Altered Hippocampal Epigenetic Regulation Underlying Reduced Cognitive Development in Response to Early Life Environmental Insults
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection
2.3. DNA Isolation
2.4. RRBS and Targeted Control Library Preparation
2.5. Illumina Sequencing
2.6. RRBS Data Analysis
2.7. Targeted Control Data Analysis
2.8. RNA-Seq Analysis
2.9. Machine Learning
2.10. Statistics
2.11. GO Term Enrichment Analysis
3. Results
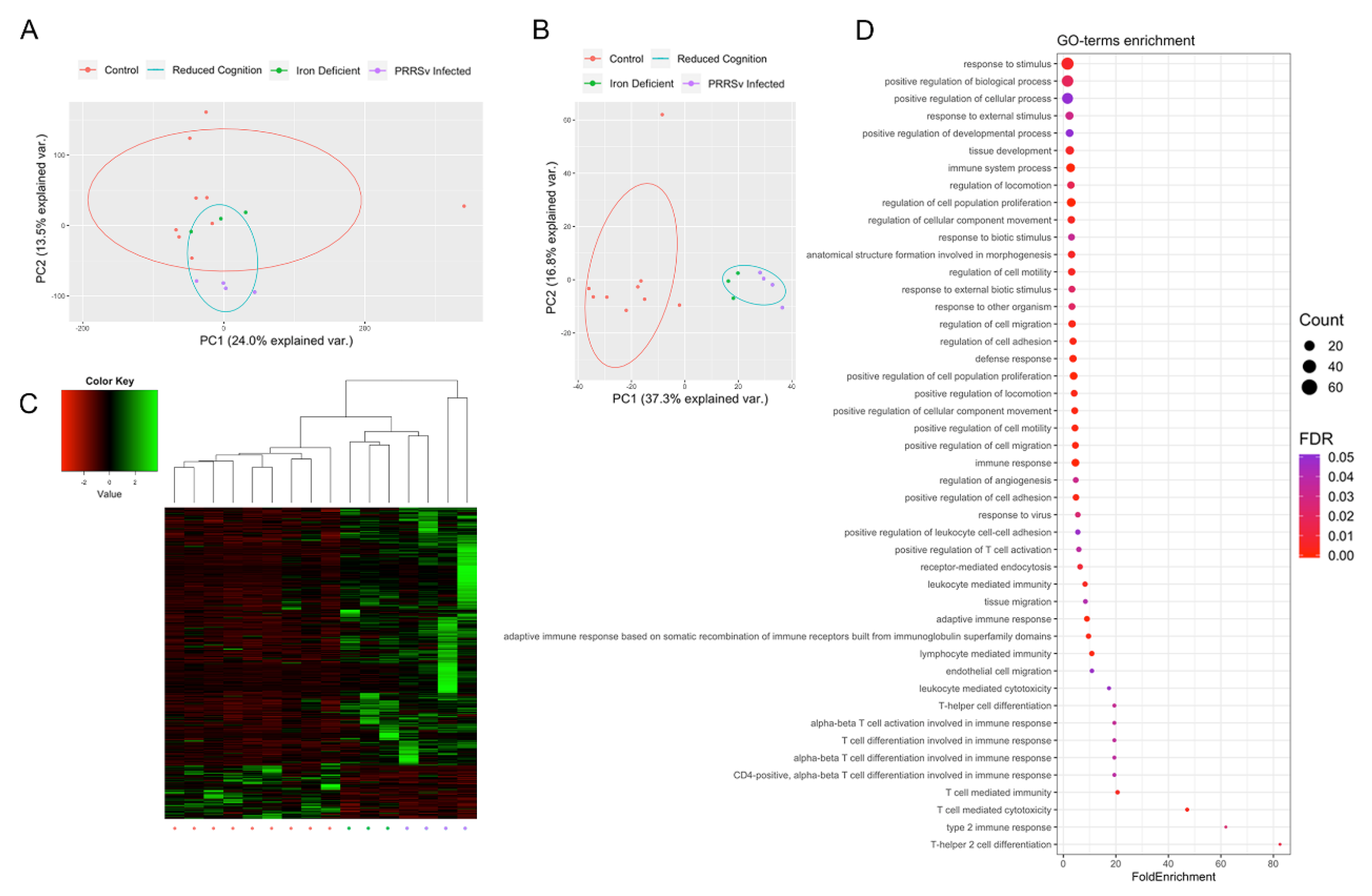
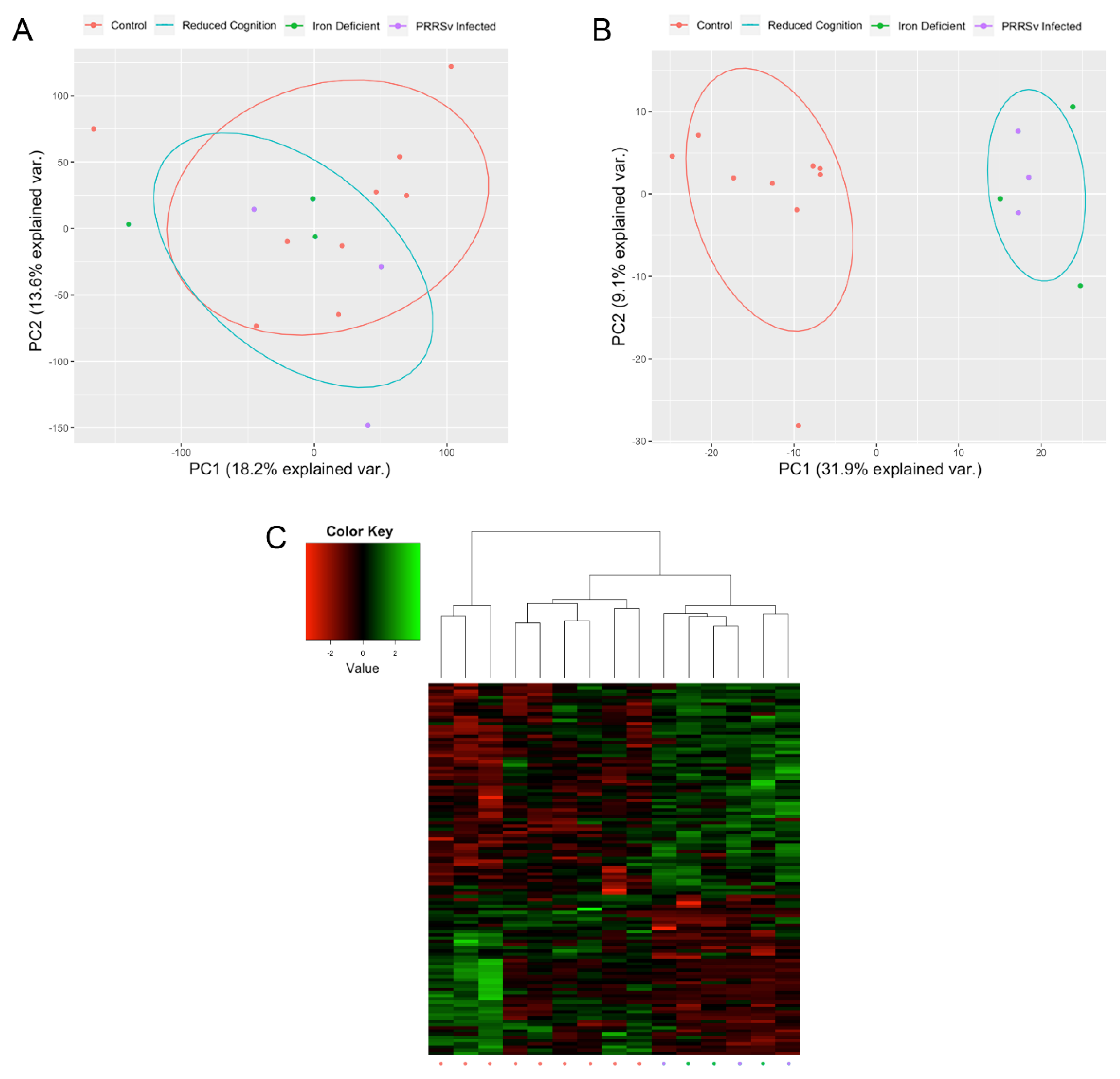
3.1. Early Life Environmental Insults Result in Altered Epigenetic and Transcriptional Patterns
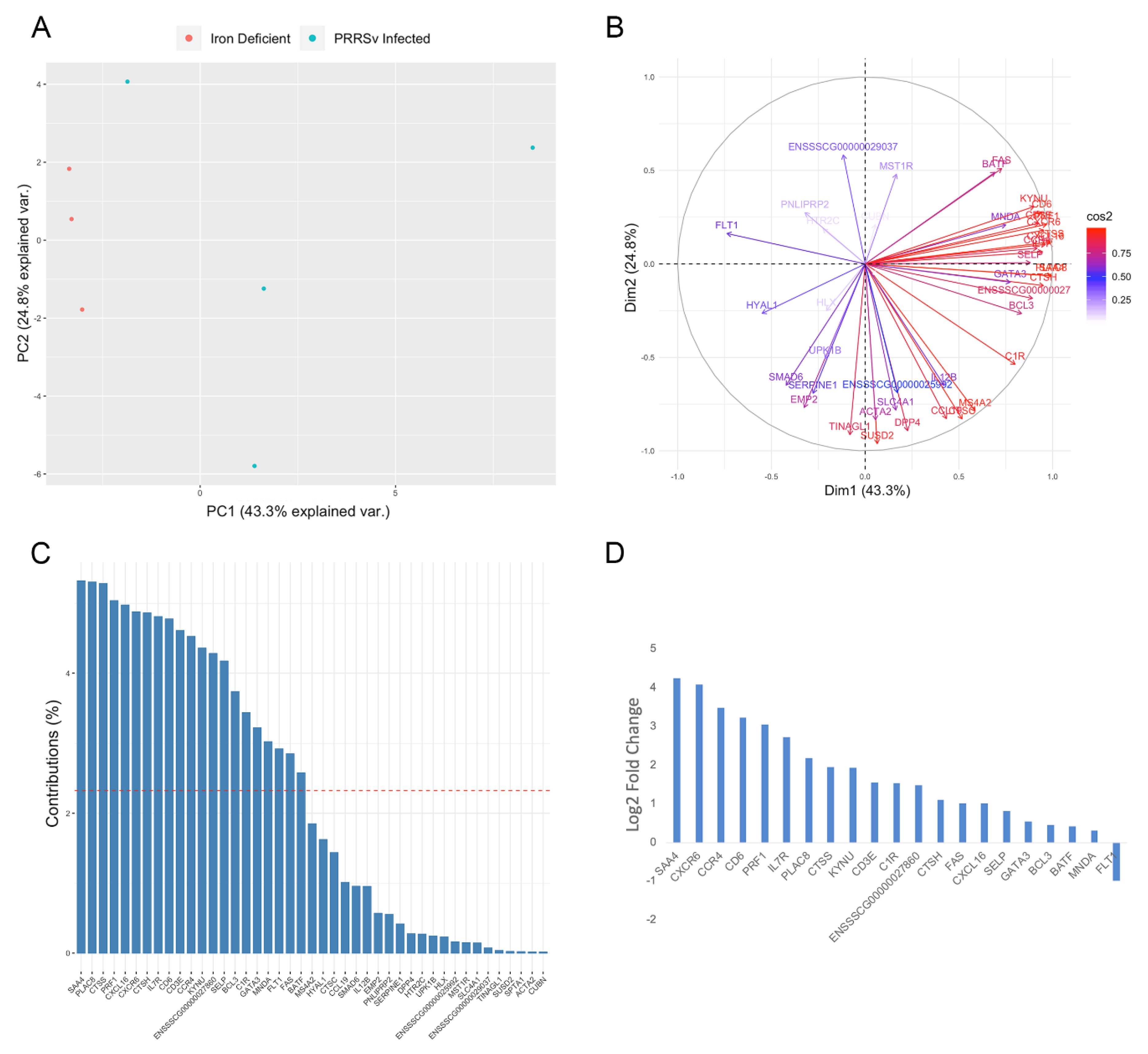
3.2. Activation of Immune Responses
3.3. Increased Angiogenesis and Blood Brain Barrier Permeability
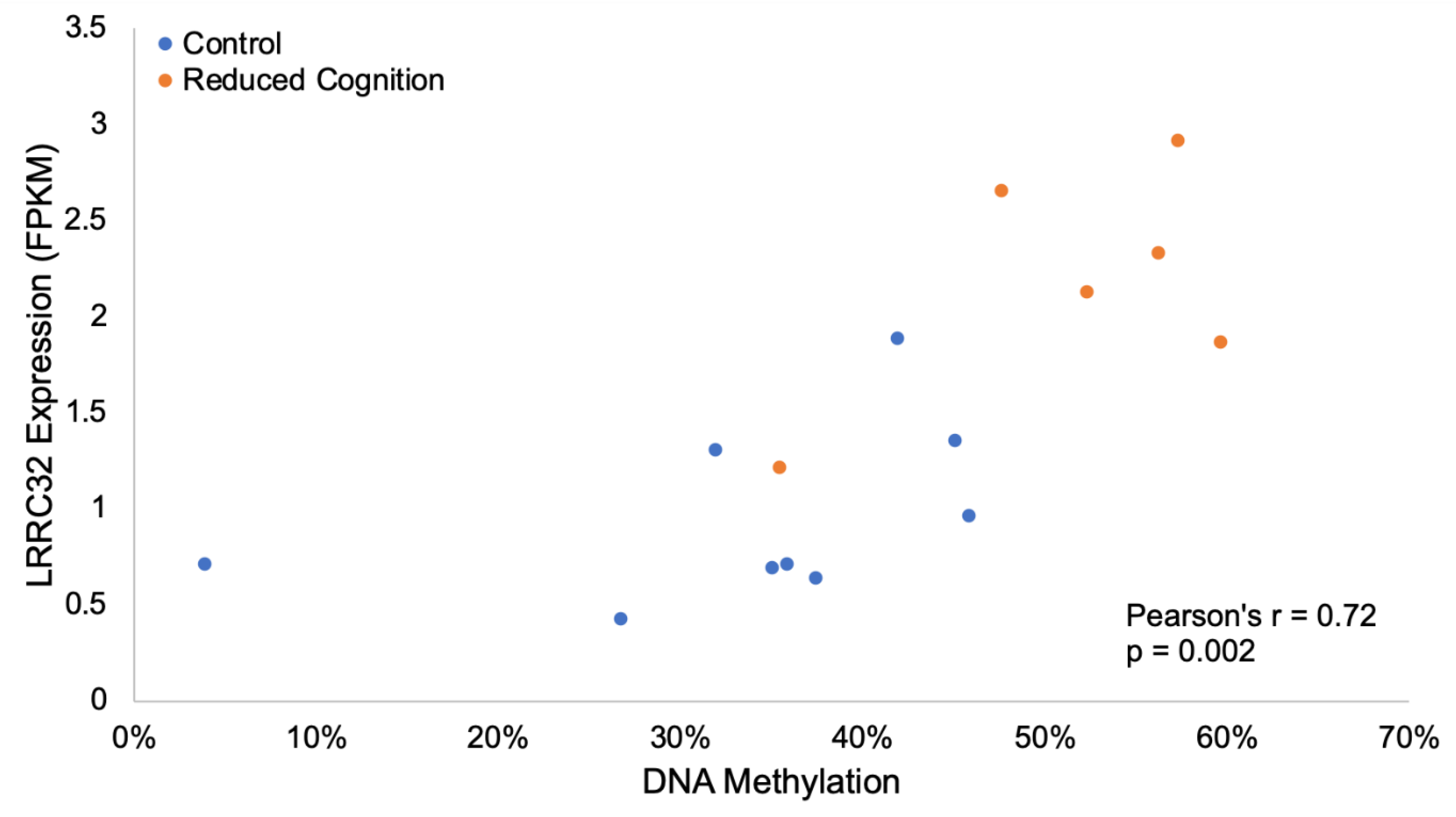
3.4. Altered Neurodevelopment and Function
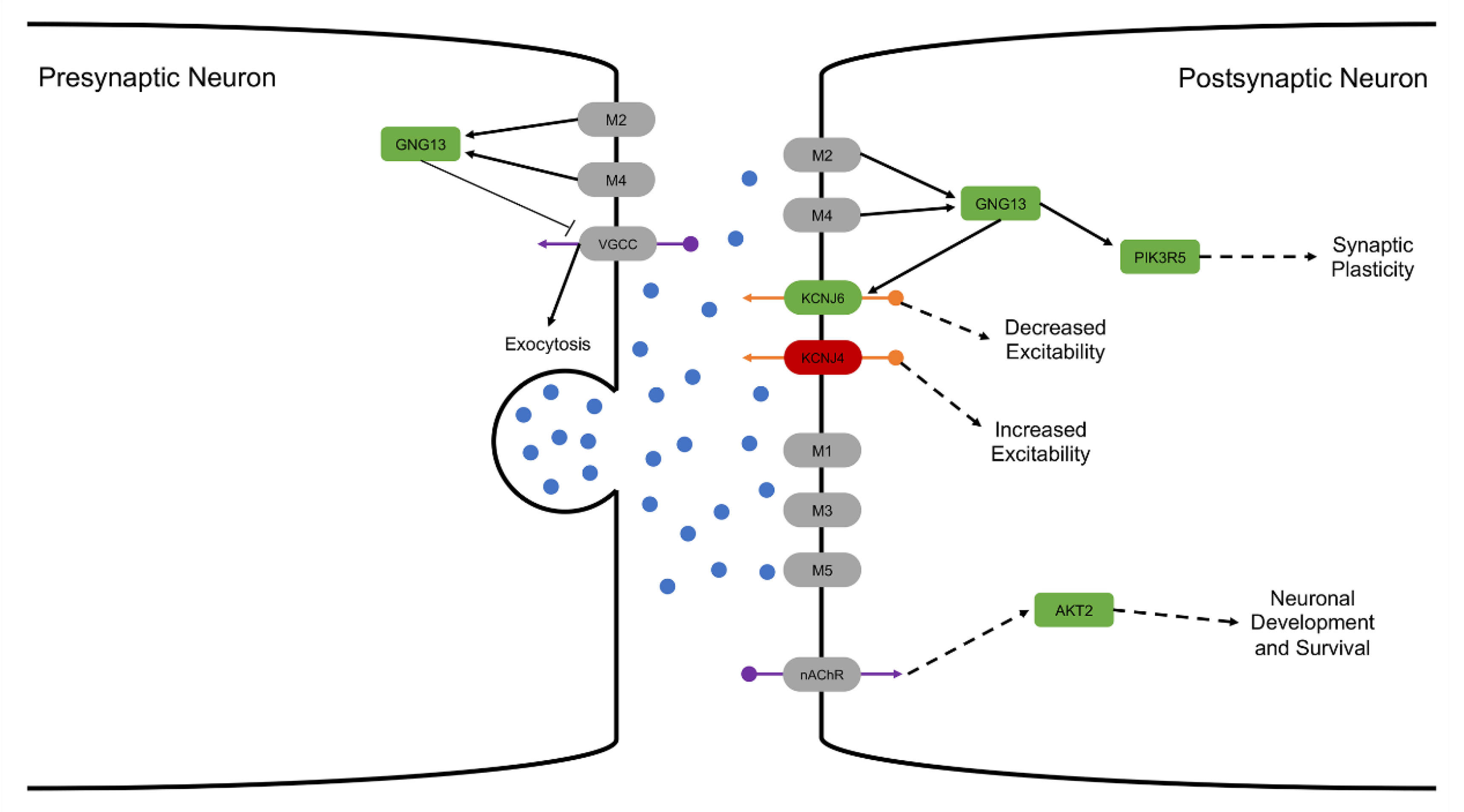
3.4.1. Altered Cholinergic Signaling
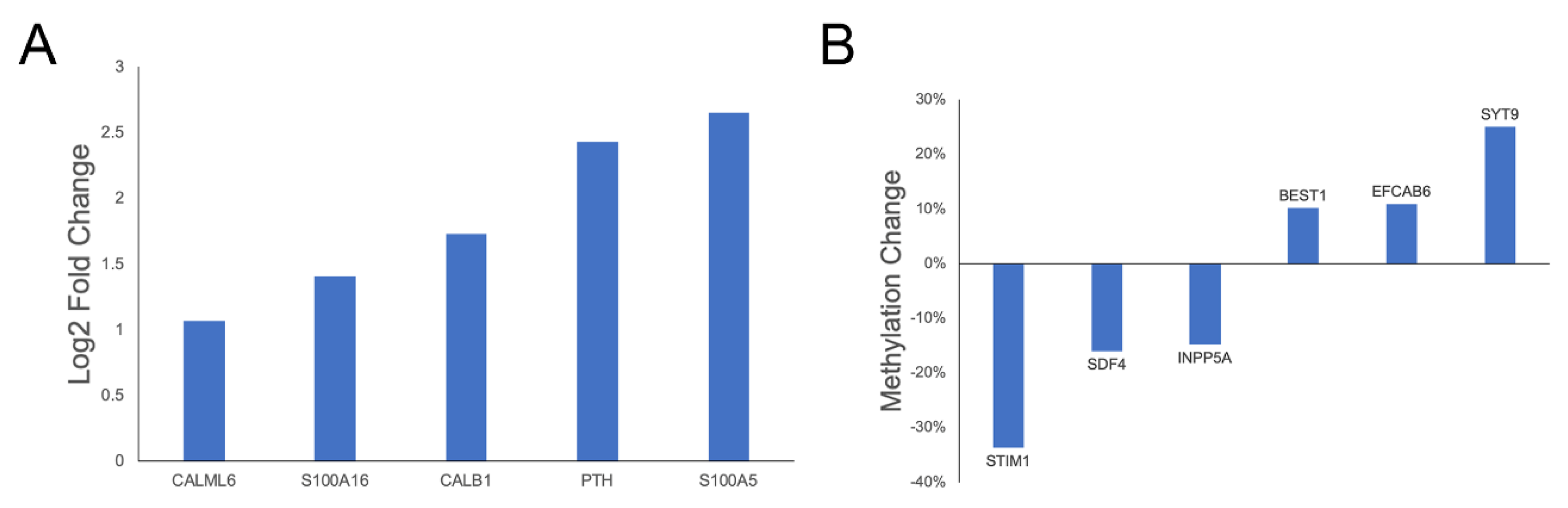
3.4.2. Glutamate Transport and Calcium Regulation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gogtay, N.; Nugent, T.F.; Herman, D.H.; Ordonez, A.; Greenstein, D.; Hayashi, K.M.; Clasen, L.; Toga, A.W.; Giedd, J.N.; Rapoport, J.L.; et al. Dynamic mapping of normal human hippocampal development. Hippocampus 2006, 16, 664–672. [Google Scholar] [CrossRef]
- Courchesne, E.; Chisum, H.J.; Townsend, J.; Cowles, A.; Covington, J.; Egaas, B.; Harwood, M.; Hinds, S.; Press, G.A. Normal brain development and aging: Quantitative analysis at in vivo MR imaging in healthy volunteers. Radiology 2000, 216, 672–682. [Google Scholar] [CrossRef]
- Brunette, K.E.; Tran, P.V.; Wobken, J.D.; Carlson, E.S.; Georgieff, M.K. Gestational and neonatal iron deficiency alters apical dendrite structure of CA1 pyramidal neurons in adult rat hippocampus. Dev. Neurosci. 2010, 32, 238–248. [Google Scholar] [CrossRef] [Green Version]
- Kohman, R.A.; Tarr, A.J.; Sparkman, N.L.; Bogale, T.M.H.; Boehm, G.W. Neonatal endotoxin exposure impairs avoidance learning and attenuates endotoxin-induced sickness behavior and central IL-1beta gene transcription in adulthood. Behav. Brain Res. 2008, 194, 25–31. [Google Scholar] [CrossRef]
- Meyer, U.; Schwendener, S.; Feldon, J.; Yee, B.K. Prenatal and postnatal maternal contributions in the infection model of schizophrenia. Exp. Brain Res. 2006, 173, 243–257. [Google Scholar] [CrossRef]
- Rao, R.; Tkac, I.; Schmidt, A.T.; Georgieff, M.K. Fetal and neonatal iron deficiency causes volume loss and alters the neurochemical profile of the adult rat hippocampus. Nutr. Neurosci. 2011, 14, 59–65. [Google Scholar] [CrossRef]
- McEwen, B.S. Stress and hippocampal plasticity. Annu. Rev. Neurosci. 1999, 22, 105–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogoch, Y.; Biala, Y.N.; Linial, M.; Weinstock, M. Anxiety induced by prenatal stress is associated with suppression of hippocampal genes involved in synaptic function. J. Neurochem. 2007, 101, 1018–1030. [Google Scholar] [CrossRef] [PubMed]
