Diagnostic and Prognostic Indications of Nasopharyngeal Carcinoma
Abstract
1. Introduction
2. Cellular Oncogenes Involved in NPC
EBV-Associated Oncogenes in NPC
3. Hypermethylation of TSGs Promoter in NPC
4. Noncoding RNAs in NPC
4.1. MicroRNA (miRNA or miR) in NPC
4.2. EBV-Encoded BART miRNAs in NPC
4.3. Long Noncoding RNAs (lncRNAs) in NPC
4.4. EBV-Encoded BART lncRNAs in NPC
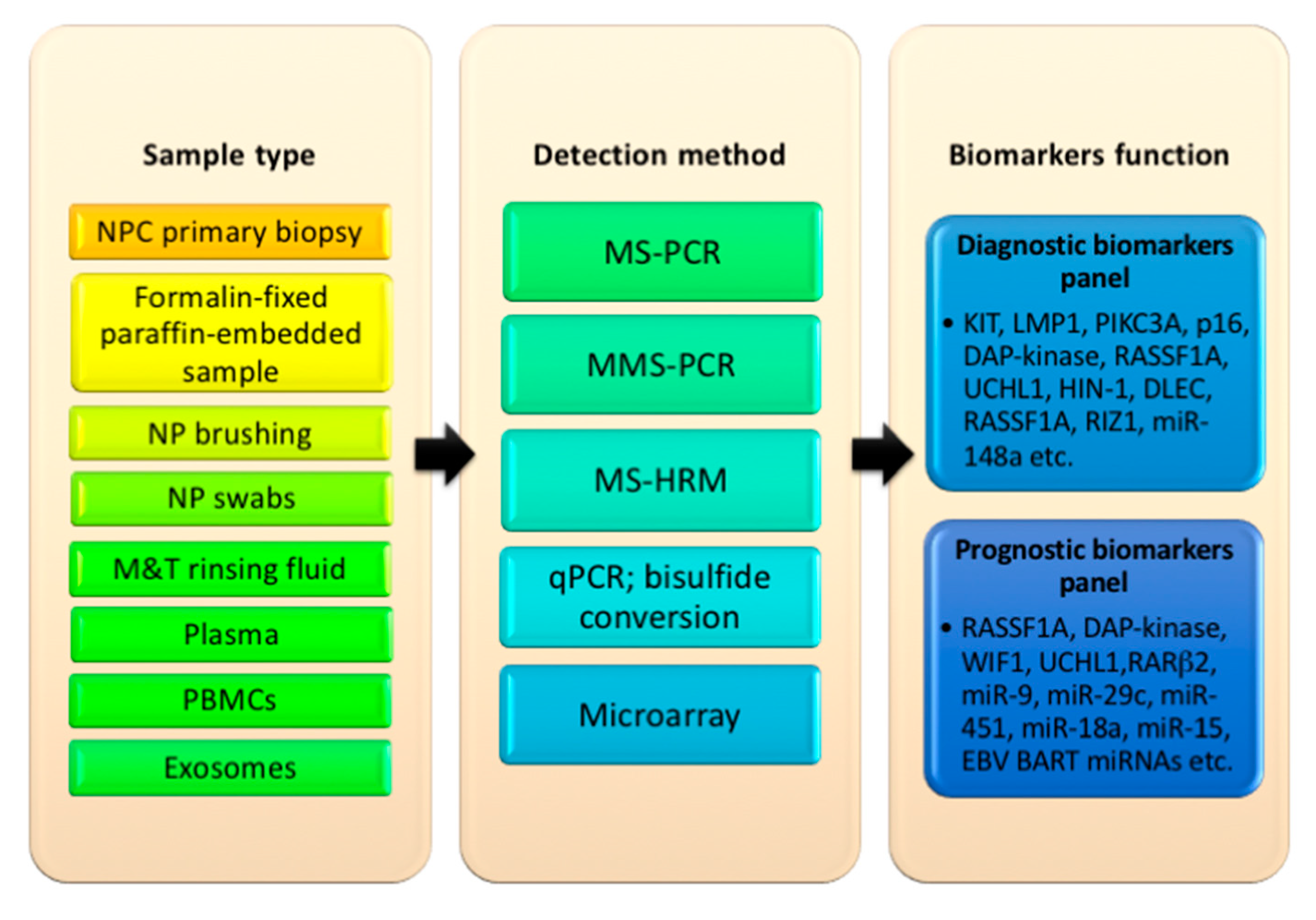
5. Types of Sample Used to Detect Oncogenes, Promoter Hypermethylation, and miRNAs in NPC
6. Proposed Selection of Oncogene-Tumor Suppressor Biomarkers for Diagnosis and Prognosis in NPC
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Young, L.S.; Dawson, C.W. Epstein-Barr virus and nasopharyngeal carcinoma. Chin. J. Cancer 2014, 33, 581–590. [Google Scholar] [CrossRef] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risk to Humans. Epstein-Barr Virus and Kaposi’s Sarcoma Herpesvirus/Human Herpesvirus 8; International Agency for Research on Cancer: Lyon, France, 1997. [Google Scholar]
- Chang, E.T.; Adami, H.-O. The Enigmatic Epidemiology of Nasopharyngeal Carcinoma. Cancer Epidemiol. Biomark. Amp. Prev. 2006, 15, 1765. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Li, C.; Pan, L. Nasopharyngeal carcinoma: A review of current updates. Exp. Ther. Med. 2018, 15, 3687–3692. [Google Scholar] [CrossRef] [PubMed]
- Sireci, F.; Speciale, R.; Sorrentino, R.; Turri-Zanoni, M.; Nicolotti, M.; Canevari, F.R. Nasal packing in sphenopalatine artery bleeding: Therapeutic or harmful? Eur. Arch. Oto-Rhino-Laryngol. 2017, 274, 1501–1505. [Google Scholar] [CrossRef] [PubMed]
- Chua, M.L.K.; Wee, J.T.S.; Hui, E.P.; Chan, A.T.C. Nasopharyngeal carcinoma. Lancet 2016, 387, 1012–1024. [Google Scholar] [CrossRef]
- Wang, K.H.; Austin, S.A.; Chen, S.H.; Sonne, D.C.; Gurushanthaiah, D. Nasopharyngeal Carcinoma Diagnostic Challenge in a Nonendemic Setting: Our Experience with 101 Patients. Perm. J. 2017, 21, 16–180. [Google Scholar] [CrossRef]
- Tabuchi, K.; Nakayama, M.; Nishimura, B.; Hayashi, K.; Hara, A. Early detection of nasopharyngeal carcinoma. Int. J. Otolaryngol. 2011, 2011, 638058. [Google Scholar] [CrossRef]
- Tay, J.K.; Lim, M.Y.; Kanagalingam, J. Screening in Nasopharyngeal Carcinoma: Current Strategies and Future Directions. Curr. Otorhinolaryngol. Rep. 2014, 2, 1–7. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, W.; Lin, L.; Xiao, X.; Zhou, X.; Ming, H.; Huang, T.; Liao, J.; Li, Y.; Zeng, X.; et al. Nasopharyngeal Epstein-Barr Virus Load: An Efficient Supplementary Method for Population-Based Nasopharyngeal Carcinoma Screening. PLoS ONE 2015, 10, e0132669. [Google Scholar] [CrossRef]
- Minamoto, T.; Mai, M.; Ronai, Z.E. Environmental factors as regulators and effectors of multistep carcinogenesis. Carcinogenesis 1999, 20, 519–527. [Google Scholar] [CrossRef]
- Broustas, C.G.; Lieberman, H.B. DNA damage response genes and the development of cancer metastasis. Radiat. Res. 2014, 181, 111–130. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Liu, M.; Cao, Y. New Insight into microRNA Functions in Cancer: Oncogene-microRNA-Tumor Suppressor Gene Network. Front. Mol. Biosci. 2017, 4, 46. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.M.; Thomas, S.D.; Islam, A.; Muench, D.; Sedoris, K. c-Myc and cancer metabolism. Clin. Cancer Res. 2012, 18, 5546–5553. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, T.; Nakagawara, A. Role of p53 in Cell Death and Human Cancers. Cancers 2011, 3, 994–1013. [Google Scholar] [CrossRef] [PubMed]
- Giudice, A.; D’Arena, G.; Crispo, A.; Tecce, M.F.; Nocerino, F.; Grimaldi, M.; Rotondo, E.; D’Ursi, A.M.; Scrima, M.; Galdiero, M.; et al. Role of Viral miRNAs and Epigenetic Modifications in Epstein-Barr Virus-Associated Gastric Carcinogenesis. Oxid. Med. Cell Longev. 2016, 2016, 6021934. [Google Scholar] [CrossRef]
- Jiang, N.; Liu, N.; Yang, F.; Zhou, Q.; Cui, R.; Jiang, W.; He, Q.; Li, W.; Guo, Y.; Zeng, J.; et al. Hotspot mutations in common oncogenes are infrequent in nasopharyngeal carcinoma. Oncol. Rep. 2014, 32, 1791–2431. [Google Scholar] [CrossRef]
- Zhang, Z.-C.; Fu, S.; Wang, F.; Wang, H.-Y.; Zeng, Y.-X.; Shao, J.-Y. Oncogene mutational profile in nasopharyngeal carcinoma. OncoTargets Ther. 2014, 7, 457–467. [Google Scholar] [CrossRef][Green Version]
- Zhang, J.-W.; Qin, T.; Hong, S.-D.; Zhang, J.; Fang, W.-F.; Zhao, Y.-Y.; Yang, Y.-P.; Xue, C.; Huang, Y.; Zhao, H.-Y.; et al. Multiple oncogenic mutations related to targeted therapy in nasopharyngeal carcinoma. Chin. J. Cancer 2015, 34, 177–183. [Google Scholar] [CrossRef]
- Chai, S.J.; Ahmad Zabidi, M.M.; Gan, S.P.; Rajadurai, P.; Lim, P.V.H.; Ng, C.C.; Yap, L.F.; Teo, S.H.; Lim, K.P.; Patel, V.; et al. An Oncogenic Role for Four-Jointed Box 1 (FJX1) in Nasopharyngeal Carcinoma. Dis. Markers 2019, 2019, 60803. [Google Scholar] [CrossRef]
- Hu, C.; Wei, W.; Chen, X.; Woodman, C.B.; Yao, Y.; Nicholls, J.M.; Joab, I.; Sihota, S.K.; Shao, J.-Y.; Derkaoui, K.D.; et al. A Global View of the Oncogenic Landscape in Nasopharyngeal Carcinoma: An Integrated Analysis at the Genetic and Expression Levels. PLoS ONE 2012, 7, e41055. [Google Scholar] [CrossRef]
- Scholle, F.; Bendt, K.M.; Raab-Traub, N. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J. Virol. 2000, 74, 10681–10689. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Ooka, T.; Middeldorp, J.; Takada, K. Reconstitution of Nasopharyngeal Carcinoma–Type EBV Infection Induces Tumorigenicity. Cancer Res. 2008, 68, 1030. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Zhang, W.; Jin, M.; Zhang, J.; Li, S.; Tong, F.; Zhou, Y. Differential expression of EBV proteins LMP1 and BHFR1 in EBV-associated gastric and nasopharyngeal cancer tissues. Mol. Med. Rep. 2016, 13, 4151–4158. [Google Scholar] [CrossRef] [PubMed]
- Khabir, A.; Karray, H.; Rodriguez, S.; Rosé, M.; Daoud, J.; Frikha, M.; Boudawara, T.; Middeldorp, J.; Jlidi, R.; Busson, P. EBV latent membrane protein 1 abundance correlates with patient age but not with metastatic behavior in north African nasopharyngeal carcinomas. Virol. J. 2005, 2, 39. [Google Scholar] [CrossRef]
- Huang, D.P.; Lo, K.-W.; van Hasselt, C.A.; Woo, J.K.S.; Choi, P.H.K.; Leung, S.-F.; Cheung, S.-T.; Cairns, P.; Sidransky, D.; Lee, J.C.K. A Region of Homozygous Deletion on Chromosome 9p21–22 in Primary Nasopharyngeal Carcinoma. Cancer Res. 1994, 54, 4003. [Google Scholar]
