Comprehensive Serum Glycopeptide Spectra Analysis Combined with Machine Learning for Early Detection of Lung Cancer: A Case–Control Study
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
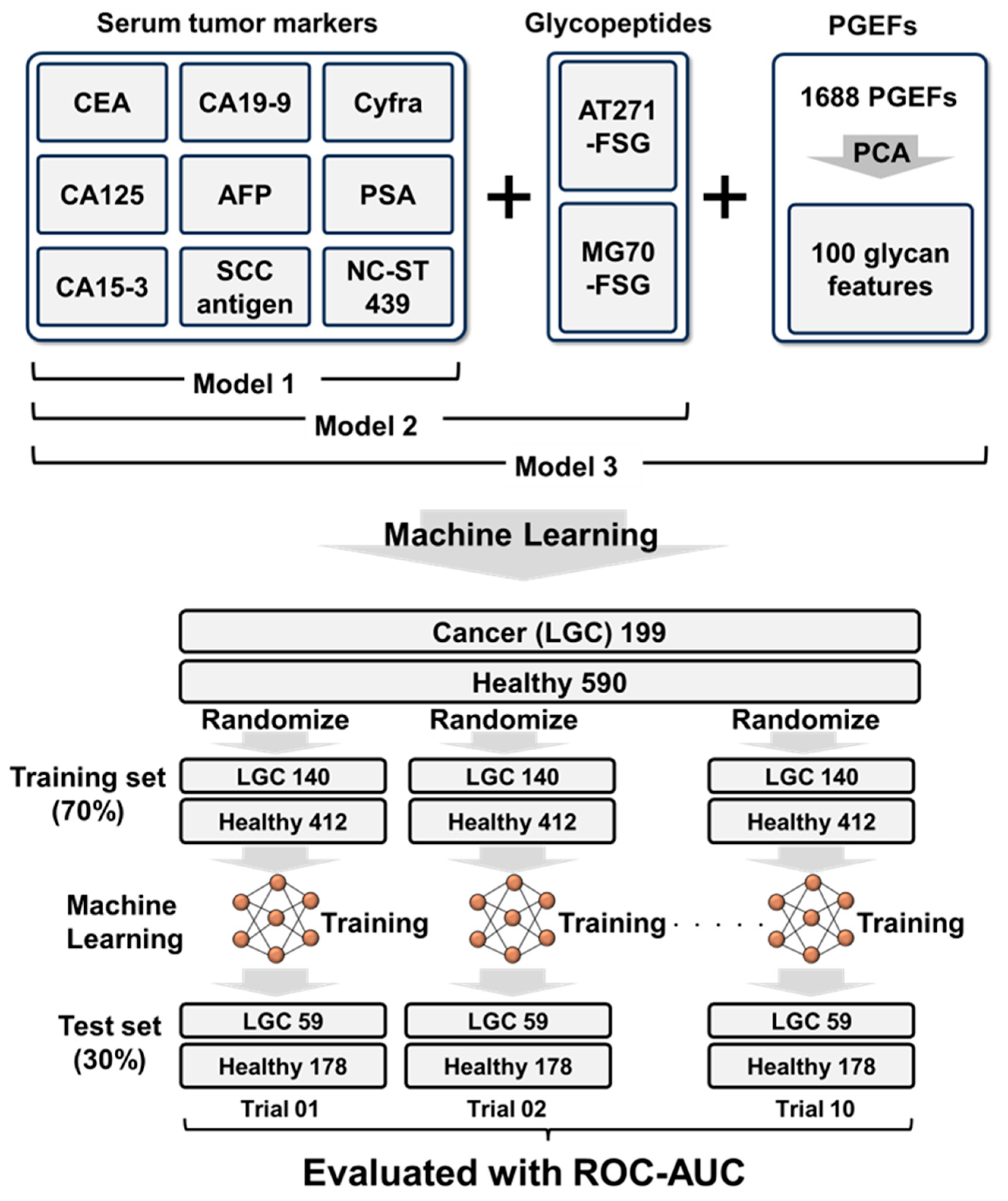
2.1. Study Design
2.2. Tumor Marker Analyses
2.3. Sample Preparation and Liquid Chromatography–Tandem Mass Spectrometry
2.4. Data Processing
2.5. Identification of the Glycopeptides Contributing to Lung Cancer Discrimination