- Guedj, F.; Pennings, J.L.A.; Wick, H.C.; Bianchi, D.W. Analysis of adult cerebral cortex and hippocampus transcriptomes reveals unique molecular changes in the Ts1Cje mouse model of down syndrome. Brain Pathol. 2015, 25, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Meng, Y.; Hanna, A.; Janus, C.; Jia, Z. Abnormal long-lasting synaptic plasticity and cognition in mice lacking the mental retardation gene Pak3. J. Neurosci. 2005, 25, 6641–6650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsui, S.; Osako, Y.; Yokoi, F.; Dang, M.T.; Yuri, K.; Li, Y.; Yamaguchi, N. A mental retardation gene, motopsin/neurotrypsin/prss12, modulates hippocampal function and social interaction. Eur. J. Neurosci. 2009, 30, 2368–2378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanguilder, H.D.; Bixler, G.V.; Sonntag, W.E.; Freeman, W.M. Hippocampal expression of myelin-associated inhibitors is induced with age-related cognitive decline and correlates with deficits of spatial learning and memory. J. Neurochem. 2012, 121, 77–98. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.D.; Allis, C.D.; Bernstein, E. Epigenetics: A landscape takes shape. Cell 2007, 128, 635–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bureau, A.; Dupuis, J.; Falls, K.; Lunetta, K.L.; Hayward, B.; Keith, T.P.; Van Eerdewegh, P. Identifying SNPs predictive of phenotype using random forests. Genet. Epidemiol. 2005, 28, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Heidema, A.G.; Boer, J.M.; Nagelkerke, N.; Mariman, E.C.; van der, A.D.L.; Feskens, E.J. The challenge for genetic epidemiologists: How to analyze large numbers of SNPs in relation to complex diseases. BMC Genet. 2006, 7, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Austin, P.C.; Tu, J.V. Automated variable selection methods for logistic regression produced unstable models for predicting acute myocardial infarction mortality. J. Clin. Epidemiol. 2004, 57, 1138–1146. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Bogdan, M.; Candes, E. False Discoveries Occur Early on the Lasso Path. Ann. Stat. 2015, arXiv:1511.01957. [Google Scholar] [CrossRef] [Green Version]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. B 2005, 67, 301–320. [Google Scholar] [CrossRef] [Green Version]
- Hemphill, E.; Lindsay, J.; Lee, C.; Măndoiu, I.I.; Nelson, C.E. Feature selection and classifier performance on diverse bio- logical datasets. BMC Bioinform. 2014, 15, S4. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Szymczak, S.; Biernacka, J.M.; Cordell, H.J.; González-Recio, O.; König, I.R.; Zhang, H.; Sun, Y.V. Machine learning in genome-wide association studies. Genet. Epidemiol. 2009, 33, S51–S57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexe, G.; Monaco, J.; Doyle, S.; Basavanhally, A.; Reddy, A.; Seiler, M.; Ganesan, S.; Bhanot, G.; Madabhushi, A. Towards Improved Cancer Diagnosis and Prognosis Using Analysis of Gene Expression Data and Computer Aided Imaging. Exp. Biol. Med. 2009, 234, 860–879. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, T. Phenotype prediction based on genome-wide DNA methylation data. BMC Bioinform. 2014, 15, 193. [Google Scholar] [CrossRef] [Green Version]
- Swan, A.L.; Mobasheri, A.; Allaway, D.; Liddell, S.; Bacardit, J. Application of machine learning to proteomics data: Classification and biomarker identification in postgenomics biology. Omics J Integr. Biol. 2013, 17, 595–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smolinska, A.; Hauschild, A.-C.; Fijten, R.R.R.; Dallinga, J.W.; Baumbach, J.; van Schooten, F.J. Current breathomics—A review on data pre-processing techniques and machine learning in metabolomics breath analysis. J. Breath Res. 2014, 8, 027105. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, F.; Seifert, S.; Szymczak, S. Evaluation of variable selection methods for random forests and omics data sets. Brief. Bioinform. 2019, 20, 492–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kursa, M.B.; Rudnicki, W.R. Feature selection with the boruta package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kursa, M.B. Robustness of Random Forest-based gene selection methods. BMC Bioinform. 2014, 15, 8. [Google Scholar] [CrossRef] [Green Version]
