AI-Enabled Algorithm for Automatic Classification of Sleep Disorders Based on Single-Lead Electrocardiogram
Abstract
:1. Introduction
2. Materials and Methods
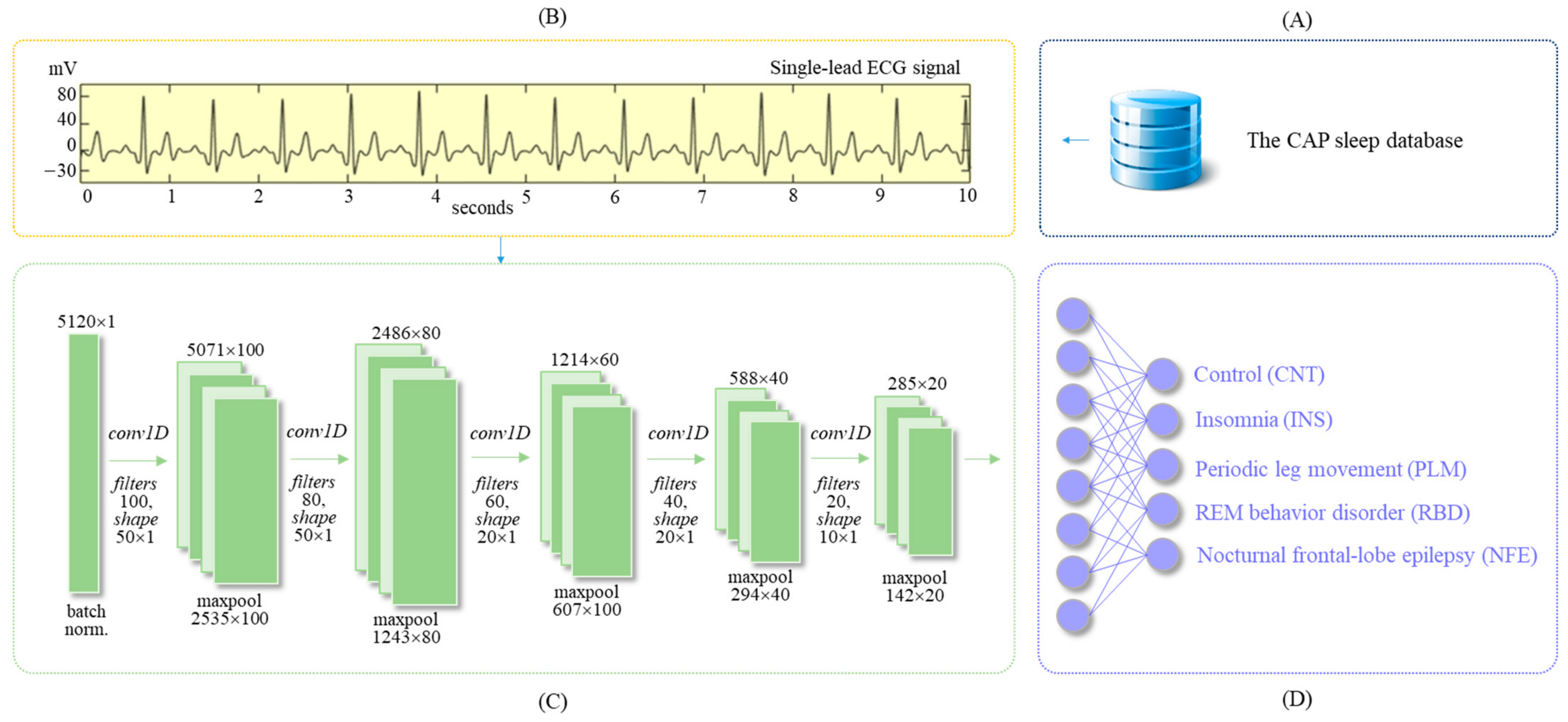
2.1. The CAP Sleep Database
2.2. ECG Dataset
2.3. Deep Learning Algorithm
2.4. Implementation
2.5. Evaluation Index
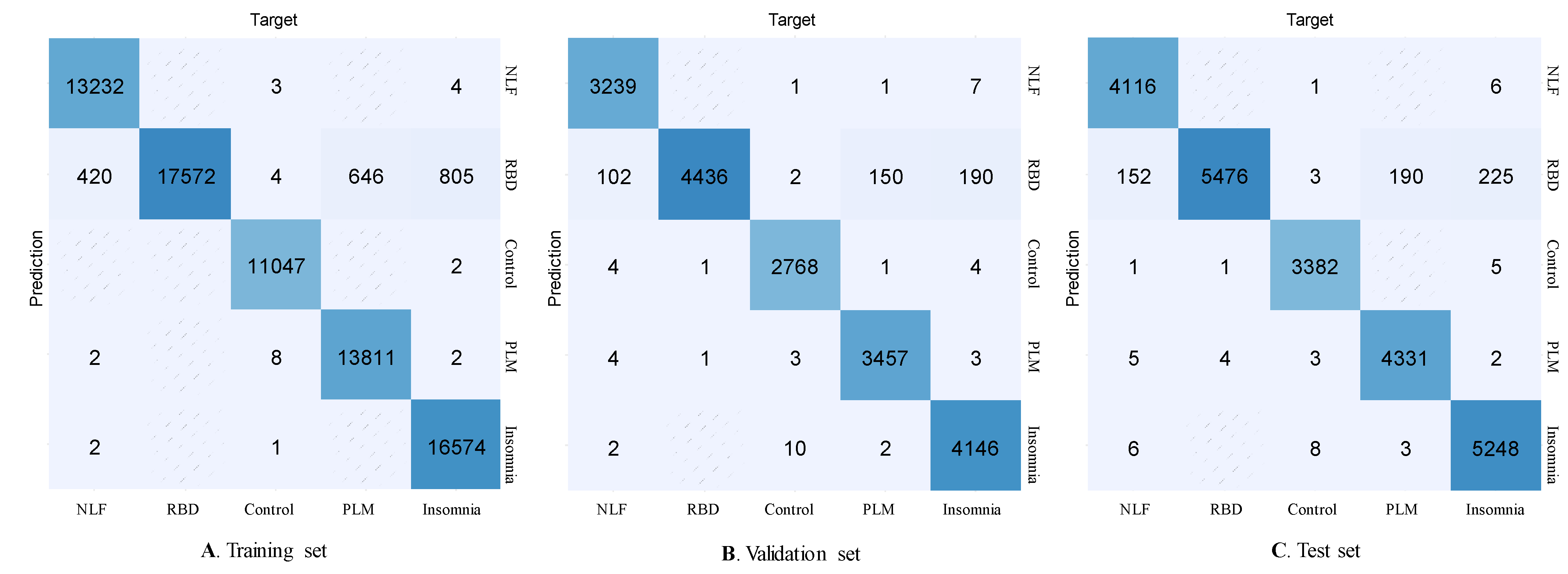
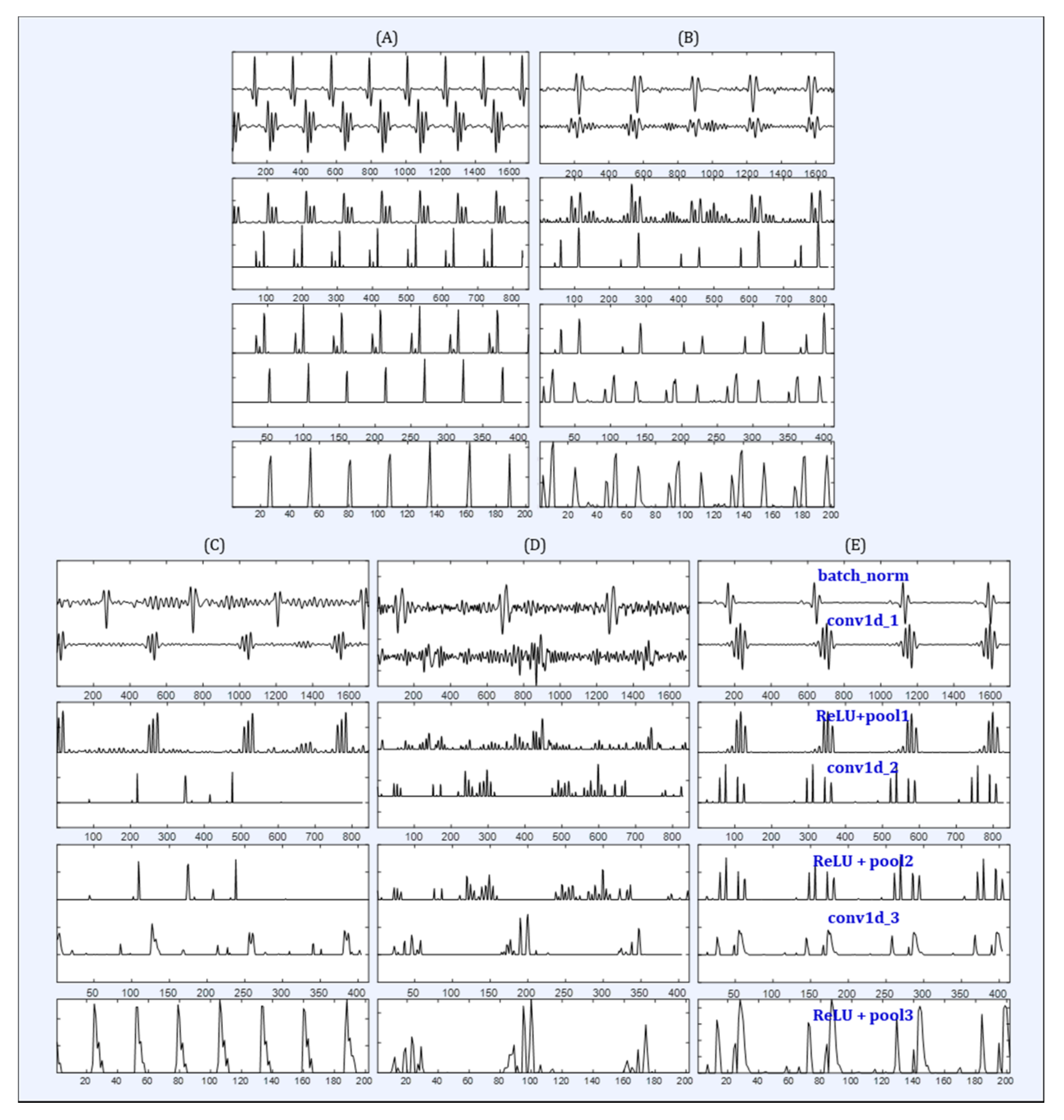
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thorpy, M. International classification of sleep disorders. In Sleep Disorders Medicine; Springer: New York, NY, USA, 2017; pp. 475–484. [Google Scholar]
- Šušmáková, K. Human Sleep and Sleep EEG. Meas. Sci. Rev. 2004, 4, 59–74. [Google Scholar]
- Banno, K.; Kryger, M.H. Sleep apnea: Clinical investigations in humans. Sleep Med. 2007, 8, 400–426. [Google Scholar]
- Morin, C.M.; Benca, R. Chronic insomnia. Lancet 2012, 379, 1129–1141. [Google Scholar] [PubMed]
- Moore, H., IV; Leary, E.; Lee, S.Y.; Carrillo, O.; Stubbs, R.; Peppard, P.; Young, T.; Widrow, B.; Mignot, E. Design and validation of a periodic leg movement detector. PLoS ONE 2014, 9, e114565. [Google Scholar] [CrossRef] [PubMed]
- McCarter, S.J.; Boswell, C.L.; Louis, E.K.S.; Dueffert, L.G.; Slocumb, N.; Boeve, B.F.; Silber, M.H.; Olson, E.J.; Tippmann-Peikert, M. Treatment outcomes in REM sleep behavior disorder. Sleep Med. 2013, 14, 237–242. [Google Scholar] [PubMed] [Green Version]
- Chervin, R.D. Sleepiness, fatigue, tiredness, and lack of energy in obstructive sleep apnea. Chest 2000, 118, 372–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graff-Radford, S.B.; Newman, A. Obstructive sleep apnea and cluster headache. Headae 2004, 44, 607–610. [Google Scholar] [CrossRef]