- Hui, A.B.; Lo, K.W.; Leung, S.F.; Choi, P.H.; Fong, Y.; Lee, J.C.; Huang, D.P. Loss of heterozygosity on the long arm of chromosome 11 in nasopharyngeal carcinoma. Cancer Res. 1996, 56, 0008–5472. [Google Scholar]
- Lo, K.-W.; Kwong, J.; Hui, A.B.-Y.; Chan, S.Y.-Y.; To, K.-F.; Chan, A.S.-C.; Chow, L.S.-N.; Teo, P.M.L.; Johnson, P.J.; Huang, D.P. High Frequency of Promoter Hypermethylation of RASSF1A in Nasopharyngeal Carcinoma. Cancer Res. 2001, 61, 3877. [Google Scholar]
- Tsang, Y.S.; Lo, K.W.; Leung, S.-F.; Choi, P.H.K.; Fong, Y.; Lee, J.C.K.; Huang, D.P. Two distinct regions of deletion on chromosome 13q in primary nasopharyngeal carcinoma. Int. J. Cancer 1999, 83, 305–308. [Google Scholar] [CrossRef]
- Shao, Y.; Jiang, H.; Wu, X.; Luo, Y.; Tang, W. p16 promoter hypermethylation is associated with increased risk of nasopharyngeal carcinoma. Mol. Clin. Oncol. 2014, 2, 1121–1124. [Google Scholar] [CrossRef]
- Wong, T.-S.; Kwong, D.L.-W.; Sham, J.S.-T.; Wei, W.I.; Kwong, Y.-L.; Yuen, A.P.-W. Quantitative Plasma Hypermethylated DNA Markers of Undifferentiated Nasopharyngeal Carcinoma. Clin. Cancer Res. 2004, 10, 2401. [Google Scholar] [CrossRef]
- Tian, F.; Yip, S.; Kwong, D.; Lin, Z.; Yang, Z.; Wu, V. Promoter hypermethylation of tumor suppressor genes in serum as potential biomarker for the diagnosis of nasopharyngeal carcinoma. Cancer Epidemiol. 2013, 37, 708–713. [Google Scholar] [CrossRef]
- Ayadi, W.; Karray-Hakim, H.; Khabir, A.; Feki, L.; Charfi, S.; Boudawara, T.; Ghorbel, A.; Daoud, J.; Frikha, M.; Busson, P.; et al. Aberrant methylation of p16, DLEC1, BLU and E-cadherin gene promoters in nasopharyngeal carcinoma biopsies from Tunisian patients. Anticancer Res. 2008, 28, 2161–2167. [Google Scholar]
- Challouf, S.; Ziadi, S.; Zaghdoudi, R.; Ksiaa, F.; Ben Gacem, R.; Trimeche, M. Patterns of aberrant DNA hypermethylation in nasopharyngeal carcinoma in Tunisian patients. Clin. Chim. Acta 2012, 413, 795–802. [Google Scholar] [CrossRef]
- Tong, J.H.M.; Tsang, R.K.Y.; Lo, K.-W.; Woo, J.K.S.; Kwong, J.; Chan, M.W.Y.; Chang, A.R.; van Hasselt, C.A.; Huang, D.P.; To, K.-F. Quantitative Epstein-Barr Virus DNA Analysis and Detection of Gene Promoter Hypermethylation in Nasopharyngeal (NP) Brushing Samples from Patients with NP Carcinoma. Clin. Cancer Res. 2002, 8, 2612. [Google Scholar] [PubMed]
- Xiao, L.; Jiang, L.; Hu, Q.; Li, Y. Promoter methylation of p16 and DAPK genes in brushing, blood, and tissue samples from patients with nasopharyngeal carcinoma: A systematic meta-analysis. Transl. Cancer Res. 2016, 5, 827–837. [Google Scholar] [CrossRef]
- Lo, K.-W.; Cheung, S.-T.; Leung, S.-F.; van Hasselt, A.; Tsang, Y.-S.; Mak, K.-F.; Chung, Y.-F.; Woo, J.K.S.; Lee, J.C.K.; Huang, D.P. Hypermethylation of the p16 Gene in Nasopharyngeal Carcinoma. Cancer Res. 1996, 56, 2721. [Google Scholar] [PubMed]
- Chang, H.W.; Chan, A.; Kwong, D.L.W.; Wei, W.I.; Sham, J.S.T.; Yuen, A.P.W. Evaluation of hypermethylated tumor suppressor genes as tumor markers in mouth and throat rinsing fluid, nasopharyngeal swab and peripheral blood of nasopharygeal carcinoma patient. Int. J. Cancer 2003, 105, 851–855. [Google Scholar] [CrossRef]
- Wong, T.S.; Tang, K.C.; Kwong, D.L.; Sham, J.S.; Wei, W.I.; Kwong, Y.L.; Yuen, A.P. Differential gene methylation in undifferentiated nasopharyngeal carcinoma. Int. J. Oncol. 2003, 22, 1019–6439. [Google Scholar] [CrossRef]
- Fendri, A.; Masmoudi, A.; Khabir, A.; Sellami-Boudawara, T.; Daoud, J.; Frikha, M.; Ghorbel, A.; Gargouri, A.; Mokdad-Gargouri, R. Inactivation of RASSF1A, RARβ2 and DAP-kinase by promoter methylation correlates with lymph node metastasis in nasopharyngeal carcinoma. Cancer Biol. Ther. 2009, 8, 444–451. [Google Scholar] [CrossRef]
- Kwong, J.; Lo, K.-W.; To, K.-F.; Teo, P.M.L.; Johnson, P.J.; Huang, D.P. Promoter Hypermethylation of Multiple Genes in Nasopharyngeal Carcinoma. Clin. Cancer Res. 2002, 8, 131. [Google Scholar]
- Jiang, W.; Liu, N.; Chen, X.-Z.; Sun, Y.; Li, B.; Ren, X.-Y.; Qin, W.-F.; Jiang, N.; Xu, Y.-F.; Li, Y.-Q.; et al. Genome-Wide Identification of a Methylation Gene Panel as a Prognostic Biomarker in Nasopharyngeal Carcinoma. Mol. Cancer Ther. 2015, 14, 2864. [Google Scholar] [CrossRef] [PubMed]
- Sze Wong, T.; Wen Chang, H.; Chi Tang, K.; Ignace Wei, W.; Lai Wen Kwong, D.; Sham, J.S.T.; Yuen, A.P.W.; Lam Kwong, Y. High Frequency of Promoter Hypermethylation of the DAP-kinase Gene in Nasopharyngeal Carcinoma and Its Detection in the Peripheral Blood of Patients. Clin. Cancer Res. 2002, 8, 433. [Google Scholar]
- Nawaz, I.; Moumad, K.; Martorelli, D.; Ennaji, M.M.; Zhou, X.; Zhang, Z.; Dolcetti, R.; Khyatti, M.; Ernberg, I.; Hu, L.-F. Detection of nasopharyngeal carcinoma in Morocco (North Africa) using a multiplex methylation-specific PCR biomarker assay. Clin. Epigenetics 2015, 7, 89. [Google Scholar] [CrossRef]
- Yang, X.; Dai, W.; Kwong, D.L.-W.; Szeto, C.Y.Y.; Wong, E.H.-W.; Ng, W.T.; Lee, A.W.M.; Ngan, R.K.C.; Yau, C.C.; Tung, S.Y.; et al. Epigenetic markers for noninvasive early detection of nasopharyngeal carcinoma by methylation-sensitive high resolution melting. Int. J. Cancer 2015, 136, E127–E135. [Google Scholar] [CrossRef] [PubMed]
- Ran, Y.; Wu, S.; You, Y. Demethylation of E-cadherin gene in nasopharyngeal carcinoma could serve as a potential therapeutic strategy. J. Biochem. 2010, 149, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Tsao, S.W.; Liu, Y.; Wang, X.; Yuen, P.W.; Leung, S.Y.; Yuen, S.T.; Pan, J.; Nicholls, J.M.; Cheung, A.L.M.; Wong, Y.C. The association of E-cadherin expression and the methylation status of the E-cadherin gene in nasopharyngeal carcinoma cells. Eur. J. Cancer 2003, 39, 524–531. [Google Scholar] [CrossRef]
- Wong, T.S.; Kwong, D.L.-W.; Sham, J.S.-T.; Tsao, S.W.; Wei, W.I.; Kwong, Y.L.; Yuen, A.P.-W. Promoter Hypermethylation of High-in-normal 1 Gene in Primary Nasopharyngeal Carcinoma. Clin. Cancer Res. 2003, 9, 3042. [Google Scholar]
- Hutajulu, S.H.; Indrasari, S.R.; Indrawati, L.P.L.; Harijadi, A.; Duin, S.; Haryana, S.M.; Steenbergen, R.D.M.; Greijer, A.E.; Middeldorp, J.M. Epigenetic markers for early detection of nasopharyngeal carcinoma in a high risk population. Mol. Cancer 2011, 10, 48. [Google Scholar] [CrossRef]
