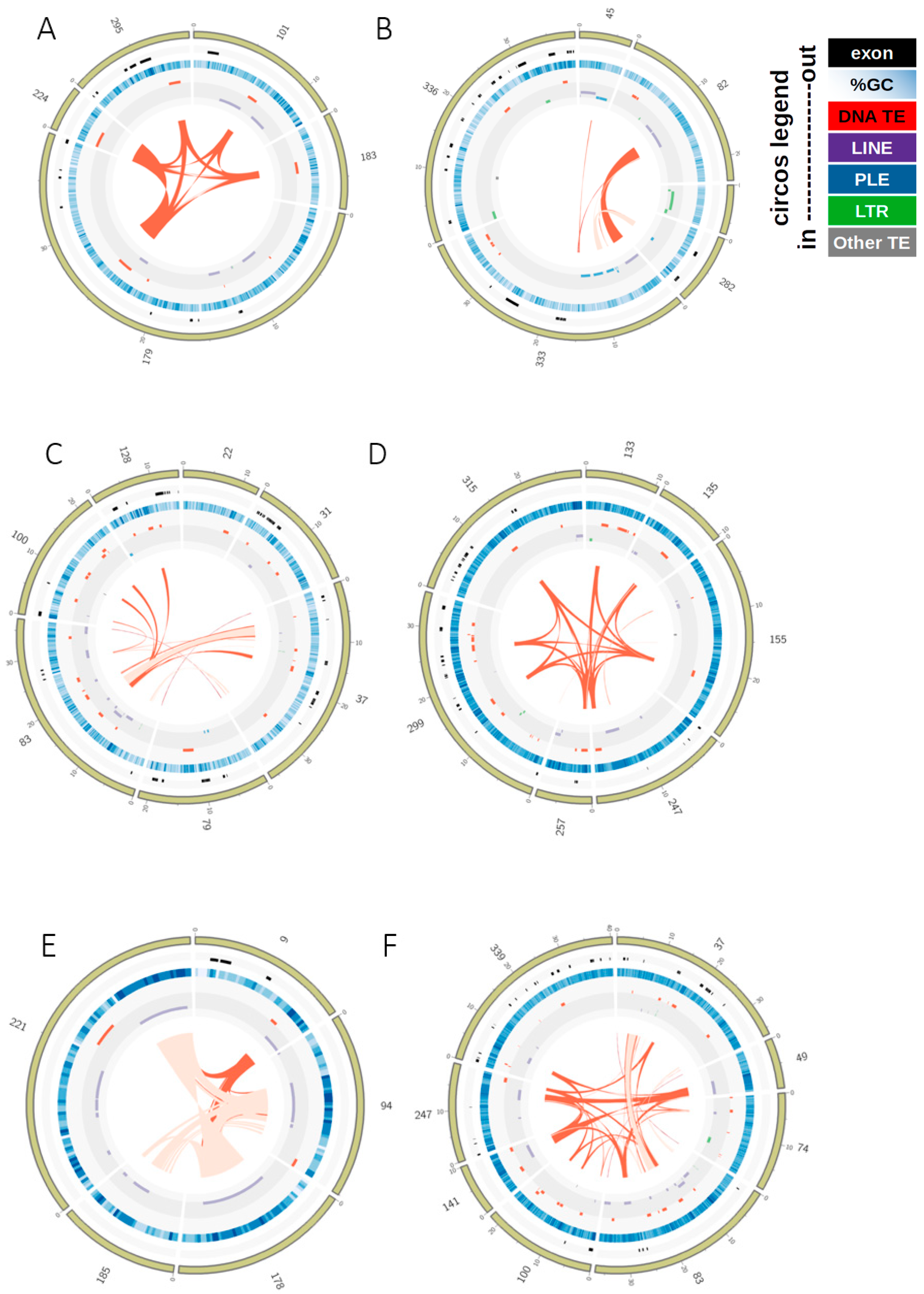
3.2. TE Insertion Sites in Fosmids and Their Relationship to Telomeres and Coding Regions
3.3. Additional Domains in R4/Dong and Gypsy Retrotransposons
4. Discussion
4.1. Comparing Results from the Draft Genome and the Fosmid Library
4.2. TE Diversity and Substitution Levels
4.3. TE abundance
4.4. TE Insertion Sites
4.5. Assessing the Impact of Putatively Ancient Asexual Reproduction on TE Landscapes in Non-Marine Ostracods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hickey, D.A. Selfish DNA: A sexually-transmitted nuclear parasite. Genetics 1982, 101, 519–531. [Google Scholar] [PubMed]
- Zeyl, C.; Bell, G.; Green, D.M. Sex and the spread of retrotransposon Ty3 in experimental populations of Saccharomyces cerevisiae. Genetics 1996, 143, 1567–1577. [Google Scholar] [CrossRef]
- Bast, J.; Jaron, K.S.; Schuseil, D.; Roze, D.; Schwander, T. Asexual reproduction reduces transposable element load in experimental yeast populations. eLife 2019, 8, e48548. [Google Scholar] [CrossRef] [PubMed]
- Kondrashov, A.S. Deleterious mutations and the evolution of sexual reproduction. Nature 1988, 336, 435–440. [Google Scholar] [CrossRef]
- Muller, H.J. The relation of recombination to mutational advance. Mutat. Res. 1964, 1, 2–9. [Google Scholar] [CrossRef]
- Dolgin, E.S.; Charlesworth, B. The fate of transposable elements in asexual populations. Genetics 2006, 174, 817–827. [Google Scholar] [CrossRef]
- Bachtrog, D. Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 2013, 14, 113–124. [Google Scholar] [CrossRef]
- Leung, W.; Shaffer, C.D.; Reed, L.K.; Smith, S.T.; Barshop, W.; Dirkes, W.; Dothager, M.; Lee, P.; Wong, J.; Xiong, D.; et al. Drosophila Muller F elements maintain a distinct set of genomic properties over 40 million years of evolution. G3 Genes Genomes Genet. 2015, 5, 719–7440. [Google Scholar] [CrossRef]
- Lynch, M.; Blanchard, J.L. Deleterious mutation accumulation in organelle genomes. Genetica 1998, 102–103, 29–39. [Google Scholar] [CrossRef]
- Arkhipova, I.; Meselson, M. Transposable elements in sexual and ancient asexual taxa. Proc. Natl. Acad. Sci. USA 2000, 97, 14473–14477. [Google Scholar] [CrossRef]
- Arkhipova, I.; Meselson, M. Deleterious transposable elements and the extinction of asexuals. Bioessays 2005, 27, 76–85. [Google Scholar] [CrossRef]
- Nuzhdin, S.V.; Petrov, D.A. Transposable elements in clonal lineages: Lethal hangover from sex. Biol. J. Linn. Soc. 2003, 79, 33–41. [Google Scholar] [CrossRef]
- Flot, J.F.; Hespeels, B.; Li, X.; Noel, B.; Arkhipova, I.; Danchin, E.G.; Hejnol, A.; Henrissat, B.; Koszul, R.; Aury, J.M.; et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 2013, 500, 453–457. [Google Scholar] [CrossRef]
- Arkhipova, I.R.; Meselson, M. Diverse DNA transposons in rotifers of the class Bdelloidea. Proc. Natl. Acad. Sci. USA 2005, 102, 11781–11786. [Google Scholar] [CrossRef] [PubMed]
- Schaack, S.; Pritham, E.J.; Wolf, A.; Lynch, M. DNA transposon dynamics in populations of Daphnia pulex with and without sex. Proc. R. Soc. B 2010, 277, 2381–2387. [Google Scholar] [CrossRef]