- Lattimore, J.D.L.; Celermajer, D.S.; Wilcox, I. Obstructive sleep apnea and cardiovascular disease. J. Am. Coll. Cardiol. 2003, 41, 1429–1437. [Google Scholar] [CrossRef] [Green Version]
- Lal, C.; Strange, C.; Bachman, D. Neurocognitive impairment in obstructive sleep apnea. Chest 2012, 141, 1601–1610. [Google Scholar] [CrossRef]
- Gamaldo, C.E.; Shaikh, A.K.; McArthur, J.C. The sleep-immunity relationship. Neurol. Clin. 2012, 30, 1313–1343. [Google Scholar] [CrossRef]
- Kapur, V.; Strohl, K.P.; Redline, S.; Iber, C.; O’connor, G.; Nieto, J. Underdiagnosis of sleep apnea syndrome in US communities. Sleep Breath 2002, 6, 49–54. [Google Scholar] [CrossRef]
- Douglas, N.J.; Thomas, S.; Jan, M.A. Clinical value of polysomnography. Lancet 1992, 339, 347–350. [Google Scholar] [CrossRef]
- Penzel, T.; Moody, G.B.; Mark, R.G.; Goldberger, A.L.; Peter, J.H. The apnea-ECG database. In Computers in Cardiology; IEEE: Cambridge, MA, USA, 2000; Volume 27, pp. 255–258. [Google Scholar]
- Faust, O.; Hagiwara, Y.; Hong, T.J.; Lih, O.S.; Acharya, U.R. Deep learning for healthcare applications based on physiological signals: A review. Comput. Methods Programs Biomed. 2018, 161, 1–13. [Google Scholar]
- Stein, P.K.; Pu, Y. Heart rate variability, sleep and sleep disorders. Sleep Med. Rev. 2012, 16, 47–66. [Google Scholar] [PubMed]
- Spiegelhalder, K.A.I.; Fuchs, L.; Ladwig, J.; Kyle, S.D.; Nissen, C.; Voderholzer, U.; Feige, B.; Riemann, D. Heart rate and heart rate variability in subjectively reported insomnia. J. Sleep Res. 2011, 20, 137–145. [Google Scholar] [CrossRef]