- Zhang, S.; Li, S.; Gao, J.-L. Promoter methylation status of the tumor suppressor gene SOX11 is associated with cell growth and invasion in nasopharyngeal carcinoma. Cancer Cell Int. 2013, 13, 109. [Google Scholar] [CrossRef]
- Yi, B.; Tan, S.-X.; Tang, C.-E.; Huang, W.-G.; Cheng, A.-L.; Li, C.; Zhang, P.-F.; Li, M.-Y.; Li, J.-L.; Yi, H.; et al. Inactivation of 14-3-3 σ by promoter methylation correlates with metastasis in nasopharyngeal carcinoma. J. Cell. Biochem. 2009, 106, 858–866. [Google Scholar] [CrossRef]
- Zhang, D.; Zhou, J.; Gao, J.; Wu, R.-Y.; Huang, Y.-L.; Jin, Q.-W.; Chen, J.-S.; Tang, W.-Z.; Yan, L.-H. Targeting snoRNAs as an emerging method of therapeutic development for cancer. Am. J. Cancer Res. 2019, 9, 1504–1516. [Google Scholar] [PubMed]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.-P.; Zhang, B.-X.; Chu, L. Small Nucleolar RNAs: Insight into Their Function in Cancer. Front. Oncol. 2019, 9, 587. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, Y.; Zhao, Y.; Gu, X. LncRNA SNHG5 promotes nasopharyngeal carcinoma progression by regulating miR-1179/HMGB3 axis. BMC Cancer 2020, 20, 178. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Zhang, X.; Xing, H.; Zhang, Y.; Cao, H.; Sang, J.; Gao, L.; Wang, L. Downregulated long non-coding RNA SNHG7 restricts proliferation and boosts apoptosis of nasopharyngeal carcinoma cells by elevating microRNA-140-5p to suppress GLI3 expression. Cell Cycle 2020, 19, 448–463. [Google Scholar] [CrossRef]
- Xu, W.; Sun, X.; Zang, C.; Jiang, Y. lncRNA SNHG7 promotes tumorigenesis of nasopharyngeal carcinoma via epithelial-to-mesenchymal transition. Oncol. Lett. 2020, 19, 2721–2726. [Google Scholar] [CrossRef]
- Liu, Z.-B.; Tang, C.; Jin, X.; Liu, S.-H.; Pi, W. Increased expression of lncRNA SNHG12 predicts a poor prognosis of nasopharyngeal carcinoma and regulates cell proliferation and metastasis by modulating Notch signal pathway. Cancer Biomark. 2018, 23, 603–613. [Google Scholar] [CrossRef]
- Sun, C.; Sun, Y.; Zhang, E. Long non-coding RNA SNHG20 promotes nasopharyngeal carcinoma cell migration and invasion by upregulating TGF-β1. Exp. Ther. Med. 2018, 16, 4967–4974. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. microRNA involvement in human cancer. Carcinogenesis 2012, 33, 1126–1133. [Google Scholar] [CrossRef]
- Li, Y.; Yan, L.; Zhang, W.; Wang, H.; Chen, W.; Hu, N.; Ou, H. miR-21 inhibitor suppresses proliferation and migration of nasopharyngeal carcinoma cells through down-regulation of BCL2 expression. Int. J. Clin. Exp. Pathol. 2014, 7, 3478–3487. [Google Scholar]
- Tan, G.; Tang, X.; Tang, F. The role of microRNAs in nasopharyngeal carcinoma. Tumour Biol. 2015, 36, 69–79. [Google Scholar] [CrossRef]
- Spence, T.; Bruce, J.; Yip, K.W.; Liu, F.-F. MicroRNAs in nasopharyngeal carcinoma. Chin. Clin. Oncol. 2016, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Claret, F.-X.; Wu, W. MicroRNAs as Therapeutic Targets in Nasopharyngeal Carcinoma. Front. Oncol. 2019, 9, 756. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Lu, J.; Zhang, B.; Liu, X.; Wang, L.; Li, S.-Y.; Peng, X.-H.; Xu, X.; Tian, W.-D.; Li, X.-P. miR-26a inhibits invasion and metastasis of nasopharyngeal cancer by targeting EZH2. Oncol. Lett. 2013, 5, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Alajez, N.M.; Shi, W.; Hui, A.B.Y.; Bruce, J.; Lenarduzzi, M.; Ito, E.; Yue, S.; O’Sullivan, B.; Liu, F.F. Enhancer of Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell Death Dis. 2010, 1, e85. [Google Scholar] [CrossRef]
- Liu, N.; Ling-Long, T.; Yung, S.; Rui-Xue, C.; Hui-Yun, W.; Bi-Jun, H.; Qing-Mei, H.; Wei, J.; Jun, M. MiR-29c suppresses invasion and metastasis by targeting TIAM1 in nasopharyngeal carcinoma. Cancer Lett. 2013, 392, 1872–7980. [Google Scholar] [CrossRef]
- Jia-Xing, Z.; D’ong, Q.; Feng-Wei, W.; D’ing-Zhun, L.; Jin-Huan, W.; Zhu-Ting, T.; Jia, F.; Xiao-Xia, H.; Yi-Ji, L.; Hai-Xia, D.; et al. MicroRNA-29c enhances the sensitivities of human nasopharyngeal carcinoma to cisplatin-based chemotherapy and radiotherapy. Cancer Lett. 2013, 329, 1872–7980. [Google Scholar]
- Xia, H.; Samuel, S.N.; Songshan, J.; William, K.C.C.; Johny, S.; Xiu-Wi, B.; Hsiang-Fu, K.; Marie, C.L. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem. Biophys. Res. Commun. 2010, 391, 1090–2104. [Google Scholar] [CrossRef]
- Wong, T.S.; Man, O.Y.; Tsang, C.M.; Tsao, S.W.; Tsang, R.K.Y.; Chan, J.Y.W.; Ho, W.K.; Wei, W.I.; To, V.S.H. MicroRNA let-7 suppresses nasopharyngeal carcinoma cells proliferation through downregulating c-Myc expression. J. Cancer Res. Clin. Oncol. 2011, 137, 415–422. [Google Scholar] [CrossRef]
- Wu, A.; Wu, K.; Li, J.; Mo, Y.; Lin, Y.; Wang, Y.; Shen, X.; Li, S.; Li, L.; Yang, Z. Let-7a inhibits migration, invasion and epithelial-mesenchymal transition by targeting HMGA2 in nasopharyngeal carcinoma. J. Transl. Med. 2015, 13, 105. [Google Scholar] [CrossRef]
- Peng, X.H.; Huang, H.R.; Lu, J.; Liu, X.; Zhao, F.P.; Zhang, B.; Lin, S.X.; Wang, L.; Chen, H.H.; Xu, X.; et al. MiR-124 suppresses tumor growth and metastasis by targeting Foxq1 in nasopharyngeal carcinoma. Mol. Cancer 2014, 13, 186. [Google Scholar] [CrossRef]
- Liu, N.; Jiang, N.; Guo, R.; Jiang, W.; He, Q.-M.; Xu, Y.-F.; Li, Y.-Q.; Tang, L.-L.; Mao, Y.-P.; Sun, Y.; et al. MiR-451 inhibits cell growth and invasion by targeting MIF and is associated with survival in nasopharyngeal carcinoma. Mol. Cancer 2013, 12, 123. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Liu, J.F.; Gu, Y.X.; Zheng, G.P.; He, Z.M. miR-216b suppresses cell proliferation and invasion by targeting PKCα in nasopharyngeal carcinoma cells. Zhonghua Zhong Liu Za Zhi 2013, 35, 645–650. [Google Scholar] [PubMed]
- Deng, M.; Tang, H.; Zhou, Y.; Zhou, M.; Xiong, W.; Zheng, Y.; Ye, Q.; Zeng, X.; Liao, Q.; Guo, X.; et al. miR-216b suppresses tumor growth and invasion by targeting KRAS in nasopharyngeal carcinoma. J. Cell Sci. 2011, 124, 2997. [Google Scholar] [CrossRef] [PubMed]
- Hui, A.B.Y.; Bruce, J.P.; Alajez, N.M.; Shi, W.; Yue, S.; Perez-Ordonez, B.; Xu, W.; Sullivan, B.; Waldron, J.; Cummings, B.; et al. Significance of Dysregulated Metadherin and MicroRNA-375 in Head and Neck Cancer. Clin. Cancer Res. 2011, 17, 7539. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Luo, H.-N.; Tian, W.-D.; Lu, J.; Li, G.; Wang, L.; Zhang, B.; Liang, B.-J.; Peng, X.-H.; Lin, S.-X.; et al. Diagnostic and prognostic value of plasma microRNA deregulation in nasopharyngeal carcinoma. Cancer Biol. Ther. 2013, 14, 1133–1142. [Google Scholar] [CrossRef]