- Rho, M.; Schaack, S.; Gao, X.; Kim, S.; Lynch, M.; Tang, H. LTR retroelements in the genome of Daphnia pulex. BMC Genom. 2010, 11, 425. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Tang, H.; Ye, Z.; Lynch, M. Insertion polymorphisms of mobile genetic elements in sexual and asexual populations of Daphnia pulex. Genome Biol. Evol. 2017, 9, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Bast, J.; Schaefer, I.; Schwander, T.; Maraun, M.; Scheu, S.; Kraaijeveld, K. No accumulation of transposable elements in asexual arthropods. Mol. Biol. Evol. 2016, 33, 697–706. [Google Scholar] [CrossRef]
- Szitenberg, A.; Cha, S.; Opperman, C.H.; Bird, D.M.; Blaxter, M.L.; Lunt, D.H. Genetic drift, not life history or RNAi, determine long-term evolution of transposable elements. Genome Biol. Evol. 2016, 8, 2964–2978. [Google Scholar] [CrossRef] [PubMed]
- Agren, J.A.; Greiner, S.; Johnson, M.T.; Wright, S.I. No evidence that sex and transposable elements drive genome size variation in evening primroses. Evolution 2015, 69, 1053–1062. [Google Scholar] [CrossRef]
- Zeyl, C.; Bell, G.; Dasilva, J. Transposon abundance in sexual and asexual populations of Chlamydomonas reinhardtii. Evolution 1994, 48, 1406–1409. [Google Scholar] [CrossRef]
- Abad, P.; Gouzy, J.; Aury, J.-M.; Castagnone-Sereno, P.; Danchin, E.G.J.; Deleury, E.; Perfus-Barbeoch, L.; Anthouard, V.; Artiguenave, F.; Blok, V.C.; et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 2008, 26, 909–915. [Google Scholar] [CrossRef]
- Kraaijeveld, K.; Zwanenburg, B.; Hubert, B.; Vieira, C.; De Pater, S.; Van Alphen, J.J.M.; Den Dunnen, J.T.; De Knijff, P. Transposon proliferation in an asexual parasitoid. Mol. Ecol. 2012, 21, 3898–3906. [Google Scholar] [CrossRef] [PubMed]
- Simonti, C.N.; Pavlicev, M.; Capra, J.A. Transposable element exaptation into regulatory regions is rare, influenced by evolutionary age, and subject to pleiotropic constraints. Mol. Biol. Evol. 2017, 34, 2856–2869. [Google Scholar] [CrossRef]
- Glémin, S.; François, C.M.; Galtier, N. Genome Evolution in Outcrossing vs. Selfing vs. Asexual Species. Methods Mol. Biol. 2019, 1910, 331–369. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B.; Langley, C. The evolution of self-regulated transposition of transposable elements. Genetics 1986, 112, 359–383. [Google Scholar] [CrossRef] [PubMed]
- Startek, M.; Le Rouzic, A.; Capy, P.; Grzebelu, D.; Gambin, A. Genomic parasites or symbionts? Modeling the effects of environmental pressure on transposition activity in asexual populations. Theor. Popul. Biol. 2013, 90, 145–151. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arkhipova, I.R. Using bioinformatic and phylogenetic approaches to classify transposable elements and understand their complex evolutionary histories. Mob. DNA 2017, 8, 19. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.R.; Burns, K.H.; Boeke, J.D. Active transposition in genomes. Ann. Rev. Genet. 2012, 46, 651–675. [Google Scholar] [CrossRef]