- Díaz-Uriarte, R.; Alvarez de Andrés, S. Gene selection and classification of microarray data using random forest. BMC Bioinform. 2006, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Rudnicki, W.R.; Kierczak, M.; Koronacki, J.; Komorowski, J. A statistical method for determining importance of variables in an information system. In Lecture Notes in Computer Science Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics; Springer: Berlin, Germany, 2006; pp. 557–566. [Google Scholar]
- Strobl, C.; Zeileis, A. Danger: High Power! – Exploring the Statistical Properties of a Test for Random Forest Variable Importance. In Proceedings of the 18th International Conference on Computational Statistics, Porto, Portugal, 19–21 June 2008. [Google Scholar]
- Guo, P.; Luo, Y.; Mai, G.; Zhang, M.; Wang, G.; Zhao, M.; Gao, L.; Li, F.; Zhou, F. Gene expression profile based classification models of psoriasis. Genomics 2014, 103, 48–55. [Google Scholar] [CrossRef] [Green Version]
- Saulnier, D.M.; Riehle, K.; Mistretta, T.A.; Diaz, M.A.; Mandal, D.; Raza, S.; Weidler, E.M.; Qin, X.; Coarfa, C.; Petrosino, J.F.; et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 2011, 141, 1782–1791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braundmeier-Fleming, A.; Russell, N.T.; Yang, W.; Nas, M.Y.; Yaggie, R.E.; Berry, M.; Bachrach, L.; Flury, S.C.; Marko, D.S.; Bushell, C.B.; et al. Stool-based biomarkers of interstitial cystitis/bladder pain syndrome. Sci. Rep. 2016, 6, 26083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Candel, S.; Karr, M.; Brard, L.; Thomson, J.A.; Groesch, K.; Braundmeier-Fleming, A. Microbial Profiles and Tumor Markers From Culdocentesis. Obstet. Gynecol. 2017, 129, 82S. [Google Scholar] [CrossRef]
- Hagler, M.A.; Thalji, N.M.; Russell, N.; Welge, M.; Bushell, C.; Berry, M.; White, B.A.; Suri, R.M.; Miller, J.D. Abstract 19746: Identification of Novel microRNA Profiles in Patients With Myxomatous Mitral Valve Disease. Circulation 2015, 132, A19746. [Google Scholar]
- Chen, K.L.A.; Liu, X.; Zhao, Y.C.; Hieronymi, K.; Rossi, G.; Auvil, L.S.; Welge, M.; Bushell, C.; Smith, R.L.; Kim, S.H.; et al. Long-Term Administration of Conjugated Estrogen and Bazedoxifene Decreased Murine Fecal β-Glucuronidase Activity Without Impacting Overall Microbiome Community. Sci. Rep. 2018, 8, 8166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corrigan, A.; Russell, N.; Welge, M.; Auvil, L.; Bushell, C.; White, B.A.; Murphy, R.A. The use of random forests modelling to detect yeast-mannan sensitive bacterial changes in the broiler cecum. Sci. Rep. 2018, 8, 13270. [Google Scholar] [CrossRef]
- Elmore, M.R.P.; Burton, M.D.; Conrad, M.S.; Rytych, J.L.; Van Alstine, W.G.; Johnson, R.W. Respiratory viral infection in neonatal piglets causes marked microglia activation in the hippocampus and deficits in spatial learning. J. Neurosci. 2014, 34, 2120–2129. [Google Scholar] [CrossRef] [Green Version]
- Rytych, J.J.L.; Elmore, M.R.P.M.; Burton, M.M.D.; Conrad, M.M.S.; Donovan, S.S.M.; Dilger, R.N.R.; Johnson, R.W.R. Early life iron deficiency impairs spatial cognition in neonatal piglets. J. Nutr. 2012, 142, 2050–2056. [Google Scholar] [CrossRef] [Green Version]