- De Chazal, P.; Heneghan, C.; Sheridan, E.; Reilly, R.; Nolan, P.; O’Malley, M. Automated processing of the single-lead electrocardiogram for the detection of obstructive sleep apnoea. IEEE Trans. Biomed. Eng. 2003, 50, 686–696. [Google Scholar]
- Mendez, M.O.; Bianchi, A.M.; Matteucci, M.; Cerutti, S.; Penzel, T. Sleep apnea screening by autoregressive models from a single ECG lead. IEEE Trans. Biomed. Eng. 2009, 56, 2838–2850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Zhang, X.; Song, C. An automatic screening approach for obstructive sleep apnea diagnosis based on single-lead electrocardiogram. IEEE Trans. Autom. Sci. Eng. 2015, 12, 106–115. [Google Scholar] [CrossRef]
- Adnane, M.; Jiang, Z.; Yan, Z. Sleep–wake stages classification and sleep efficiency estimation using single-lead electrocardiogram. Expert Syst. Appl. 2012, 39, 1401–1413. [Google Scholar]
- Xiao, M.; Yan, H.; Song, J.; Yang, Y.; Yang, X. Sleep stages classification based on heart rate variability and random forest. Biomed. Signal Process. Control 2013, 8, 624–633. [Google Scholar]
- Singh, J.; Sharma, R.K.; Gupta, A.K. A method of REM-NREM sleep distinction using ECG signal for unobtrusive personal monitoring. Comput. Biol. Med. 2016, 78, 138–143. [Google Scholar] [CrossRef]
- Yücelbaş, Ş.; Yücelbaş, C.; Tezel, G.; Özşen, S.; Yosunkaya, Ş.; Yosunkaya, S. Automatic sleep staging based on SVD, VMD, HHT and morphological features of single-lead ECG signal. Expert Syst. Appl. 2018, 102, 193–206. [Google Scholar]
- Wei, R.; Zhang, X.; Wang, J.; Dang, X. The research of sleep staging based on single-lead electrocardiogram and deep neural network. Biomed. Eng. Lett. 2018, 8, 87–93. [Google Scholar] [CrossRef]