2.6. Statistical Analysis
3. Results
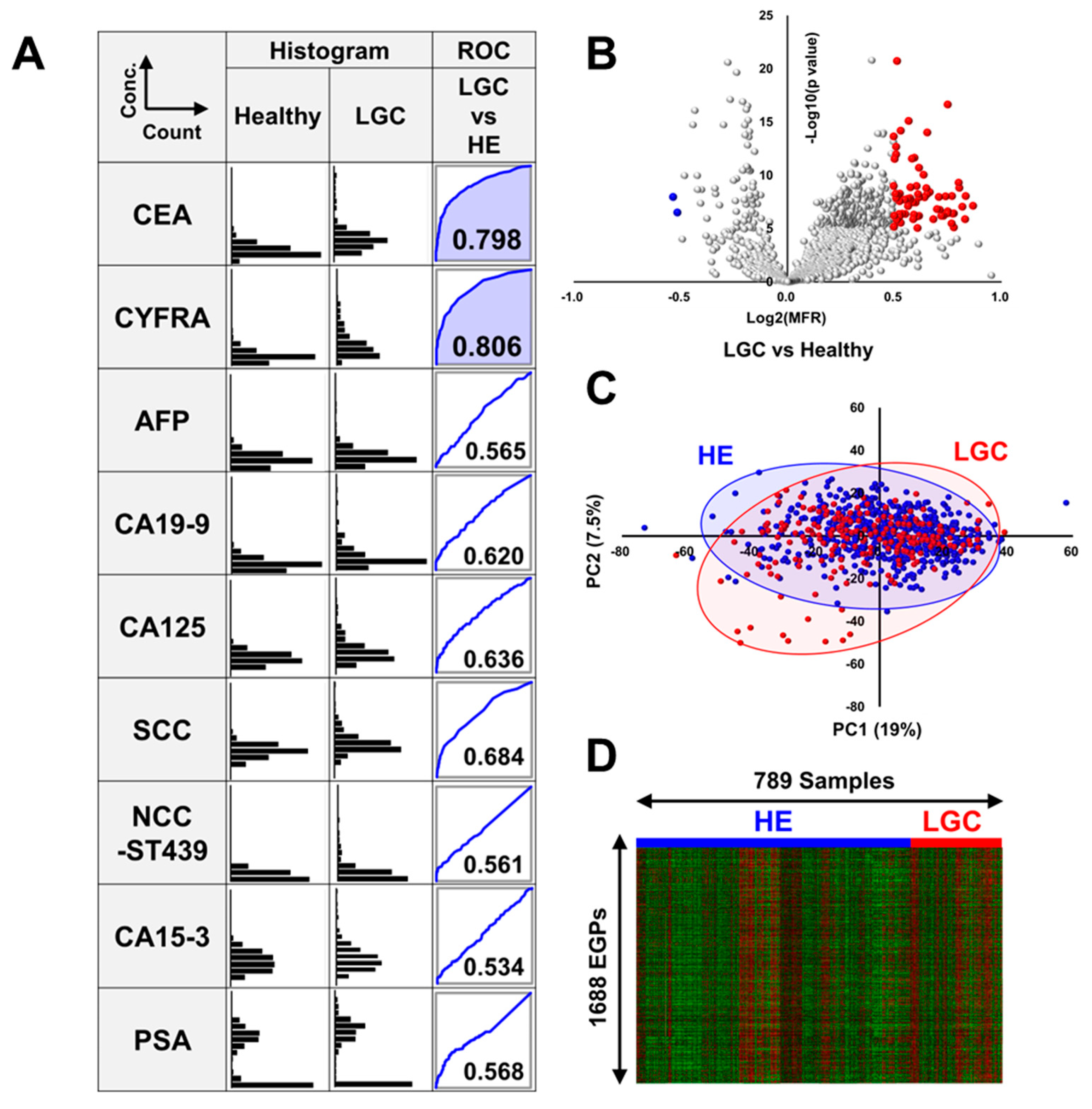
3.1. Comparison of the Levels of Tumor Markers in Patients with LGC and Healthy Volunteers
3.2. Volcano Plot Analysis, Principal Component Analysis, and Heatmap Analysis of EGPs in LGC
3.3. Identification of Novel LGC-Specific Biomarkers in EGPs
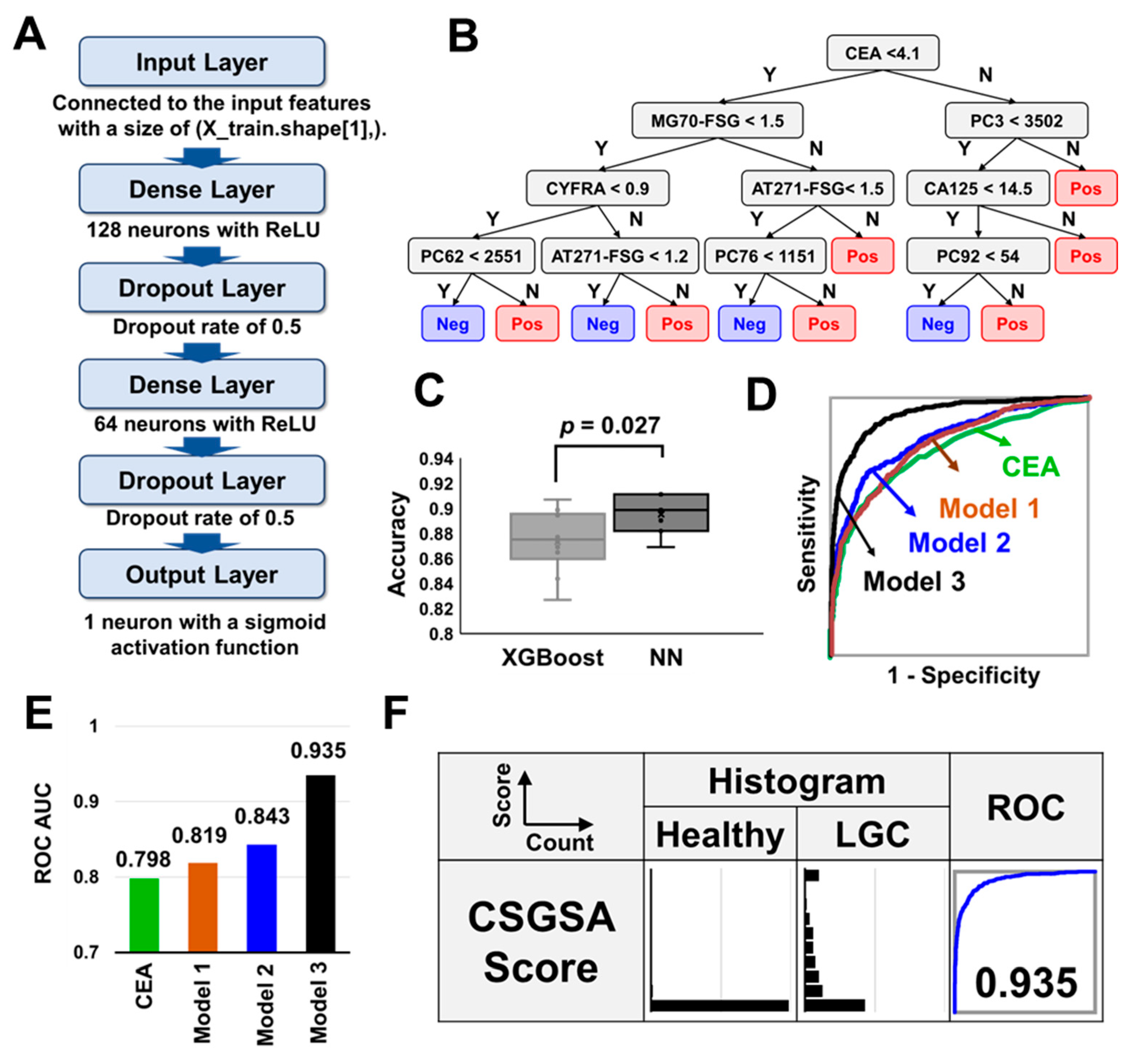
3.4. Combination Analysis of Tumor Markers, AT271-FSG, MG70-FSG, and 1688 EPGs