- Dickerson, J.W.T.; Dobbing, J. Prenatal and Postnatal Growth and Development of the Central Nervous System of the Pig. Proc. R. Soc. B Biol. Sci. 1967, 166, 384–395. [Google Scholar]
- Thibault, K.L.; Margulies, S.S. Age-dependent material properties of the porcine cerebrum: Effect on pediatric inertial head injury criteria. J. Biomech. 1998, 31, 1119–1126. [Google Scholar] [CrossRef]
- Dilger, R.N.; Johnson, R.W. Behavioral assessment of cognitive function using a translational neonatal piglet model. Brain. Behav. Immun. 2010, 24, 1156–1165. [Google Scholar] [CrossRef] [PubMed]
- Elmore, M.R.P.; Dilger, R.N.; Johnson, R.W. Place and direction learning in a spatial T-maze task by neonatal piglets. Anim. Cogn. 2012, 15, 667–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schachtschneider, K.M.; Madsen, O.; Park, C.; Rund, L.A.; Groenen, M.A.; Schook, L.B. Adult porcine genome-wide DNA methylation patterns support pigs as a biomedical model. Bmc Genom. 2015, 16, 743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, M.; Lee, J.; Le, M.T.; Nguyen, D.T.; Park, S.; Soundrarajan, N.; Schachtschneider, K.M.; Kim, J.; Park, J.-K.; Kim, J.-H.; et al. Genome-wide analysis of DNA methylation in pigs using reduced representation bisulfite sequencing. DNA Res. 2015, 22, 343–355. [Google Scholar] [CrossRef] [Green Version]
- Ji, P.; Schachtschneider, K.M.; Schook, L.B.; Walker, F.R.; Johnson, R.W. Peripheral viral infection induced microglial sensome genes and enhanced microglial cell activity in the hippocampus of neonatal piglets. Brain. Behav. Immun. 2016, 54, 243–251. [Google Scholar] [CrossRef] [Green Version]
- Schachtschneider, K.M.; Liu, Y.; Rund, L.A.; Madsen, O.; Johnson, R.W.; Groenen, M.A.M.; Schook, L.B. Impact of neonatal iron deficiency on hippocampal DNA methylation and gene transcription in a porcine biomedical model of cognitive development. BMC Genomics 2016, 17, 856. [Google Scholar] [CrossRef] [Green Version]
- Groenen, M.A.M.; Archibald, A.L.; Uenishi, H.; Tuggle, C.K.; Takeuchi, Y.; Rothschild, M.F.; Rogel-Gaillard, C.; Park, C.; Milan, D.; Megens, H.-J.; et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 2012, 491, 393–398. [Google Scholar] [CrossRef]
- Guo, W.; Fiziev, P.; Yan, W.; Cokus, S.; Sun, X.; Zhang, M.Q.; Chen, P.-Y.; Pellegrini, M. BS-Seeker2: A versatile aligning pipeline for bisulfite sequencing data. BMC Genom. 2013, 14, 774. [Google Scholar] [CrossRef] [Green Version]
- Gaspar, J.M.; Hart, R.P. DMRfinder: Efficiently identifying differentially methylated regions from MethylC-seq data. BMC Bioinform. 2017, 18, 528. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. R Found. Stat. Comput. Vienna Austria 2014. Available online: http://www.R-project.org/ (accessed on 13 December 2019).
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017, 45, D331–D338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef] [Green Version]
- Sawano, A.; Iwai, S.; Sakurai, Y.; Ito, M.; Shitara, K.; Nakahata, T.; Shibuya, M. Flt-1, vascular endothelial growth factor receptor 1, is a novel cell surface marker for the lineage of monocyte-macrophages in humans. Blood 2001, 97, 785–791. [Google Scholar] [CrossRef] [Green Version]
- Probst-Kepper, M.; Buer, J. FOXP3 and GARP (LRRC32): The master and its minion. Biol. Direct. 2010, 5, 8. [Google Scholar] [CrossRef] [Green Version]