- Luo, Z.; Dai, Y.; zhang, L.; Jiang, C.; Li, Z.; Yang, J.; McCarthy, J.B.; She, X.; Zhang, W.; Ma, J.; et al. miR-18a promotes malignant progression by impairing microRNA biogenesis in nasopharyngeal carcinoma. Carcinogenesis 2012, 34, 415–425. [Google Scholar] [CrossRef]
- Yu, X.; Zhen, Y.; Yang, H.; Wang, H.; Zhou, Y.; Wang, E.; Marincola, F.M.; Mai, C.; Chen, Y.; Wei, H.; et al. Loss of connective tissue growth factor as an unfavorable prognosis factor activates miR-18b by PI3K/AKT/C-Jun and C-Myc and promotes cell growth in nasopharyngeal carcinoma. Cell Death Dis. 2013, 4, e634. [Google Scholar] [CrossRef]
- Zhang, L.; Deng, T.; Li, X.; Liu, H.; Zhou, H.; Ma, J.; Wu, M.; Zhou, M.; Shen, S.; Li, X.; et al. microRNA-141 is involved in a nasopharyngeal carcinoma-related genes network. Carcinogenesis 2010, 31, 559–566. [Google Scholar] [CrossRef]
- Deng, M.; Ye, Q.; Qin, Z.; Zheng, Y.; He, W.; Tang, H.; Zhou, Y.; Xiong, W.; Zhou, M.; Li, X.; et al. miR-214 promotes tumorigenesis by targeting lactotransferrin in nasopharyngeal carcinoma. Tumor Biol. 2013, 34, 1793–1800. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Li, Y.-Y.; Fu, S.; Wang, X.-P.; Huang, M.-Y.; Zhang, X.; Shao, Q.; Deng, L.; Zeng, M.-S.; Zeng, Y.-X.; et al. MicroRNA-30a promotes invasiveness and metastasis in vitro and in vivo through epithelial–mesenchymal transition and results in poor survival of nasopharyngeal carcinoma patients. Exp. Biol. Med. 2014, 239, 891–898. [Google Scholar] [CrossRef]
- Luo, Z.; Zhang, L.; Li, Z.; Jiang, C.; Dai, Y.; Liu, X.; Zheng, Y.; Yu, H.; Xiang, J.; Li, G. miR-149 promotes epithelial-mesenchymal transition and invasion in nasopharyngeal carcinoma cells. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2011, 36, 604–609. [Google Scholar] [PubMed]
- Du, Z.-M.; Hu, L.-F.; Wang, H.-Y.; Yan, L.-X.; Zeng, Y.-X.; Shao, J.-Y.; Ernberg, I. Upregulation of MiR-155 in Nasopharyngeal Carcinoma is Partly Driven by LMP1 and LMP2A and Downregulates a Negative Prognostic Marker JMJD1A. PLoS ONE 2011, 6, e19137. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Tang, M.; Hu, Z.; Yan, B.; Pi, W.; Li, Z.; Zhang, J.; Zhang, L.; Jiang, W.; Li, G.; et al. miR-504 mediated down-regulation of nuclear respiratory factor 1 leads to radio-resistance in nasopharyngeal carcinoma. Oncotarget 2015, 6, 15995–16018. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, Z.; Shu, Y.; Zhou, H.; Wang, H.; Zhang, W. BART miRNAs: An unimaginable force in the development of nasopharyngeal carcinoma. Eur. J. Cancer Prev. 2017, 26, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lu, J.; Wang, F.; Liu, X.; Peng, X.; Yu, B.; Zhao, F.; Li, X. Dynamic Changes in Plasma MicroRNAs Have Potential Predictive Values in Monitoring Recurrence and Metastasis of Nasopharyngeal Carcinoma. BioMed Res. Int. 2018, 2018, 7329195. [Google Scholar] [CrossRef]
- Lu, J.; Xu, X.; Liu, X.; Peng, Y.; Zhang, B.; Wang, L.; Luo, H.; Peng, X.; Li, G.; Tian, W.; et al. Predictive value of miR-9 as a potential biomarker for nasopharyngeal carcinoma metastasis. Br. J. Cancer 2014, 110, 392–398. [Google Scholar] [CrossRef]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenetics 2018, 10, 59. [Google Scholar] [CrossRef]
- Zeng, X.; Xiang, J.; Wu, M.; Xiong, W.; Tang, H.; Deng, M.; Li, X.; Liao, Q.; Su, B.; Luo, Z.; et al. Circulating miR-17, miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. PLoS ONE 2012, 7, e46367. [Google Scholar] [CrossRef]
- Liu, N.; Cui, R.-X.; Sun, Y.; Guo, R.; Mao, Y.-P.; Tang, L.-L.; Jiang, W.; Liu, X.; Cheng, Y.-K.; He, Q.-M.; et al. A four-miRNA signature identified from genome-wide serum miRNA profiling predicts survival in patients with nasopharyngeal carcinoma. Int. J. Cancer 2014, 134, 1359–1368. [Google Scholar] [CrossRef]
- He, Y.; Zhang, L.; Cheng, G.; Yuan, R.; Zhuang, Y.; Zhang, D.; Zhou, D.; Xu, X. Upregulation of circulating miR-21 is associated with poor prognosis of nasopharyngeal carcinoma. Int. J. Clin. Exp. Pathol. 2017, 10, 7362–7368. [Google Scholar]
- Miao, B.-P.; Zhang, R.-S.; Li, M.; Fu, Y.-T.; Zhao, M.; Liu, Z.-G.; Yang, P.-C. Nasopharyngeal cancer-derived microRNA-21 promotes immune suppressive B cells. Cell. Mol. Immunol. 2015, 12, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Marquitz, A.R.; Raab-Traub, N. The role of miRNAs and EBV BARTs in NPC. Semin. Cancer Biol. 2012, 22, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Zhou, Y.; Zhang, L.; Chen, Y.; Lyu, X.; Cai, L.; Lu, Y.; Deng, Y.; Wang, J.; Yao, K.; et al. EBV-miR-BART1 is involved in regulating metabolism-associated genes in nasopharyngeal carcinoma. Biochem. Biophys. Res. Commun. 2013, 436, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-J.; Chen, G.-H.; Chen, Y.-H.; Liu, C.-Y.; Chang, K.-P.; Chang, Y.-S.; Chen, H.-C. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS ONE 2010, 5, e12745. [Google Scholar] [CrossRef] [PubMed]
- Gourzones, C.; Ferrand, F.-R.; Amiel, C.; Vérillaud, B.; Barat, A.; Guérin, M.; Gattolliat, C.-H.; Gelin, A.; Klibi, J.; Chaaben, A.B.; et al. Consistent high concentration of the viral microRNA BART17 in plasma samples from nasopharyngeal carcinoma patients-evidence of non-exosomal transport. Virol. J. 2013, 10, 119. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.Y.; Gao, W.; Ho, W.K.; Wei, W.I.; Wong, T.S. Overexpression of Epstein-Barr virus-encoded microRNA-BART7 in undifferentiated nasopharyngeal carcinoma. Anticancer Res. 2012, 32, 1791–7530. [Google Scholar]
- Zhang, G.; Zong, J.; Lin, S.; Verhoeven, R.J.A.; Tong, S.; Chen, Y.; Ji, M.; Cheng, W.; Tsao, S.-W.; Lung, M.; et al. Circulating Epstein–Barr virus microRNAs miR-BART7 and miR-BART13 as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int. J. Cancer 2015, 136, E301–E312. [Google Scholar] [CrossRef]
- Zhang, J.; Li, X.; Hu, J.; Cao, P.; Yan, Q.; Zhang, S.; Dang, W.; Lu, J. Long noncoding RNAs involvement in Epstein-Barr virus infection and tumorigenesis. Virol. J. 2020, 17, 51. [Google Scholar] [CrossRef]
- Zhang, W.; Huang, C.; Gong, Z.; Zhao, Y.; Tang, K.; Li, X.; Fan, S.; Shi, L.; Li, X.; Zhang, P.; et al. Expression of LINC00312, a long intergenic non-coding RNA, is negatively correlated with tumor size but positively correlated with lymph node metastasis in nasopharyngeal carcinoma. J. Mol. Histol. 2013, 44, 545–554. [Google Scholar] [CrossRef]