- Peccoud, J.; Loiseau, V.; Cordaux, R.; Gilbert, C. Horizontal transfer of transposons in insects. Proc. Natl. Acad. Sci. USA 2017, 114, 4721–4726. [Google Scholar] [CrossRef]
- Schön, I.; Rossetti, G.; Martens, K. Ancient asexual darwinulids: Ancient scandals or scandalous gossip? In Lost Sex: The Evolutionary Biology of Parthenogenesis; Schön, I., Martens, K., Van Dijk, P., Eds.; Springer Academic Publishers: Dordrecht, The Netherlands, 2009; pp. 217–240. [Google Scholar]
- Heethoff, M.; Norton, R.A.; Scheu, S.; Maraun, M. Parthenogenesis in oribatid mites (Acari, Oribatida): Evolution without sex. In Lost Sex: The Evolutionary Biology of Parthenogenesis; Schön, I., Martens, K., Van Dijk, P., Eds.; Springer Academic Publishers: Dordrecht, The Netherlands, 2009; pp. 241–258. [Google Scholar]
- Schwander, T. Evolution: The end of an ancient asexual scandal. Curr. Biol. 2016, 26, R233–R235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schwander, T.; Lee, H.; Crespi, B.J. Molecular evidence for ancient asexuality in Timema stick insects. Curr. Biol. 2011, 21, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Mark Welch, D.; Ricci, C.; Meselson, M. Bdelloid rotifers: Progress in understanding the success of an evolutionary scandal. In Lost Sex: The Evolutionary Biology of Parthenogenesis; Schön, I., Martens, K., Van Dijk, P., Eds.; Springer Academic Publishers: Dordrecht, The Netherlands, 2009; pp. 259–280. [Google Scholar]
- Martens, K.; Rossetti, G.; Horne, D. How ancient are ancient asexuals? Proc. R. Soc. B 2003, 270, 723–729. [Google Scholar] [CrossRef]
- Straub, E.B. Mikropaläontologische Untersuchungen im Tertiär zwischen Ehingen und Ulm a.d. Donau. Geol. Jahrb. 1952, 66, 433–523. [Google Scholar]
- Schön, I.; Arkhipova, I.R. Two families of non-LTR retrotransposons, Syrinx and Daphne, from the Darwinulid ostracod, Darwinula stevensoni. Gene 2006, 371, 296–307. [Google Scholar] [CrossRef][Green Version]
- Shribak, M. Polychromatic polarization microscope: Bringing colors to a colorless world. Sci. Rep. 2015, 5, 17340. [Google Scholar] [CrossRef]
- Tétart, J. Les garnitures chromosomiques des Ostracodes d’eau douce. Trav. Lab. Hydrobiol. Univ. Grenoble 1978, 69–70, 113–140. [Google Scholar]
- Frydrychová, R.; Grossmann, P.; Trubac, P.; Vítková, M.; Marec, F.E. Phylogenetic distribution of TTAGG telomeric repeats in insects. Genome 2004, 47, 163–178. [Google Scholar] [CrossRef]
- Schön, I.; Martens, K. No slave to sex. Proc. R. Soc. Lond. B 2003, 270, 827–833. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tran Van, P.; Anselmetti, Y.; Bast, J.; Dumas, Z.; Galtier, N.; Jaron, K.; Martens, K.; Parker, D.; Robinson-Rechavi, M.; Schwander, T.; et al. First annotated draft genomes of non-marine ostracods (Ostracoda, Crustacea) with different reproductive modes. G3 Genes Genomes Genet. 2021. [Google Scholar] [CrossRef]
- Flutre, T.; Duprat, E.; Feuillet, C.; Quesneville, H. Considering transposable element diversification in de novo annotation approaches. PLoS ONE 2011, 6, e16526. [Google Scholar] [CrossRef]