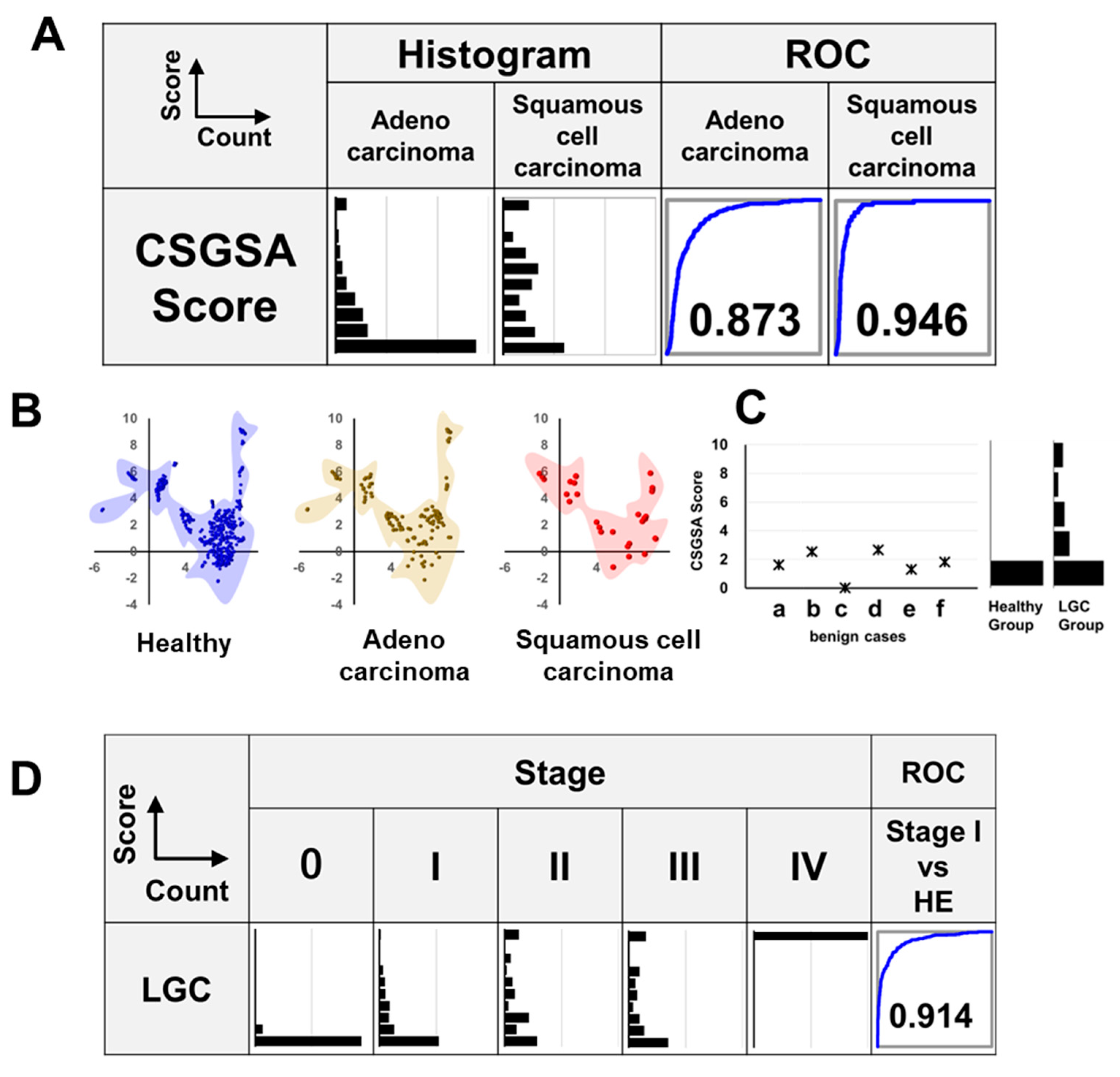
3.5. Relationship Between LGC Histological Type and CSGSA Score
3.6. CSGSA Determination for Patients with Benign Lung Disease
3.7. Relationship Between CSGSA Score and Cancer Development (Stage)
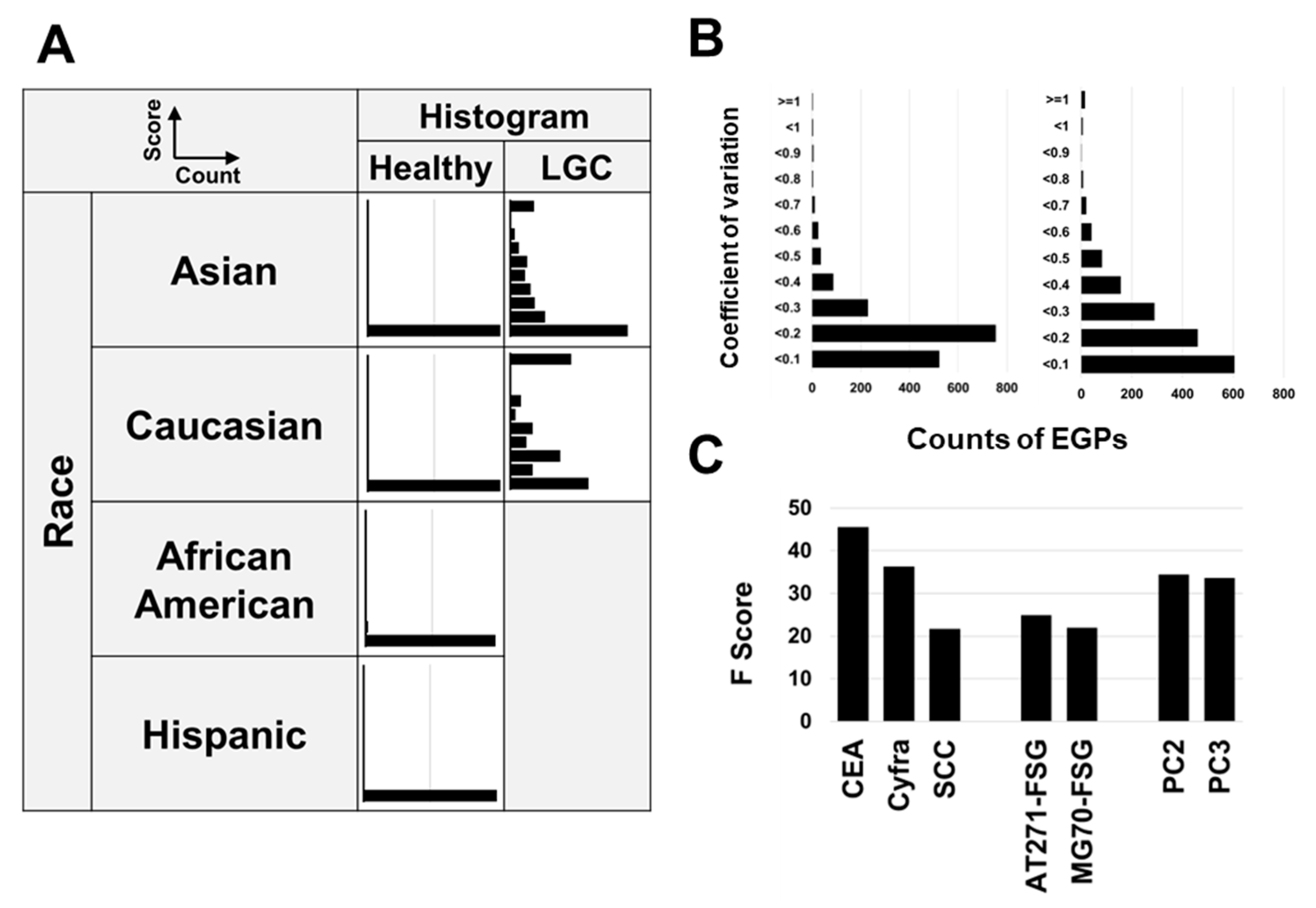
3.8. Relationship Between CSGSA Scores and Ethnicity
3.9. Method Validation
3.10. Key Contributors to Model Efficacy
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LGC | Lung cancer |
| EGP | Enriched glycopeptide |
| LC-MS | liquid chromatography–mass spectrometry |
| MS/MS | Tandem mass spectrometry |
| AFP | Alpha-fetoprotein |
| PSA | Prostate specific antigen |
| CEA | Carcinoembryonic antigen |
| CA125 | Cancer antigen 125 |
| CSGSA | Comprehensive Serum Glycopeptide Spectra Analysis |
| QC | quality control |
| PPV | positive predictive value |
| MFR | mean-fold ratio |
References
- Li, C.; Lei, S.; Ding, L.; Xu, Y.; Wu, X.; Wang, H.; Zhang, Z.; Gao, T.; Zhang, Y.; Li, L. Global burden and trends of lung cancer incidence and mortality. Chin. Med. J. 2023, 136, 1583–1590. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Center Research Institute Japan, Cancer Statistics in Japan. 2024. Available online: https://ganjoho.jp/public/qa_links/report/statistics/en.html (accessed on 31 May 2024).