- Baek, H.; Ye, M.; Kang, G.-H.; Lee, C.; Lee, G.; Choi, D.B.; Jung, J.; Kim, H.; Lee, S.; Kim, J.S.; et al. Neuroprotective effects of CD4+CD25+Foxp3+ regulatory T cells in a 3xTg-AD Alzheimer’s disease model. Oncotarget 2016, 7, 69347–69357. [Google Scholar] [CrossRef] [Green Version]
- Gentil, B.J.; Benaud, C.; Delphin, C.; Remy, C.; Berezowski, V.; Cecchelli, R.; Feraud, O.; Vittet, D.; Baudier, J. Specific AHNAK expression in brain endothelial cells with barrier properties. J. Cell. Physiol. 2005, 203, 362–371. [Google Scholar] [CrossRef]
- Suidan, G.L.; Brill, A.; De Meyer, S.F.; Voorhees, J.R.; Cifuni, S.M.; Cabral, J.E.; Wagner, D.D. Endothelial Von Willebrand factor promotes blood-brain barrier flexibility and provides protection from hypoxia and seizures in mice. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2112–2120. [Google Scholar] [CrossRef] [Green Version]
- Fischer, S.; Clauss, M.; Wiesnet, M.; Renz, D.; Schaper, W.; Karliczek, G.F. Hypoxia induces permeability in brain microvessel endothelial cells via VEGF and NO. Am. J. Physiol. 1999, 276, C812–C820. [Google Scholar] [CrossRef]
- Sia, G.M.; Clem, R.L.; Huganir, R.L. The human language-associated gene SRPX2 regulates synapse formation and vocalization in mice. Science 2013, 342, 987–991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, C.S.L.; Fisher, S.E.; Hurst, J.A.; Vargha-Khadem, F.; Monaco, A.P. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 2001, 413, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Fisher, S.E.; Scharff, C. FOXP2 as a molecular window into speech and language. Trends Genet. 2009, 25, 166–177. [Google Scholar] [CrossRef] [PubMed]
- Cavallaro, S. Genomic analysis of serotonin receptors in learning and memory. Behav. Brain Res. 2008, 195, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Pang, T.Y.C.; Du, X.; Zajac, M.S.; Howard, M.L.; Hannan, A.J. Altered serotonin receptor expression is associated with depression-related behavior in the R6/1 transgenic mouse model of Huntington’s disease. Hum. Mol. Genet. 2009, 18, 753–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duric, V.; Banasr, M.; Stockmeier, C.A.; Simen, A.A.; Newton, S.S.; Overholser, J.C.; Jurjus, G.J.; Dieter, L.; Duman, R.S. Altered expression of synapse and glutamate related genes in post-mortem hippocampus of depressed subjects. Int. J. Neuropsychopharmacol. 2013, 16, 69–82. [Google Scholar] [CrossRef] [Green Version]
- Nahon, J.-L.; Joly, C.; Levan, G.; Szpirer, J.; Szpirer, C. Pro-melanin-concentrating hormone gene (PMCH) is localized on human chromosome 12q and rat chromosome 7. Genomics 1992, 12, 846–848. [Google Scholar] [CrossRef]
- Homberg, J.R.; Olivier, J.D.A.; VandenBroeke, M.; Youn, J.; Ellenbroek, A.K.; Karel, P.; Shan, L.; van Boxtel, R.; Ooms, S.; Balemans, M.; et al. The role of the dopamine D1 receptor in social cognition: Studies using a novel genetic rat model. Dis. Model. Mech. 2016, 9, 1147–1158. [Google Scholar] [CrossRef] [Green Version]
- Paul, M.L.; Graybiel, A.M.; David, J.C.; Robertson, H.A. D1-like and D2-like dopamine receptors synergistically activate rotation and c-fos expression in the dopamine-depleted striatum in a rat model of Parkinson’s disease. J. Neurosci. 1992, 12, 3729–3742. [Google Scholar] [CrossRef] [Green Version]
- St-Gelais, F.; Jomphe, C.; Trudeau, L.-E. The role of neurotensin in central nervous system pathophysiology: What is the evidence? J. Psychiatry Neurosci. 2006, 31, 229–245. [Google Scholar]