- Li, Q.; Li, Q.; Liu, C.; Shashikumar, S.P.; Nemati, S.; Clifford, G.D. Deep learning in the cross-time frequency domain for sleep staging from a single-lead electrocardiogram. Physiol. Meas. 2018, 39, 124005. [Google Scholar]
- Radha, M.; Fonseca, P.; Ross, M.; Cerny, A.; Anderer, P.; Aarts, R.M. LSTM knowledge transfer for HRV-based sleep staging. arXiv 2018, arXiv:1809.06221v1. [Google Scholar]
- Erdenebayar, U.; Park, J.U.; Lee, S.; Joo, E.Y.; Lee, K.J. Prediction Method of Periodic Limb Movements Based on Deep Learning Using ECG Signal. Int. J. Fuzzy Log. Intell. Syst. 2020, 20, 138–144. [Google Scholar] [CrossRef]
- Dodds, K.L.; Miller, C.B.; Kyle, S.D.; Marshall, N.S.; Gordon, C.J. Heart rate variability in insomnia patients: A critical review of the literature. Sleep Med. Rev. 2017, 33, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Benjamens, S.; Dhunnoo, P.; Meskó, B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: An online database. NPJ Digit. Med. 2020, 3, 118. [Google Scholar] [PubMed]
- Mincholé, A.; Rodriguez, B. Artificial intelligence for the electrocardiogram. Nat. Med. 2019, 25, 22–23. [Google Scholar] [CrossRef] [PubMed]
- Castelletti, S.; Dagradi, F.; Goulene, K.; Danza, A.I.; Baldi, E.; Stramba-Badiale, M.; Schwartz, P.J. A wearable remote monitoring system for the identification of subjects with a prolonged QT interval or at risk for drug-induced long QT syndrome. Int. J. Cardiol. 2018, 266, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Torres-Soto, J.; Ashley, E.A. Multi-task deep learning for cardiac rhythm detection in wearable devices. NPJ Digit. Med. 2020, 3, 116. [Google Scholar] [CrossRef] [PubMed]