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics. 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Leiter, A.; Veluswamy, R.R.; Wisnivesky, J.P. The global burden of lung cancer: Current status and future trends. Nat. Rev. Clin. Oncol. 2023, 20, 624–639. [Google Scholar] [CrossRef] [PubMed]
- Grunnet, M.; Sorensen, J.B. Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer 2012, 76, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhang, G.; Yang, M.; Zhang, S.; Zhao, B.; Shen, G.; Chai, Y. Systematic review of CYFRA 21-1 as a prognostic indicator and its predictive correlation with clinicopathological features in Non-small Cell Lung Cancer: A meta-analysis. Oncotarget 2017, 8, 4043–4050. [Google Scholar] [CrossRef]
- Seijo, L.M.; Peled, N.; Ajona, D.; Boeri, M.; Field, J.K.; Sozzi, G.; Pio, R.; Zulueta, J.J.; Spira, A.; Massion, P.P.; et al. Biomarkers in Lung Cancer Screening: Achievements, Promises, and Challenges. J. Thorac. Oncol. 2019, 14, 343–357. [Google Scholar] [CrossRef]
- Chu, G.C.W.; Lazare, K.; Sullivan, F. Serum and blood based biomarkers for lung cancer screening: A systematic review. BMC Cancer 2018, 18, 181. [Google Scholar] [CrossRef]
- Dantas, E.; Murthy, A.; Ahmed, T.; Ahmed, M.; Ramsamooj, S.; Hurd, M.A.; Lam, T.; Malbari, M.; Agrusa, C.; Elemento, O.; et al. TIMP1 is an early biomarker for detection and prognosis of lung cancer. Clin. Transl. Med. 2023, 13, e1391. [Google Scholar] [CrossRef]
- Saviana, M.; Romano, G.; McElroy, J.; Nigita, G.; Distefano, R.; Toft, R.; Calore, F.; Le, P.; Morales, D.D.V.; Atmajoana, S.; et al. A plasma miRNA-based classifier for small cell lung cancer diagnosis. Front. Oncol. 2023, 13, 1255527. [Google Scholar] [CrossRef]
- Zhao, Y.; O’Keefe, C.M.; Hsieh, K.; Cope, L.; Joyce, S.C.; Pisanic, T.R.; Herman, J.G.; Wang, T.H. Multiplex Digital Methylation-Specific PCR for Noninvasive Screening of Lung Cancer. Adv. Sci. 2023, 10, e2206518. [Google Scholar] [CrossRef]
- Smolarz, M.; Widlak, P. Serum Exosomes and Their miRNA Load-A Potential Biomarker of Lung Cancer. Cancers 2021, 13, 1373. [Google Scholar] [CrossRef] [PubMed]
- Fahrmann, J.F.; Marsh, T.; Irajizad, E.; Patel, N.; Murage, E.; Vykoukal, J.; Dennison, J.B.; Do, K.A.; Ostrin, E.; Spitz, M.R.; et al. Blood-Based Biomarker Panel for Personalized Lung Cancer Risk Assessment. J. Clin. Oncol. 2022, 40, 876–883. [Google Scholar] [CrossRef]
- Xie, Y.; Meng, W.Y.; Li, R.Z.; Wang, Y.W.; Qian, X.; Chan, C.; Yu, Z.F.; Fan, X.X.; Pan, H.D.; Xie, C.; et al. Early lung cancer diagnostic biomarker discovery by machine learning methods. Transl. Oncol. 2021, 14, 100907. [Google Scholar] [CrossRef]
- Cancer Research UK, Breaking Down Barriers to Cancer Screening. 2024. Available online: https://news.cancerresearchuk.org/2022/09/23/health-inequalities-breaking-down-barriers-to-cancer-screening/ (accessed on 13 July 2024).
- Flevaris, K.; Davies, J.; Nakai, S.; Vučković, F.; Lauc, G.; Dunlop, M.G.; Kontoravdi, C. Machine learning framework to extract the biomarker potential of plasma IgG N-glycans towards disease risk stratification. Comput. Struct. Biotechnol. J. 2024, 23, 1234–1243. [Google Scholar] [CrossRef]
- Lazar, J.; Antal-Szalmas, P.; Kurucz, I.; Ferenczi, A.; Jozsi, M.; Tornyi, I.; Muller, M.; Fekete, J.T.; Lamont, J.; FitzGerald, P.; et al. Large-Scale Plasma Proteome Epitome Profiling is an Efficient Tool for the Discovery Cancer Biomarkers. Mol. Cell. Proteom. 2023, 22, 100580. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, Z.; Yang, Y.; Han, C.; Wang, L.; Kang, K.; Zhao, A. Exploration of potential novel drug targets and biomarkers for small cell lung cancer by plasma proteome screening. Front. Pharmacol. 2023, 14, 1266782. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; An, R.; Wang, L.; Shan, J.; Wang, X. Specific Gut Microbiome and Serum Metabolome Changes in Lung Cancer Patients. Front. Cell. Infect. Microbiol. 2021, 11, 725284. [Google Scholar] [CrossRef]
- Yao, Y.; Wang, X.; Guan, J.; Xie, C.; Zhang, H.; Yang, J.; Luo, Y.; Chen, L.; Zhao, M.; Huo, B.; et al. Metabolomic differentiation of benign vs malignant pulmonary nodules with high specificity via high-resolution mass spectrometry analysis of patient sera. Nat. Commun. 2023, 14, 2339. [Google Scholar] [CrossRef]