- Hoischen, A.; Krumm, N.; Eichler, E.E. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat. Neurosci. 2014, 17, 764–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marseglia, G.; Scordo, M.R.; Pescucci, C.; Nannetti, G.; Biagini, E.; Scandurra, V.; Gerundino, F.; Magi, A.; Benelli, M.; Torricelli, F. 372 kb microdeletion in 18q12.3 causing SETBP1 haploinsufficiency associated with mild mental retardation and expressive speech impairment. Eur. J. Med. Genet. 2012, 55, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Potts, R.C.; Zhang, P.; Wurster, A.L.; Precht, P.; Mughal, M.R.; Wood, W.H.; Zhang, Y.; Becker, K.G.; Mattson, M.P.; Pazin, M.J. CHD5, a Brain-Specific Paralog of Mi2 Chromatin Remodeling Enzymes, Regulates Expression of Neuronal Genes. PLoS ONE 2011, 6, e24515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martiskainen, H.; Paldanius, K.M.A.; Natunen, T.; Takalo, M.; Marttinen, M.; Leskelä, S.; Huber, N.; Mäkinen, P.; Bertling, E.; Dhungana, H.; et al. DHCR24 exerts neuroprotection upon inflammation-induced neuronal death. J. Neuroinflammation 2017, 14, 215. [Google Scholar] [CrossRef] [Green Version]
- Shih, D.-F.; Hsiao, C.-D.; Min, M.-Y.; Lai, W.-S.; Yang, C.-W.; Lee, W.-T.; Lee, S.-J. Aromatic L-Amino Acid Decarboxylase (AADC) Is Crucial for Brain Development and Motor Functions. PLoS ONE 2013, 8, e71741. [Google Scholar] [CrossRef] [Green Version]
- Hamada, N.; Ogaya, S.; Nakashima, M.; Nishijo, T.; Sugawara, Y.; Iwamoto, I.; Ito, H.; Maki, Y.; Shirai, K.; Baba, S.; et al. De novo PHACTR1 mutations in West syndrome and their pathophysiological effects. Brain 2018, 141, 3098–3114. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, M.A.; Dwyer, B.E.; Franco, S.J. Csmd2 Is a Synaptic Transmembrane Protein that Interacts with PSD-95 and Is Required for Neuronal Maturation. eNeuro 2019, 6, ENEURO.0434-18.2019. [Google Scholar] [CrossRef] [Green Version]
- Krupp, M.; Weinmann, A.; Galle, P.R.; Teufel, A. Actin binding LIM protein 3 (abLIM3). Int. J. Mol. Med. 2006, 17, 129–133. [Google Scholar] [CrossRef]
- Bowden, N.A.; Scott, R.J.; Tooney, P.A. Altered gene expression in the superior temporal gyrus in schizophrenia. BMC Genom. 2008, 9, 199. [Google Scholar] [CrossRef] [Green Version]
- Martinowich, K.; Hattori, D.; Wu, H.; Fouse, S.; He, F.; Hu, Y.; Fan, G.; Sun, Y.E. DNA Methylation-Related Chromatin Remodeling in Activity-Dependent Bdnf Gene Regulation. Science 2003, 302, 890–893. [Google Scholar] [CrossRef] [Green Version]
- Luchicchi, A.; Bloem, B.; Viaña, J.N.M.; Mansvelder, H.D.; Role, L.W. Illuminating the role of cholinergic signaling in circuits of attention and emotionally salient behaviors. Front. Synaptic Neurosci. 2014, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.M.; Franks, K.M.; Titterness, A.K.; Russell, K.; Merrill, D.A.; Christie, B.R.; Sejnowski, T.J.; Tuszynski, M.H. NGF Is Essential for Hippocampal Plasticity and Learning. J. Neurosci. 2009, 29, 10883–10889. [Google Scholar] [CrossRef] [PubMed]
- Large, T.H.; Bodary, S.C.; Clegg, D.O.; Weskamp, G.; Otten, U.; Reichardt, L.F. Nerve growth factor gene expression in the developing rat brain. Science 1986, 234, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Whittemore, S.R.; Ebendal, T.; Lärkfors, L.; Olson, L.; Seiger, A.; Strömberg, I.; Persson, H. Development and regional expression of beta nerve growth factor messenger RNA and protein in the rat central nervous system. Proc. Natl. Acad. Sci. USA 1986, 83, 817–821. [Google Scholar] [CrossRef] [Green Version]