- Quesneville, H.; Bergman, C.M.; Andrieu, O.; Autard, D.; Nouaud, D.; Ashburner, M.; Anxolabehere, D. Combined evidence annotation of transposable elements in genome sequences. PLoS Computat. Biol. 2005, 1, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. Available online: http://www.repeatmasker.org (accessed on 21 June 2017).
- Stanke, M.; Schöffmann, O.; Morgenstern, B.; Waack, S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinform. 2006, 7, 62. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kubler, J.; Lozajic, M.; Gabler, F.; Soding, J.; Lupas, A.N.; Alva, V. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Burke, W.D.; Malik, H.S.; Jones, J.P.; Eickbush, T.H. The domain structure and retrotransposition mechanism of R2 elements are conserved throughout arthropods. Mol. Biol. Evol. 1999, 16, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Volff, J.N.; Körting, C.; Froschauer, A.; Sweeney, K.; Schartl, M. Non-LTR retrotransposons encoding a restriction enzyme-like endonuclease in vertebrates. J. Mol. Evol. 2001, 52, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Biedler, J.K.; Chen, X.; Tu, Z. Horizontal transmission of an R4 clade non-long terminal repeat retrotransposon between the divergent Aedes and Anopheles mosquito genera. Insect Mol. Biol. 2015, 24, 331–337. [Google Scholar] [CrossRef]
- Shivram, H.; Cawley, D.; Christensen, S.M. Targeting novel sites: The N-terminal DNA binding domain of non-LTR retrotransposons is an adaptable module that is implicated in changing site specificities. Mob. Genet. Elem. 2011, 1, 169–178. [Google Scholar] [CrossRef]
- Kojima, K.K.; Fujiwara, H. Cross-genome screening of novel sequence-specific non-LTR retrotransposons: Various multicopy RNA genes and microsatellites are selected as targets. Mol. Biol. Evol. 2004, 21, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Liu, J.; Zheng, Q.; Yong, W.; Lu, M. Structures and polymorphic interactions of two heptad-repeat regions of the SARS virus S2 protein. Structure 2006, 14, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhang, J.; Xiao, T.; Peng, H.; Sterling, S.M.; Walsh, R.M., Jr.; Rawson, S.; Rits-Volloch, S.; Chen, B. Distinct conformational states of SARS-CoV-2 spike protein. Science 2020, 369, 1586–1592. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.H.; Ogden, K.M.; Long, J.M.; Ebenhoch, R.; Thor, A.; Dermody, T.S.; Stehle, T. Structural and functional features of the reovirus σ1 tail. J. Virol. 2018, 92, 14. [Google Scholar] [CrossRef]
- Malik, H.S.; Henikoff, S.; Eickbush, T.H. Poised for contagion: Evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000, 10, 1307–1318. [Google Scholar] [CrossRef]
- Rodriguez, F.; Kenefick, A.W.; Arkhipova, I.R. LTR-retrotransposons from bdelloid rotifers capture additional ORFs shared between highly diverse retroelement types. Viruses 2017, 9, 78. [Google Scholar] [CrossRef]