- de Oliveira, R.M.; Ornelas Ricart, C.A.; Araujo Martins, A.M. Use of Mass Spectrometry to Screen Glycan Early Markers in Hepatocellular Carcinoma. Front. Oncol. 2017, 7, 328. [Google Scholar] [CrossRef]
- He, K.; Baniasad, M.; Kwon, H.; Caval, T.; Xu, G.; Lebrilla, C.; Hommes, D.W.; Bertozzi, C. Decoding the glycoproteome: A new frontier for biomarker discovery in cancer. J. Hematol. Oncol. 2024, 17, 12. [Google Scholar] [CrossRef]
- Ruhaak, L.R.; Stroble, C.; Dai, J.; Barnett, M.; Taguchi, A.; Goodman, G.E.; Miyamoto, S.; Gandara, D.; Feng, Z.; Lebrilla, C.B.; et al. Serum Glycans as Risk Markers for Non-Small Cell Lung Cancer. Cancer Prev. Res. 2016, 9, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; He, C.; Zhang, M.; Li, X.; Liu, X.; Liu, Y.; Zhang, D.; Li, Z. Disease-specific haptoglobin-β chain N-glycosylation as biomarker to differentiate non-small cell lung cancer from benign lung diseases. J. Cancer 2019, 10, 5628–5637. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, K.; Kitagawa, K.; Kojima, N.; Iijima, S. Multifucosylated Alpha-1-acid Glycoprotein as a Novel Marker for Hepatocellular Carcinoma. J. Proteome Res. 2016, 15, 2935–2944. [Google Scholar] [CrossRef]
- Mikami, M.; Tanabe, K.; Matsuo, K.; Miyazaki, Y.; Miyazawa, M.; Hayashi, M.; Asai, S.; Ikeda, M.; Shida, M.; Hirasawa, T.; et al. Fully-sialylated alpha-chain of complement 4-binding protein: Diagnostic utility for ovarian clear cell carcinoma. Gynecol. Oncol. 2015, 139, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Mikami, M.; Tanabe, K.; Imanishi, T.; Ikeda, M.; Hirasawa, T.; Yasaka, M.; Machida, H.; Yoshida, H.; Hasegawa, M.; Shimada, M.; et al. Comprehensive serum glycopeptide spectra analysis to identify early-stage epithelial ovarian cancer. Sci. Rep. 2024, 14, 20000. [Google Scholar] [CrossRef]
- Matsuo, K.; Tanabe, K.; Hayashi, M.; Ikeda, M.; Yasaka, M.; Machida, H.; Shida, M.; Sato, K.; Yoshida, H.; Hirasawa, T.; et al. Utility of Comprehensive Serum Glycopeptide Spectra Analysis (CSGSA) for the Detection of Early Stage Epithelial Ovarian Cancer. Cancers 2020, 12, 2374. [Google Scholar] [CrossRef]
- Hayashi, M.; Matsuo, K.; Tanabe, K.; Ikeda, M.; Miyazawa, M.; Yasaka, M.; Machida, H.; Shida, M.; Imanishi, T.; Grubbs, B.H.; et al. Comprehensive Serum Glycopeptide Spectra Analysis (CSGSA): A Potential New Tool for Early Detection of Ovarian Cancer. Cancers 2019, 11, 591. [Google Scholar] [CrossRef]
- Tanabe, K.; Ikeda, M.; Hayashi, M.; Matsuo, K.; Yasaka, M.; Machida, H.; Shida, M.; Katahira, T.; Imanishi, T.; Hirasawa, T.; et al. Comprehensive Serum Glycopeptide Spectra Analysis Combined with Artificial Intelligence (CSGSA-AI) to Diagnose Early-Stage Ovarian Cancer. Cancers 2020, 12, 2373. [Google Scholar] [CrossRef]
- Union for International Cancer Control, TNM Classification of Malignant Tumours. 2023. Available online: https://www.uicc.org/what-we-do/sharing-knowledge/tnm (accessed on 31 May 2024).
- Du, Y.; Wen, Y.; Huang, J. Analysis of variation of serum CEA, SCC, CYFRA21-1 in patients with lung cancer and their diagnostic value with EBUS-TBNA. J. Med. Biochem. 2024, 43, 363–371. [Google Scholar] [CrossRef]
- Ravi Kumar, G.; Nagamani, K.; Anjan Babu, G. A Framework of Dimensionality Reduction Utilizing PCA for Neural Network Prediction. In Advances in Data Science and Management; Springer: Singapore, 2020; pp. 173–180. [Google Scholar]
- Wu, P.; Zhang, C.; Tang, X.; Li, D.; Zhang, G.; Zi, X.; Liu, J.; Yin, E.; Zhao, J.; Wang, P.; et al. Pan-cancer characterization of cell-free immune-related miRNA identified as a robust biomarker for cancer diagnosis. Mol. Cancer 2024, 23, 31. [Google Scholar] [CrossRef]
- Fan, Z.; Guo, Y.; Gu, X.; Huang, R.; Miao, W. Development and validation of an artificial neural network model for non-invasive gastric cancer screening and diagnosis. Sci. Rep. 2022, 12, 21795. [Google Scholar] [CrossRef] [PubMed]
- Cancer Research UK, Types of Lung Cancer. Available online: https://www.cancerresearchuk.org/about-cancer/lung-cancer/stages-types-grades/types (accessed on 15 August 2024).