- Sofroniew, M.V.; Galletly, N.P.; Isacson, O.; Svendsen, C.N. Survival of adult basal forebrain cholinergic neurons after loss of target neurons. Science 1990, 247, 338–342. [Google Scholar] [CrossRef]
- Freeman, R.S.; Burch, R.L.; Crowder, R.J.; Lomb, D.J.; Schoell, M.C.; Straub, J.A.; Xie, L. NGF deprivation-induced gene expression: After ten years, where do we stand? In Progress in Brain Research; Elsevier: Amsterdam, The Netherlands, 2004; Volume 146, pp. 111–126. [Google Scholar]
- Fischer, W.; Wictorin, K.; Björklund, A.; Williams, L.R.; Varon, S.; Gage, F.H. Amelioration of cholinergic neuron atrophy and spatial memory impairment in aged rats by nerve growth factor. Nature 1987, 329, 65–68. [Google Scholar] [CrossRef]
- Schliebs, R.; Arendt, T. The cholinergic system in aging and neuronal degeneration. Behav. Brain Res. 2011, 221, 555–563. [Google Scholar] [CrossRef]
- Scheiderer, C.L.; McCutchen, E.; Thacker, E.E.; Kolasa, K.; Ward, M.K.; Parsons, D.; Harrell, L.E.; Dobrunz, L.E.; McMahon, L.L. Sympathetic sprouting drives hippocampal cholinergic reinnervation that prevents loss of a muscarinic receptor-dependent long-term depression at CA3-CA1 synapses. J. Neurosci. 2006, 26, 3745–3756. [Google Scholar] [CrossRef] [Green Version]
- Taniguchi, M.; Masuda, T.; Fukaya, M.; Kataoka, H.; Mishina, M.; Yaginuma, H.; Watanabe, M.; Shimizu, T. Identification and characterization of a novel member of murine semaphorin family. Genes Cells 2005, 10, 785–792. [Google Scholar] [CrossRef]
- Frere, S.G.; Chang-Ileto, B.; Di Paolo, G. Role of phosphoinositides at the neuronal synapse. Subcell. Biochem. 2015, 59, 131–175. [Google Scholar]
- Mattson, M.P. Calcium and neurodegeneration. Aging Cell 2007, 6, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Traynelis, S.F.; Wollmuth, L.P.; McBain, C.J.; Menniti, F.S.; Vance, K.M.; Ogden, K.K.; Hansen, K.B.; Yuan, H.; Myers, S.J.; Dingledine, R. Glutamate Receptor Ion Channels: Structure, Regulation, and Function. Pharmacol. Rev. 2010, 62, 405–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, A.S.; Gill, M.B.; Ho, M.T.; Yu, H.; Tu, Y.; Siuda, E.R.; Wang, H.; Qian, Y.W.; Nisenbaum, E.S.; Tomita, S.; et al. Hippocampal AMPA Receptor Gating Controlled by Both TARP and Cornichon Proteins. Neuron 2010, 68, 1082–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sturchler, E.; Cox, J.A.; Durussel, I.; Weibel, M.; Heizmann, C.W. S100A16, a novel calcium-binding protein of the EF-hand superfamily. J. Biol. Chem. 2006, 281, 38905–38917. [Google Scholar] [CrossRef] [Green Version]
- Soontornniyomkij, V.; Risbrough, V.B.; Young, J.W.; Soontornniyomkij, B.; Jeste, D.V.; Achim, C.L. Hippocampal calbindin-1 immunoreactivity correlate of recognition memory performance in aged mice. Neurosci. Lett. 2012, 516, 161–165. [Google Scholar] [CrossRef] [Green Version]
- Molinari, S.; Battini, R.; Ferrari, S.; Pozzi, L.; Killcross, A.S.; Robbins, T.W.; Jouvenceau, A.; Billard, J.M.; Dutar, P.; Lamour, Y.; et al. Deficits in memory and hippocampal long-term potentiation in mice with reduced calbindin D28K expression. Proc. Natl. Acad. Sci. USA 1996, 93, 8028–8033. [Google Scholar] [CrossRef] [Green Version]
- Dumas, T.C.; Powers, E.C.; Tarapore, P.E.; Sapolsky, R.M. Overexpression of calbindin D(28k) in dentate gyrus granule cells alters mossy fiber presynaptic function and impairs hippocampal-dependent memory. Hippocampus 2004, 14, 701–709. [Google Scholar] [CrossRef]