- Colbourne, J.K.; Pfrender, M.E.; Gilbert, D.; Thomas, W.K.; Tucker, A.; Oakley, T.H.; Tokishita, S.; Aerts, A.; Arnold, G.J.; Basu, M.K.; et al. The ecoresponsive genome of Daphnia pulex. Science 2011, 331, 555–561. [Google Scholar] [CrossRef]
- Ye, Z.; Xu, S.; Spitze, K.; Asselman, J.; Jiang, X.; Ackerman, M.S.; Lopez, J.; Harker, B.; Raborn, R.T.; Thomas, W.K.; et al. A new reference genome assembly for the microcrustacean Daphnia pulex. G3 Genes Genomes Genet. 2017, 7, 1405–1416. [Google Scholar] [CrossRef]
- Blanc-Mathieu, R.; Perfus-Barbeoch, L.; Aury, J.M.; Da Rocha, M.; Gouzy, J.; Sallet, E.; Martin-Jimenez, C.; Bailly-Bechet, M.; Castagnone-Sereno, P.; Flot, J.F.; et al. Hybridization and polyploidy enable genomic plasticity without sex in the most devastating plant-parasitic nematodes. PLoS Genet. 2017, 13, e1006777. [Google Scholar] [CrossRef]
- Schaack, S.; Gilbert, C.; Feschotte, C. Promiscuous DNA: Horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol. Evol. 2010, 25, 537–546. [Google Scholar] [CrossRef]
- Dupeyron, M.; Leclercq, S.; Cerveau, N.; Bouchon, D.; Gilbert, C. Horizontal transfer of transposons between and within crustaceans and insects. Mob. DNA 2014, 5, 4. [Google Scholar] [CrossRef]
- Wallau, G.L.; Vieira, C.; Loreto, É. Genetic exchange in eukaryotes through horizontal transfer: Connected by the mobilome. Mob. DNA 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Metzger, M.J.; Paynter, A.N.; Siddall, M.E.; Goff, S.P. Horizontal transfer of retrotransposons between bivalves and other aquatic species of multiple phyla. Proc. Natl. Acad. Sci. USA 2018, 115, E4227–E4235. [Google Scholar] [CrossRef]
- Arkhipova, I.R.; Rodriguez, F. Genetic and epigenetic changes involving (retro)transposons in animal hybrids and polyploids. Cytogenet. Genome Res. 2013, 140, 295–311. [Google Scholar] [CrossRef]
- Petersen, M.; Armisén, D.; Gibbs, R.A.; Hering, L.; Khila, A.; Mayer, G.; Richards, S.; Niehuis, O.; Misof, B. Diversity and evolution of the transposable element repertoire in arthropods with particular reference to insects. BMC Evol. Biol. 2019, 19, 11. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Bian, C.; Luo, Y.; Wang, L.; You, X.; Li, J.; Qiu, Y.; Ma, X.; Zhu, Z.; Ma, L.; et al. Draft genome of the Chinese mitten crab, Eriocheir sinensis. GigaScience 2016, 5, 5. [Google Scholar] [CrossRef]
- Yuan, J.; Gao, Y.; Zhang, X.; Wei, J.; Liu, C.; Li, F.; Xiang, J. Genome sequences of marine shrimp Exopalaemon carinicauda Holthuis provide insights into genome size evolution of Caridea. Mar. Drugs 2017, 15, 213. [Google Scholar] [CrossRef] [PubMed]
- Kao, D.; Lai, A.G.; Stamataki, E.; Rosic, S.; Konstantinides, N.; Jarvis, E.; Di Donfrancesco, A.; Pouchkina-Stancheva, N.; Semon, M.; Grillo, M.; et al. The genome of the crustacean Parhyale hawaiensis, a model for animal development, regeneration, immunity and lignocellulose digestion. Elife 2016, 5, e20062. [Google Scholar] [CrossRef]
- Muszewska, A.; Steczkiewicz, K.; Stepniewska-Dziubinska, M.; Ginalski, K. Transposable elements contribute to fungal genes and impact fungal lifestyle. Sci. Rep. 2019, 9, 4307. [Google Scholar] [CrossRef]