- Zhang, W.; Du, M.; Wang, T.; Chen, W.; Wu, J.; Li, Q.; Tian, X.; Qian, L.; Wang, Y.; Peng, F.; et al. Long non-coding RNA LINC01133 mediates nasopharyngeal carcinoma tumorigenesis by binding to YBX1. Am. J. Cancer Res. 2019, 9, 779–790. [Google Scholar]
- Kong, J.; Sun, W.; Li, C.; Wan, L.; Wang, S.; Wu, Y.; Xu, E.; Zhang, H.; Lai, M. Long non-coding RNA LINC01133 inhibits epithelial–mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016, 380, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Sun, W.; Zhu, W.; Liu, C.; Zhang, H.; Wang, H. Long noncoding RNA LINC01133 inhibits oral squamous cell carcinoma metastasis through a feedback regulation loop with GDF15. J. Surg. Oncol. 2018, 118. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-Z.; Cheng, T.-T.; He, Q.-J.; Lei, Z.-Y.; Chi, J.; Tang, Z.; Liao, Q.-X.; Zhang, H.; Zeng, L.-S.; Cui, S.-Z. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol. Cancer 2018, 17, 126. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Jing, Y.; Wei, F.; Tang, Y.; Yang, L.; Luo, J.; Yang, P.; Ni, Q.; Pang, J.; Liao, Q.; et al. Long non-coding RNA PVT1 predicts poor prognosis and induces radioresistance by regulating DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell Death Dis. 2018, 9, 235. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Li, M.; Liu, J. Long Noncoding RNA HOTTIP Promotes Nasopharyngeal Cancer Cell Proliferation, Migration, and Invasion by Inhibiting miR-4301. Med. Sci. Monit. 2019, 25, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Liu, X.; Qu, S.; Song, E.; Zou, H.; Gong, C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013, 104, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Gu, M.; You, B.; Shi, S.; Shan, Y.; Bao, L.; You, Y. Long non-coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinoma. Cancer Sci. 2016, 107, 1215–1222. [Google Scholar] [CrossRef]
- Song, P.; Ye, L.-F.; Zhang, C.; Peng, T.; Zhou, X.-H. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene 2016, 592, 8–14. [Google Scholar] [CrossRef]
- Zhuang, K.; Wu, Q.; Jin, C.-S.; Yuan, H.-J.; Cheng, J.-Z. Long non-coding RNA HNF1A-AS is upregulated and promotes cell proliferation and metastasis in nasopharyngeal carcinoma. Cancer Biomark. 2016, 16, 291–300. [Google Scholar] [CrossRef]
- Liao, B.; Wang, Z.; Zhu, Y.; Wang, M.; Liu, Y. Long noncoding RNA DRAIC acts as a microRNA-122 sponge to facilitate nasopharyngeal carcinoma cell proliferation, migration and invasion via regulating SATB1. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3585–3597. [Google Scholar] [CrossRef]
- Su, H.; Liu, L.; Zhang, Y.; Wang, J.; Zhao, Y. Long noncoding RNA NPCCAT1 promotes nasopharyngeal carcinoma progression via upregulating YY1. Biochimie 2019, 157, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-H.; Tang, J.-M.; Li, J.; Li, X.-W. Upregulation of SOX2-activated lncRNA ANRIL promotes nasopharyngeal carcinoma cell growth. Sci. Rep. 2018, 8, 3333. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lin, Y.; Yang, X.; Wu, X.; He, X. Long noncoding RNA H19 regulates EZH2 expression by interacting with miR-630 and promotes cell invasion in nasopharyngeal carcinoma. Biochem. Biophys. Res. Commun. 2016, 473, 913–919. [Google Scholar] [CrossRef] [PubMed]
- He, S.-W.; Xu, C.; Li, Y.-Q.; Li, Y.-Q.; Zhao, Y.; Zhang, P.-P.; Lei, Y.; Liang, Y.-L.; Li, J.-Y.; Li, Q.; et al. AR-induced long non-coding RNA LINC01503 facilitates proliferation and metastasis via the SFPQ-FOSL1 axis in nasopharyngeal carcinoma. Oncogene 2020, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.Y.; Cao, H.X. Long non-coding RNA CASC15 promotes nasopharyngeal carcinoma cell proliferation and metastasis by downregulating miR-101-3p. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8897–8904. [Google Scholar] [PubMed]
- Jia, X.; Niu, P.; Xie, C.; Liu, H. Long noncoding RNA PXN-AS1-L promotes the malignancy of nasopharyngeal carcinoma cells via upregulation of SAPCD2. Cancer Med. 2019, 8, 4278–4291. [Google Scholar] [CrossRef]
- Hu, X.; Liu, W.; Jiang, X.; Wang, B.; Li, L.; Wang, J.; Ma, J. Long noncoding RNA LINC00460 aggravates invasion and metastasis by targeting miR-30a-3p/Rap1A in nasopharyngeal carcinoma. Human Cell 2019, 32, 465–476. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, H.; Liu, H. Long Noncoding RNA UCA1 Accelerates Nasopharyngeal Carcinoma Cell Progression by Modulating miR-124-3p/ITGB1 Axis. OncoTargets Ther 2019, 12, 8455–8466. [Google Scholar] [CrossRef]
- Li, L.; Zhang, F. Novel long noncoding RNA LINC01385 promotes nasopharyngeal carcinoma proliferation via the miR-140-3p/Twist1 signaling pathway. Cell Cycle 2020, 19, 1352–1362. [Google Scholar] [CrossRef]
- Song, P.; Yin, S.-C. Long non-coding RNA 319 facilitates nasopharyngeal carcinoma carcinogenesis through regulation of miR-1207-5p/KLF12 axis. Gene 2019, 680, 51–58. [Google Scholar] [CrossRef]
- Cui, Z.; Pu, T.; Zhang, Y.; Wang, J.; Zhao, Y. Long non-coding RNA LINC00346 contributes to cisplatin resistance in nasopharyngeal carcinoma by repressing miR-342-5p. Open Biol. 2020, 10, 190286. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Yan, B.; Lu, Q.; Lin, Y.; Ma, L. The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumor Biol. 2016, 37, 4025–4033. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, T.; Wei, G.; Liu, L.; Chen, Q.; Xu, L.; Zhang, K.; Zeng, D.; Liao, R. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumor Biol. 2016, 37, 11733–11741. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Sun, L.; Dong, L.; Cui, P.; Xia, Z.; Li, C.; Zhu, Y. The role of long noncoding RNA-LET in cell proliferation and invasion of nasopharyngeal carcinoma and its mechanism. OncoTargets Ther. 2017, 10, 2769–2778. [Google Scholar] [CrossRef]
- Chak, W.-P.; Lung, R.W.-M.; Tong, J.H.-M.; Chan, S.Y.-Y.; Lun, S.W.-M.; Tsao, S.-W.; Lo, K.-W.; To, K.-F. Downregulation of long non-coding RNA MEG3 in nasopharyngeal carcinoma. Mol. Carcinog. 2017, 56, 1041–1054. [Google Scholar] [CrossRef]
- Guo, J.; Ma, J.; Zhao, G.; Li, G.; Fu, Y.; Luo, Y.; Gui, R. Long Noncoding RNA LINC0086 Functions as a Tumor Suppressor in Nasopharyngeal Carcinoma by Targeting miR-214. Oncol. Res. 2017, 25, 1189–1197. [Google Scholar] [CrossRef]