- Lyell, D.; Coiera, E.; Chen, J.; Shah, P.; Magrabi, F. How machine learning is embedded to support clinician decision making: An analysis of FDA-approved medical devices. BMJ Health Care Inform. 2021, 28, e100301. [Google Scholar] [CrossRef] [PubMed]
- Terzano, M.G.; Parrino, L.; Sherieri, A.; Chervin, R.; Chokroverty, S.; Guilleminault, C.; Hirshkowitz, M.; Mahowald, M.; Moldofsky, H.; Rosa, A.; et al. Atlas, rules, and recording techniques for the scoring of cyclic alternating pattern (CAP) in human sleep. Sleep Med. 2001, 2, 537–553. [Google Scholar] [CrossRef]
- Berry, R.B.; Brooks, R.; Gamaldo, C.E.; Harding, S.M.; Marcus, C.; Vaughn, B.V. The AASM manual for the scoring of sleep and associated events. Rules, terminology and technical specifications, Darien, Illinois. Am. Acad. Sleep Med. 2012, 176, 2012. [Google Scholar]
- Chung, J.; Gulcehre, C.; Cho, K.; Bengio, Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv 2014, arXiv:1412.3555. Available online: https://arxiv.org/abs/1412.3555 (accessed on 25 November 2015).
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. Int. Conf. Mach. Learn. PMLR 2015, 37, 448–456. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Nair, V.; Hinton, G.E. Rectified linear units improve restricted boltzmann machines. In Proceedings of the 27th International Conference on Machine Learning, Haifa, Israel, 21–24 June 2010; pp. 807–814. [Google Scholar]
- Chollet, F. Keras. 2015. Available online: http://keras.io/ (accessed on 7 June 2016).