- Milošević, D.; Medeiros, A.S.; Stojković Piperac, M.; Cvijanović, D.; Soininen, J.; Milosavljević, A.; Predić, B. The application of Uniform Manifold Approximation and Projection (UMAP) for unconstrained ordination and classification of biological indicators in aquatic ecology. Sci. Total Environ. 2022, 815, 152365. [Google Scholar] [CrossRef] [PubMed]
- Šunderić, M.; Šedivá, A.; Robajac, D.; Miljuš, G.; Gemeiner, P.; Nedić, O.; Katrlík, J. Lectin-based protein microarray analysis of differences in serum alpha-2-macroglobulin glycosylation between patients with colorectal cancer and persons without cancer. Biotechnol. Appl. Biochem. 2016, 63, 457–464. [Google Scholar] [CrossRef]
- Mondal, G.; Saroha, A.; Bose, P.P.; Chatterjee, B.P. Altered glycosylation, expression of serum haptoglobin and alpha-1-antitrypsin in chronic hepatitis C, hepatitis C induced liver cirrhosis and hepatocellular carcinoma patients. Glycoconj. J. 2016, 33, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Mikami, H.; Kimura, O.; Yamamoto, H.; Kikuchi, S.; Nakamura, Y.; Ando, T.; Yamakado, M. A multicentre clinical validation of AminoIndex Cancer Screening (AICS). Sci. Rep. 2019, 9, 13831. [Google Scholar] [CrossRef]

| Condition | Age | Number | Sex (Man Ratio) | Stage | Race |
|---|---|---|---|---|---|
| Healthy Volunteers (HE) | 48.2 (±12.2) | 590 | 50.3% | Asian (297) Caucasian (114) African American (115) Hispanic (63) Mixed Ethnicities (1) | |
| Lung Cancer (LGC) | 68.8 (±9.7) | 199 | 57.4% | Stage 0 (5) Stage I (124) Stage II (19) Stage III (20) Stage IV (2) Unclassified (29) | Asian (179) Caucasian (20) |
| Total | 53.3 (±12.2) | 789 | 52.1% |
| True State | ||||||
|---|---|---|---|---|---|---|
| LGC | Healthy | Sum | PPV or NPV | |||
| Predicted State | Observed Samples | Positive | 336 | 35 | 371 | |
| Negative | 251 | 1748 | 1999 | |||
| Sum | 587 | 1783 | 2370 | |||
| Prevalence Correction | Positive | 57 | 1961 | 2018 | 2.8% | |
| Negative | 43 | 97,939 | 97,982 | 99.96% | ||
| Sum | 100 | 99,900 | 100,000 | |||
| Sensitivity or specificity | 57.2% | 98.0% | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamazaki, K.; Kawauchi, S.; Okamoto, M.; Tanabe, K.; Hayashi, C.; Mikami, M.; Kusumoto, T. Comprehensive Serum Glycopeptide Spectra Analysis Combined with Machine Learning for Early Detection of Lung Cancer: A Case–Control Study. Cancers 2025, 17, 1474. https://doi.org/10.3390/cancers17091474
Yamazaki K, Kawauchi S, Okamoto M, Tanabe K, Hayashi C, Mikami M, Kusumoto T. Comprehensive Serum Glycopeptide Spectra Analysis Combined with Machine Learning for Early Detection of Lung Cancer: A Case–Control Study. Cancers. 2025; 17(9):1474. https://doi.org/10.3390/cancers17091474
Chicago/Turabian StyleYamazaki, Koji, Shigeto Kawauchi, Masaki Okamoto, Kazuhiro Tanabe, Chihiro Hayashi, Mikio Mikami, and Tetsuya Kusumoto. 2025. "Comprehensive Serum Glycopeptide Spectra Analysis Combined with Machine Learning for Early Detection of Lung Cancer: A Case–Control Study" Cancers 17, no. 9: 1474. https://doi.org/10.3390/cancers17091474
APA StyleYamazaki, K., Kawauchi, S., Okamoto, M., Tanabe, K., Hayashi, C., Mikami, M., & Kusumoto, T. (2025). Comprehensive Serum Glycopeptide Spectra Analysis Combined with Machine Learning for Early Detection of Lung Cancer: A Case–Control Study. Cancers, 17(9), 1474. https://doi.org/10.3390/cancers17091474