- Xu, Y.-Z.; Chen, F.-F.; Zhang, Y.; Liang, H.; Li, X.-J.; He, C. Identification of potential long noncoding RNA associated with nasopharyngeal carcinoma using deep sequencing. J. Int. Med Res. 2019, 47, 3271–3281. [Google Scholar] [CrossRef]
- Chen, S.; Luo, X.; Wu, W.; Li, Y.; Yu, H.; Wang, Y.; Yan, J. The long non-coding RNA MACC1-AS1 promotes nasopharyngeal carcinoma cell stemness via suppressing miR-145-mediated inhibition on SMAD2/MACC1-AS1 axis. Biomed. Pharmacother. 2020, 125, 109986. [Google Scholar] [CrossRef]
- He, B.; Zeng, J.; Chao, W.; Chen, X.; Huang, Y.; Deng, K.; Huang, Z.; Li, J.; Dai, M.; Chen, S.; et al. Serum long non-coding RNAs MALAT1, AFAP1-AS1 and AL359062 as diagnostic and prognostic biomarkers for nasopharyngeal carcinoma. Oncotarget 2017, 8, 41166–41177. [Google Scholar] [CrossRef]
- Fu, W.-M.; Lu, Y.-F.; Hu, B.-G.; Liang, W.-C.; Zhu, X.; Yang, H.-d.; Li, G.; Zhang, J.-F. Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget 2016, 7, 4712–4723. [Google Scholar] [CrossRef]
- He, B.; Li, W.; Wu, Y.; Wei, F.; Gong, Z.; Bo, H.; Wang, Y.; Li, X.; Xiang, B.; Guo, C.; et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016, 7, e2353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, S.; Zuo, L.; Yue, W.; Li, S.; Xin, S.; Liu, L.; Lu, J. Differential expression profiling of lncRNAs related to Epstein-Barr virus infection in the epithelial cells. J. Med Virol. 2019, 91, 1845–1855. [Google Scholar] [CrossRef] [PubMed]
- Marrugo-Ramírez, J.; Mir, M.; Samitier, J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int. J. Mol. Sci. 2018, 19, 2877. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.-F.; Zheng, X.-H.; Li, X.-Z.; Tian, T.; Zhang, S.-D.; Hu, Y.-Z.; Jia, W.-H. Nasopharyngeal brushing: A convenient and feasible sampling method for nucleic acid-based nasopharyngeal carcinoma research. Cancer Commun. 2018, 38, 8. [Google Scholar] [CrossRef]
- Lao, T.D.; Nguyen, T.V.; Nguyen, D.H.; Nguyen, M.T.; Nguyen, C.H.; Le, T.H.A. miR-141 is up-regulated in biopsies from Vietnamese patients with nasopharyngeal carcinoma. Braz. Oral Res. 2018, 32, e126. [Google Scholar] [CrossRef]
- Chang, H.W.; Chan, A.; Kwong, D.L.W.; Wei, W.I.; Sham, J.S.T.; Yuen, A.P.W. Detection of Hypermethylated RIZ1 Gene in Primary Tumor, Mouth, and Throat Rinsing Fluid, Nasopharyngeal Swab, and Peripheral Blood of Nasopharyngeal Carcinoma Patient. Clin. Cancer Res. 2003, 9, 1033. [Google Scholar]
- Peng, D.; Ren, C.-P.; Yi, H.-M.; Zhou, L.; Yang, X.-Y.; Li, H.; Yao, K.-T. Genetic and Epigenetic Alterations of DLC-1, a Candidate Tumor Suppressor Gene, in Nasopharyngeal Carcinoma. Acta Biochim. Biophys. Sin. 2006, 38, 349–355. [Google Scholar] [CrossRef][Green Version]
- Li, J.; Gong, P.; Lyu, X.; Yao, K.; Li, X.; Peng, H. Aberrant CpG island methylation of PTEN is an early event in nasopharyngeal carcinoma and a potential diagnostic biomarker. Oncol. Rep. 2014, 31, 1791–2431. [Google Scholar] [CrossRef]
- Lu, J.; Luo, H.; Liu, X.; Peng, Y.; Zhang, B.; Wang, L.; Xu, X.; Peng, X.; Li, G.; Tian, W.; et al. miR-9 targets CXCR4 and functions as a potential tumor suppressor in nasopharyngeal carcinoma. Carcinogenesis 2013, 35, 554–563. [Google Scholar] [CrossRef]
- Bar-Sela, G.; Kuten, A.; Ben-Eliezer, S.; Gov-Ari, E.; Ben-Izhak, O. Expression of HER2 and C-KIT in Nasopharyngeal Carcinoma: Implications for a New Therapeutic Approach. Mod. Pathol. 2003, 16, 1035–1040. [Google Scholar] [CrossRef]
- Wang, X.; Huang, Y.; Guo, R.; Liu, Y.; Qian, Y.; Liu, D.; Dai, X.; Wei, Z.; Jin, F.; Liu, Y. Clinicopathological significance of ROCK1 and PIK3CA expression in nasopharyngeal carcinoma. Exp. Ther. Med. 2017, 13, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Xianglan, M.; Wu, Y.; Yongta, H.; Wenwen, G.; Minyan, Z.; Hongtao, Y. Expression of miR-3182 and EBV-miR-BART8-3p in nasopharyngeal carcinoma is correlated with distant metastasis. Int. J. Clin. Exp. Pathol. 2018, 11, 3134–3140. [Google Scholar]
- Chen, X.; Wang, J.; Cheng, L.; Lu, M.-P. miR-18a downregulates DICER1 and promotes proliferation and metastasis of nasopharyngeal carcinoma. Int. J. Clin. Exp. Med. 2014, 7, 847–855. [Google Scholar] [PubMed]
- Wahyuningsih, L.; Dwianingsih, E.K.; Risanti, E.D.; Tirtoprodjo, P.; Rinonce, H.T.; Hakim, F.A.; Herdini, C.; Fachiroh, J. Tissue P16 is Associated with Smoking Status among Indonesian Nasopharyngeal Carcinoma Subjects. Asian Pac. J. Cancer Prev. 2019, 20, 2125–2130. [Google Scholar] [CrossRef]
- Hutajulu, S.H.; Hoebe, E.K.; Verkuijlen, S.A.; Fachiroh, J.; Hariwijanto, B.; Haryana, S.M.; Stevens, S.J.; Greijer, A.E.; Middeldorp, J.M. Conserved mutation of Epstein-Barr virus-encoded BamHI-A Rightward Frame-1 (BARF1) gene in Indonesian nasopharyngeal carcinoma. Infect. Agents Cancer 2010, 5, 16. [Google Scholar] [CrossRef]
- Zheng, X.-H.; Lu, L.-X.; Cui, C.; Chen, M.-Y.; Li, X.-Z.; Jia, W.-H. Epstein-Barr virus mir-bart1-5p detection via nasopharyngeal brush sampling is effective for diagnosing nasopharyngeal carcinoma. Oncotarget 2016, 7, 4972–4980. [Google Scholar] [CrossRef]
- Hao, S.-P.; Tsang, N.-M.; Chang, K.-P. Screening nasopharyngeal carcinoma by detection of the latent membrane protein 1 (LMP-1) gene with nasopharyngeal swabs. Cancer 2003, 97, 1909–1913. [Google Scholar] [CrossRef]
- Chung, A.-K.; OuYang, C.-N.; Liu, H.; Chao, M.; Luo, J.-D.; Lee, C.-Y.; Lu, Y.-J.; Chung, I.C.; Chen, L.-C.; Wu, S.-M.; et al. Targeted sequencing of cancer-related genes in nasopharyngeal carcinoma identifies mutations in the TGF-β pathway. Cancer Med. 2019, 8, 5116–5127. [Google Scholar] [CrossRef]
- Aga, M.; Bentz, G.L.; Raffa, S.; Torrisi, M.R.; Kondo, S.; Wakisaka, N.; Yoshizaki, T.; Pagano, J.S.; Shackelford, J. Exosomal HIF1α supports invasive potential of nasopharyngeal carcinoma-associated LMP1-positive exosomes. Oncogene 2014, 33, 4613–4622. [Google Scholar] [CrossRef]
- Zhou, Y.; Xia, L.; Lin, J.; Wang, H.; Oyang, L.; Tan, S.; Tian, Y.; Su, M.; Wang, H.; Cao, D.; et al. Exosomes in Nasopharyngeal Carcinoma. J. Cancer 2018, 9, 767–777. [Google Scholar] [CrossRef]
- Lu, J.; Liu, Q.-H.; Wang, F.; Tan, J.-J.; Deng, Y.-Q.; Peng, X.-H.; Liu, X.; Zhang, B.; Xu, X.; Li, X.-P. Exosomal miR-9 inhibits angiogenesis by targeting MDK and regulating PDK/AKT pathway in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 147. [Google Scholar] [CrossRef] [PubMed]