- Hartley, G.; O’Neill, R.J. Centromere repeats: Hidden gems of the genome. Genes 2019, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Jiang, N.; Wing, R.A.; Jiang, J.; Jackson, S.A. Transposons play an important role in the evolution and diversification of centromeres among closely related species. Front. Plant Sci. 2015, 6, 216. [Google Scholar] [CrossRef]
- Pardue, M.-L.; DeBaryshe, P.G. Retrotransposons that maintain chromosome ends. Proc. Natl. Acad. Sci. USA 2011, 108, 20317–20324. [Google Scholar] [CrossRef]
- Gladyshev, E.A.; Arkhipova, I.R. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc. Natl. Acad. Sci. USA 2007, 104, 9352–9357. [Google Scholar] [CrossRef]
- Goodier, J.L. Restricting retrotransposons: A review. Mob. DNA 2016, 7, 16. [Google Scholar] [CrossRef]
- Rodriguez, F.; Arkhipova, I.R. Multitasking of the piRNA silencing machinery: Targeting transposable elements and foreign genes in the bdelloid rotifer Adineta vaga. Genetics 2016, 203, 255–268. [Google Scholar] [CrossRef]
- Molaro, A.; Malik, H.S. Hide and seek: How chromatin-based pathways silence retroelements in the mammalian germline. Curr. Opin. Genet. Dev. 2016, 37, 51–58. [Google Scholar] [CrossRef]
- Schön, I.; Kamiya, T.; Van den Berghe, T.; Van den Broecke, L.; Martens, K. The evolutionary history of novel Cardinium strains and their non-marine ostracod (Crustacea) hosts. Mol. Phylogenet. Evol. 2019, 130, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Martens, K.; Horne, D.J.; Griffiths, H.I. Age and diversity of non-marine ostracods. In Sex and Parthenogenesis: Evolutionary Ecology of Reproductive Modes in Non-Marine Ostracods; Martens, K., Ed.; Backhuys Publishers: Leiden, The Netherlands, 1998; pp. 37–55. [Google Scholar]
- Wolfe, J.M.; Daley, A.C.; Legg, D.A.; Edgecombe, G.D. Fossil calibrations for the arthropod Tree of Life. Earth Sci. Rev. 2016, 160, 43–110. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schön, I.; Rodriguez, F.; Dunn, M.; Martens, K.; Shribak, M.; Arkhipova, I.R. A Survey of Transposon Landscapes in the Putative Ancient Asexual Ostracod Darwinula stevensoni. Genes 2021, 12, 401. https://doi.org/10.3390/genes12030401
Schön I, Rodriguez F, Dunn M, Martens K, Shribak M, Arkhipova IR. A Survey of Transposon Landscapes in the Putative Ancient Asexual Ostracod Darwinula stevensoni. Genes. 2021; 12(3):401. https://doi.org/10.3390/genes12030401
Chicago/Turabian StyleSchön, Isa, Fernando Rodriguez, Matthew Dunn, Koen Martens, Michael Shribak, and Irina R. Arkhipova. 2021. "A Survey of Transposon Landscapes in the Putative Ancient Asexual Ostracod Darwinula stevensoni" Genes 12, no. 3: 401. https://doi.org/10.3390/genes12030401
APA StyleSchön, I., Rodriguez, F., Dunn, M., Martens, K., Shribak, M., & Arkhipova, I. R. (2021). A Survey of Transposon Landscapes in the Putative Ancient Asexual Ostracod Darwinula stevensoni. Genes, 12(3), 401. https://doi.org/10.3390/genes12030401