- TensorFlow. Available online: https://www.tensorflow.org/ (accessed on 7 April 2021).
| Groups | Number (N) | Sex (M:F) | Age (Mean ± Std.) |
|---|---|---|---|
| CNT | 7 | 3:4 | 32.6 ± 3.7 |
| INS | 7 | 2:5 | 58.9 ± 10.2 |
| PLM | 7 | 5:2 | 54.7 ± 7.0 |
| RBD | 7 | 6:1 | 72.3 ± 6.7 |
| NFE | 7 | 3:4 | 26.3 ± 8.0 |
| Total | 35 | 19:16 | 48.9 ± 2.1 |
| Groups | Training Set | Validation Set | Test Set | Total |
|---|---|---|---|---|
| CNT | 11,087 | 2713 | 3444 | 17,244 |
| INS | 17,475 | 4309 | 5439 | 27,223 |
| PLM | 14,439 | 3675 | 4478 | 22,592 |
| RBD | 17,490 | 4458 | 5543 | 27,491 |
| NFE | 13,644 | 3379 | 4264 | 21,287 |
| Total | 74,135 | 18,534 | 23,168 | 115,837 |
| No. | Layers | Filters, Kernel Size | Output Shape | Parameters |
|---|---|---|---|---|
| 1 | batchnorm_1 | = | 5120 × 1 | 4 |
| 2 | conv1d_1 maxpool1d_1 dropout_1 | 100, 50 × 1 2 × 1 p = 0.25 | 5071 × 100 2535 × 100 | 5100 |
| 3 | conv1d_2 maxpool1d_2 dropout_2 | 80, 40 × 1 2 × 1 p = 0.25 | 2496 × 80 1248 × 80 | 320,080 |
| 4 | conv1d_3 maxpool1d_3 dropout_3 | 60, 30 × 1 2 × 1 p = 0.25 | 1219 × 60 609 × 60 | 144,060 |
| 5 | conv1d_4 maxpool1d_4 droput_4 | 40, 20 × 1 2 × 1 p = 0.25 | 590 × 40 295 × 40 | 48,040 |
| 6 | gru_1 droput_5 | 40 p = 0.25 | 295 × 40 | 9840 |
| 7 | gru_2 droput_6 | 40 p = 0.25 | 295 × 40 | 9840 |
| 8 | dense_1 | 5 | 40 | 205 |
| Total | 4 conv1d layers (280 filters), 2 gru (80 units) | 537,165 | ||
| Groups | Precision | Recall | F1-Score | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Train | Valid | Test | Train | Valid | Test | Train | Valid | Test | |
| CNT | 1.00 | 1.00 | 0.99 | 1.00 | 0.99 | 0.99 | 1.00 | 1.00 | 0.99 |
| INS | 1.00 | 1.00 | 0.99 | 0.95 | 0.95 | 0.95 | 0.98 | 0.97 | 0.97 |
| PLM | 1.00 | 1.00 | 0.99 | 0.96 | 0.96 | 0.95 | 0.98 | 0.98 | 0.97 |
| RBD | 0.90 | 0.91 | 0.91 | 1.00 | 1.00 | 1.00 | 0.95 | 0.95 | 0.95 |
| NLF | 1.00 | 1.00 | 1.00 | 0.97 | 0.97 | 0.96 | 0.98 | 0.98 | 0.98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urtnasan, E.; Joo, E.Y.; Lee, K.H. AI-Enabled Algorithm for Automatic Classification of Sleep Disorders Based on Single-Lead Electrocardiogram. Diagnostics 2021, 11, 2054. https://doi.org/10.3390/diagnostics11112054
Urtnasan E, Joo EY, Lee KH. AI-Enabled Algorithm for Automatic Classification of Sleep Disorders Based on Single-Lead Electrocardiogram. Diagnostics. 2021; 11(11):2054. https://doi.org/10.3390/diagnostics11112054
Chicago/Turabian StyleUrtnasan, Erdenebayar, Eun Yeon Joo, and Kyu Hee Lee. 2021. "AI-Enabled Algorithm for Automatic Classification of Sleep Disorders Based on Single-Lead Electrocardiogram" Diagnostics 11, no. 11: 2054. https://doi.org/10.3390/diagnostics11112054
APA StyleUrtnasan, E., Joo, E. Y., & Lee, K. H. (2021). AI-Enabled Algorithm for Automatic Classification of Sleep Disorders Based on Single-Lead Electrocardiogram. Diagnostics, 11(11), 2054. https://doi.org/10.3390/diagnostics11112054