- Yue, P.Y.-K.; Ha, W.-Y.; Lau, C.-C.; Cheung, F.M.-F.; Lee, A.W.-M.; Ng, W.-T.; Ngan, R.K.-C.; Yau, C.-C.; Kwong, D.L.-W.; Lung, H.-L.; et al. MicroRNA profiling study reveals miR-150 in association with metastasis in nasopharyngeal carcinoma. Sci. Rep. 2017, 7, 12012. [Google Scholar] [CrossRef] [PubMed]
| TSG Biomarkers | Main Biological Function(s) | Method of Detection | Function of Biomarkers | Reference |
|---|---|---|---|---|
| p16 | Inhibitor of cyclin-dependent kinases (CDK) which slows down the cell cycle by hindering progression from G1 phase to S phase | MS-PCR | Diagnosis | [30,31,32,33,34,35,36,37] |
| p15 | Inhibits the growth of some kinds of tumor cells and acts as a mediator of TGF-β induced cell arrest. p15 shares extensive homology with p16 | MS-PCR | Diagnosis | [38,39] |
| RASSF1A | Regulates microtubule dynamics, cell cycle progression, and apoptosis | MS-PCR | Diagnosis, prognosis | [38,39,40,41,42] |
| DAP-kinase | Activates in various cellular activities including regulation of apoptosis, caspase-dependent death programs, cytoskeletal dynamics, and immune functions | MS-PCR | Diagnosis | [32,35,38,43,44,45] |
| CDH1 | Involved in various mechanisms like regulating cell-to-cell adhesions, mobility and proliferation of epithelial cells especially E-cadherin protein | MS-PCR | Diagnosis | [31,39,46] |
| E-cadherin | Important mediators of cell-to-cell interactions in epithelial tissues and holds cells together | MS-PCR | Diagnosis | [38,47] |
| HIN-1 | Inhibitor for cell growth, invasion, and AKT1 activation | MS-PCR | Diagnosis | [48] |
| MGMT | Has the ability to stoichiometrically repair DNA adducts and to self-inactivate | MS-PCR | Diagnosis | [39] |
| MLH1 | Provides instructions for making a protein which plays an important role in DNA repairs by fixing errors during DNA replication in preparation for cell division | MS-PCR | Diagnosis | [39] |
| RIZ1 | Induces G2-M cell cycle arrest and/or apoptosis | MS-PCR | Diagnosis | [44,49] |
| DLEC1 | Suppresses tumor growth and reduces the invasiveness of cancer cells | MS-PCR | Diagnosis | [32] |
| UCHL1 | Provides instructions for making ubiquitin carboxyl-terminal esterase L1 enzyme which is involved in cell machinery that degrades unwanted proteins | MS-PCR | Diagnosis | [32,42] |
| WIF1 | A lipid-binding protein which binds to Wnt proteins and prevents them from triggering signaling pathways | MS-PCR | Diagnosis, prognosis | [42,44,49] |
| DLC1 | Has the ability to enhance activated GTP-bound Rho-GTPases’ intrinsic ability to convert their GTP into GDP, thus rendering them inactive | MS-PCR | Diagnosis | [49] |
| SOX11 | Regulates embryonic development and determines the cell’s fate | MS-PCR | Diagnosis, prognosis | [50] |
| 14-3-3 sigma | Act as a negative regulator in the cell cycle and has been identified as p53-inducible gene product involved in cell cycle checkpoint control after DNA damage | MS-PCR | Prognosis | [51] |
| MiRNAS (Biomarkers) | Main Biological Function(s) | Validated Targets in NPC | Prognostic Association |
|---|---|---|---|
| Tumor Suppressor MiRNAs | |||
| miR-9 | Regulates cell proliferation, migration, invasion, epithelial–mesenchymal transition (EMT), metastasis, apoptosis, and tumor angiogenesis | CXCR4 [63] | Negative |
| miR-26a | Suppress cell proliferation and colony formation | EZH2 [64,65], c-Myc [61] | - |
| miR-29c | Inhibits cell migration and invasion, metastasis, associated with chemoresistance and radioresistance | TIAM1 [66], MCL-1 [67], BCL-2 [67] | Negative |
| miR-200 family | Regulates cell proliferation, migration, invasion, and EMT | ZEB2 [68], CTNNB1 [68] | - |
| Let-7 family | Inhibits cell proliferation and induces cell apoptosis | c-Myc [69], HMG2A [70] | - |
| miR-124 | Inhibits cell growth, migration, and invasion | Foxq1 [71] | - |
| miR-451 | Regulates cell proliferation, invasion and predicts outcome | MIF [72] | Negative |
| miR-216b | Suppress cell proliferation and invasion | PKCa [73], K-Ras [74] | - |
| miR-98 | Suppress NPC relapse and predicts recurrence | EZH2 [65] | Negative |
| miR-375 | Suppress NPC relapse and predicts recurrence | Metadherin [75] | Negative |
| Onco-miRNAs | |||
| miR-21 | Promotes cell proliferation and migration and high expression resulting in chemoresistance | PTEN/AKT [76], PDCD4 [76], TPM1, SPRY [76], ERCK [76], Bcl-2 [60] | Positive |
| miR-18a | Lymph node metastasis, overall downregulation of miRNA expression | Dicer1 [77], c-Jun and c-Myc [77] | Positive |
| miR-18b | Promotes cell proliferation | CTGF [78], | - |
| miR-141 | Promotes cell proliferation, migration, invasion, and cell apoptosis | BRD3 [79], PTEN [79], SPLUNC1 [79], UBAP1 [79] | - |
| miR-214 | Promotes cell proliferation, invasion, and metastasis | Lactotransferrin [80] | - |
| miR-30a | Increase cell capability of metastasis and invasion | E-cadherin [81] | - |
| miR-149 | Promotes cell migration, EMT, and invasion | E-cadherin [82] | - |
| miR-155 | Promotes cell proliferation, migration, invasion, colony formation, invasion, prognostic for tumor stage and predicts outcome | JMJD1A [83], BACH1 [83] | Positive |
| miR-504 | Predicts radioresistance outcome | NRF1 [84] | Positive |
| EBV-encoded BART miRNAs such as BART 1-3p, 5p, BART5, BART 6-5p, BART 7, BART 3, -6, -8, -16, -22 | Promotes tumor metastasis, cellular growth, and proliferation, inhibit cell apoptosis, and maintain virus latency | PTEN [85], PUMA [85], DICE1 [85], E-cadherin [85], Dicer [85], C-myc, and C-jun [85] | Positive |
| lncRNAs (Biomarkers) | Main Biological Function(s) | Method of Detection | Potential Function of lncRNAs |
|---|---|---|---|
| Tumor suppressor lncRNAs | |||
| LET | Inhibits the proliferation of NPC cells and induces cell apoptosis; transcriptional repressed by EZH2-mediated H3K27 histone methylation on the LET promoter | qRT-PCR [125] | - |
| LINC00312-NAG7 | Inhibits proliferation, induces apoptosis, and cell invasion | In situ hybridization | Diagnosis, Prognosis [100] |
| MEG3 | Repression of cell proliferation, colony formation, induction of cell cycle arrest and tumorigenicity in vitro and in vivo | Bisulfite sequencing, MS-PCR [126] | - |
| LINC0086 | Inhibits cell proliferation and promotes apoptosis. Upregulation of LINC0086 decreased miR-214 expression | In situ hybridization, qRT-PCR [127] | - |
| LINC01133 | Inhibits cell proliferation, invasion, and migration both in vitro and in vivo | qRT-PCR | Prognosis [101] |