- Roos, J.; DiGregorio, P.J.; Yeromin, A.V.; Ohlsen, K.; Lioudyno, M.; Zhang, S.; Safrina, O.; Kozak, J.A.; Wagner, S.L.; Cahalan, M.D.; et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 2005, 169, 435–445. [Google Scholar] [CrossRef] [Green Version]
- Oh, S.-J.; Lee, C.J. Distribution and Function of the Bestrophin-1 (Best1) Channel in the Brain. Exp. Neurobiol. 2017, 26, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Windhorst, S.; Minge, D.; Bähring, R.; Hüser, S.; Schob, C.; Blechner, C.; Lin, H.-Y.; Mayr, G.W.; Kindler, S. Inositol-1,4,5-trisphosphate 3-kinase A regulates dendritic morphology and shapes synaptic Ca2+ transients. Cell. Signal. 2012, 24, 750–757. [Google Scholar] [CrossRef]
- Sepp, K.J.; Hong, P.; Lizarraga, S.B.; Liu, J.S.; Mejia, L.A.; Walsh, C.A.; Perrimon, N. Identification of neural outgrowth genes using genome-wide RNAi. PLoS Genet. 2008, 4, e1000111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Südhof, T.C. Calcium Control of Neurotransmitter Release. Cold Spring Harb. Perspect. Biol. 2012, 4, a011353. [Google Scholar]
- Weiss, G. Iron, infection and anemia—A classical triad. Wien. Klin. Wochenschr. 2002, 114, 357–367. [Google Scholar] [PubMed]
- McClintick, J.N.; Xuei, X.; Tischfield, J.A.; Goate, A.; Foroud, T.; Wetherill, L.; Ehringer, M.A.; Edenberg, H.J. Stress-response pathways are altered in the hippocampus of chronic alcoholics. Alcohol 2013, 47, 505–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehedint, M.G.; Craciunescu, C.N.; Zeisel, S.H. Maternal dietary choline deficiency alters angiogenesis in fetal mouse hippocampus. Proc. Natl. Acad. Sci. USA 2010, 107, 12834–12839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meck, W.H.; Smith, R.A.; Williams, C.L. Pre- and postnatal choline supplementation produces long-term facilitation of spatial memory. Dev. Psychobiol. 1988, 21, 339–353. [Google Scholar] [CrossRef] [PubMed]
- Meck, W.H.; Williams, C.L. Perinatal choline supplementation increases the threshold for chunking in spatial memory. Neuroreport 1997, 8, 3053–3059. [Google Scholar] [CrossRef]
- Duff, M.C.; Brown-Schmidt, S. The hippocampus and the flexible use and processing of language. Front. Hum. Neurosci. 2012, 6, 69. [Google Scholar] [CrossRef] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schachtschneider, K.M.; Welge, M.E.; Auvil, L.S.; Chaki, S.; Rund, L.A.; Madsen, O.; Elmore, M.R.P.; Johnson, R.W.; Groenen, M.A.M.; Schook, L.B. Altered Hippocampal Epigenetic Regulation Underlying Reduced Cognitive Development in Response to Early Life Environmental Insults. Genes 2020, 11, 162. https://doi.org/10.3390/genes11020162
Schachtschneider KM, Welge ME, Auvil LS, Chaki S, Rund LA, Madsen O, Elmore MRP, Johnson RW, Groenen MAM, Schook LB. Altered Hippocampal Epigenetic Regulation Underlying Reduced Cognitive Development in Response to Early Life Environmental Insults. Genes. 2020; 11(2):162. https://doi.org/10.3390/genes11020162
Chicago/Turabian StyleSchachtschneider, Kyle M., Michael E. Welge, Loretta S. Auvil, Sulalita Chaki, Laurie A. Rund, Ole Madsen, Monica R.P. Elmore, Rodney W. Johnson, Martien A.M. Groenen, and Lawrence B. Schook. 2020. "Altered Hippocampal Epigenetic Regulation Underlying Reduced Cognitive Development in Response to Early Life Environmental Insults" Genes 11, no. 2: 162. https://doi.org/10.3390/genes11020162