| ENSG00000230489 – VAV3-AS1 | Downregulation of VAV3-AS1 leads to interference with natural killer cell-mediated cytotoxicity | qRT-PCR | Diagnosis [128] |
| Onco-lncRNAs | |||
| LINC01385 | Promotes cell proliferation via miR-140-3p/Twist1 signaling pathway | qRT-PCR | Prognosis [120] |
| UCA1 | Promotes cell progression by modulating miR-124-3p/ITGβ1 axis | qRT-PCR | Diagnosis [119] |
| LINC00460 | Aggravates invasion and metastasis by targeting miR-30a-3p/Rap1A axis | qRT-PCR | Prognosis [118] |
| LINC01503 | Facilitates proliferation and metastasis via SFPQ-FOSL1 axis | qRT-PCR | Prognosis [115] |
| MACC1-AS1 | Promotes cell stemness via suppressing miR-145-mediated inhibition on SMAD2/MACC1-AS1 axis | In situ hybridization, qRT-PCR | Diagnosis [129] |
| LINC00346 | Contributes to cisplastin resistance by repressing miR-342-5p | qRT-PCR | Prognosis [122] |
| HOTTIP | Promotes cell proliferation, migration and invasion by inhibiting miR-4301 | qRT-PCR | Prognosis [106] |
| DRAIC | Acts as a miR-122 sponge to facilitate cell proliferation, migration, and invasion via regulating SATB1 | qRT-PCR | Prognosis [111] |
| ENSG00000227084 | Interferes with Rap1 signaling pathway | qRT-PCR | Diagnosis [128] |
| PXN-AS1-L | Promotes cell proliferation, migration, and invasion in vitro and in vivo. It promotes NPC malignancy by upregulating SAPCD2 via direct RNA–RNA interaction | qRT-PCR | Prognosis [117] |
| CASC15 | Promotes cell proliferation and metastasis by downregulating miR-101-3p | qRT-PCR | Prognosis [116] |
| NPCCAT1 | Promotes cell growth, migration in vitro, and in vivo. Upregulates YY1 protein to promote NPC progression | qRT-PCR [112] | - |
| PVT1 | Inhibits cell proliferation, induces apoptosis | In situ hybridization, qRT-PCR | Prognosis [105] |
| ANRIL | Inhibits cell proliferation in vitro and in vivo. SOX2-ANRIL-β-catenin plays a role in NPC proliferation | qRT-PCR | Prognosis [113] |
| XIST | XIST upregulates E2F3 in part through “sponging” miR-34a-5p | qRT-PCR [109] | - |
| ROR | Suppress p53 signal pathway, promotes proliferation, migration, and chemoresistance | qRT-PCR | Diagnosis, Prognosis [108] |
| AFAP1-AS1 | Inhibits AFAP1 protein expression and affects the expression of several small GTPase family members and molecules in the actin cytokeratin signaling pathway | qRT-PCR | Diagnosis, Prognosis [130] |
| HOTAIR | Promoted angiogenesis through directly activating the transcription of angiogenic factor VEGFA as well as through GRP78-mediated upregulation of VEGFA and Ang2 expression | In situ hybridization, qRT-PCR | Diagnosis [131], Prognosis [107] |
| HNF1A-AS | Promotes proliferation, migration, and EMT | qRT-PCR [110] | Diagnosis |
| H19 | Inhibits E-cadherin expression and promotes invasion via the miR-630/EZH2 pathway | qRT-PCR [114] | - |
| NEAT1 | Regulates EMT phenotype and radioresistance by modulating the miR-204/ZEB1 axis | In situ hybridization, qRT-PCR | Diagnosis, Prognosis [124] |
| MALAT1 | Regulates CSC activity and radioresistance by modulating miR-1/slug axis | In situ hybridization, qRT-PCR [123] | Diagnosis, prognosis [130] |
| LINC00319 | Promotes cell growth in vitro. LINC00319 contributed to NPC progression by regulating miR-1207-5p/KLF12 signal pathway | qRT-PCR | Prognosis [121] |
| Type of Samples | Frequency of Oncogenes Involved (%) | Frequency of Tumor Suppressors Involved (%) |
|---|---|---|
| NPC primary biopsy | EBV-LMP1 (62.5%) [24] EBV-BARF1 (13.3%) [24] miR-141 [136] lncRNA PVT1 (64%) [105] lncRNA NPCCAT1 [112] lncRNA CASC15 [116] lncRNA PXN-AS1-L [117] lncRNA HOTTIP [106] lncRNA LINC00346 [122] lncRNA LINC01503 [115] lncRNA LINC00460 [118] lncRNA UCA1 [119] lncRNA ANRIL [113] lncRNA H19 [114] lncRNA MALAT1 [123] lncRNA NEAT1 [124] lncRNA LINC01385 [120] lncRNA HNF1A-AS [110] | p16 (23–66%) [32,33,34] p15 (50–80%) [38,39] RASSF1A (46–67%) [38,39] DAP-kinase (75–77%) [38,43] RIZ1 (60%) [137] CDH1 (50%) [39,46] E-cadherin (52–65%) [38,46] HIN-1 (77%) [48] MGMT (28%) [39] MLH1 (40%) [39] DLC1 (79%) [138] SOX11 (67.4%) [50] 14-3-3 sigma (84%) [51] PTEN (82.2%) [139] miR-9 [140] lncRNA LINC00312-NAG7 (51.4%) [100] |
| Formalin-fixed paraffin-embedded sample | ABL1 (1.6%) * AKT1 (0%) * AKT2 (0%) * BRAF (0.8%) * CDK (0%) * EGFR (0.8%) * ERBB2 (0%) * FGFR1 (0%) * FGFR3 (0%) * FLT3 (0%) * HRAS (0.8%) * JAK2 (0%) * KIT (3.3–33%) [18,141] KRAS (0%) * MET (0%) * NRAS (4.1%) * PDGFRA (1.6%) * PIKC3A (4.9–62.96%) [18,142] RET (0%) * ROCK1 (28.4%) [142] EBV-miR-BART8-3p (52%) [143] miR-3182 (51%) [143] miR-18a (71.1%) [144] miR-149 (82.4%) ** miR-141 (52.9%) ** miR-205 (94.1%) ** miR-196a (88.2%) ** miR-149 (82.4%) ** miR-183 (64.7%) ** miR-224 (58.8%) ** miR-210 (58.8%) ** miR-136 (47.1%) ** miR-200c (64.7%) ** lncRNA MACC1-AS1 [129] lncRNA HOTAIR [107] | p16 (5%) [145] miR-150 (82.4%) ** |
| NP brushing | BARF1 [146] EBV-miR-BART1–5p [147] EBV-miR-BART5 [147] EBV-miR-BART6-5p [147] EBV-miR-BART17-5p [147] | WIF1 (61.2%) [49] p16 (46.4 –66%) [35,49] RASSF1A (39.3–75.5%) [35,49] DAP-kinase (79.2%) [49] RIZ1 (56.6%) [49] DLC1 (76.9%) [49] |
| NP swabs | LMP1 [148] | p16 (17%) [38] RIZ1 (37%) [137] E-cadherin (27%) [38] |
| M&T rinsing fluid | - | p16 (17%) [38] RIZ1 (30%) [137] E-cadherin (43%) [38] |
| Plasma | EBV-miR-BART7 [98] EBV-miR-BART13 [98] miR-21 [60] | p16 (42%) [31] RIZ1 (23%) [137] miR-9 [87] |
| PBMCs | PIKC3A [149] TP53 [149] | RIZ1 (10%) [137] |
| Exosomes | HIF1α [150] LMP1 [151] | DLEC1 (25%) UCHL1 (64.9%) miR-9 [152] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
E. A. R., E.N.S.; Irekeola, A.A.; Yean Yean, C. Diagnostic and Prognostic Indications of Nasopharyngeal Carcinoma. Diagnostics 2020, 10, 611. https://doi.org/10.3390/diagnostics10090611
E. A. R. ENS, Irekeola AA, Yean Yean C. Diagnostic and Prognostic Indications of Nasopharyngeal Carcinoma. Diagnostics. 2020; 10(9):611. https://doi.org/10.3390/diagnostics10090611
Chicago/Turabian StyleE. A. R., Engku Nur Syafirah, Ahmad Adebayo Irekeola, and Chan Yean Yean. 2020. "Diagnostic and Prognostic Indications of Nasopharyngeal Carcinoma" Diagnostics 10, no. 9: 611. https://doi.org/10.3390/diagnostics10090611
APA StyleE. A. R., E. N. S., Irekeola, A. A., & Yean Yean, C. (2020). Diagnostic and Prognostic Indications of Nasopharyngeal Carcinoma. Diagnostics, 10(9), 611. https://doi.org/10.3390/diagnostics10090611







