Structural Insights into the Distortion of the Ribosomal Small Subunit at Different Magnesium Concentrations
Abstract
1. Introduction
2. Materials and Methods
2.1. E. coli Strains and Cell Culture
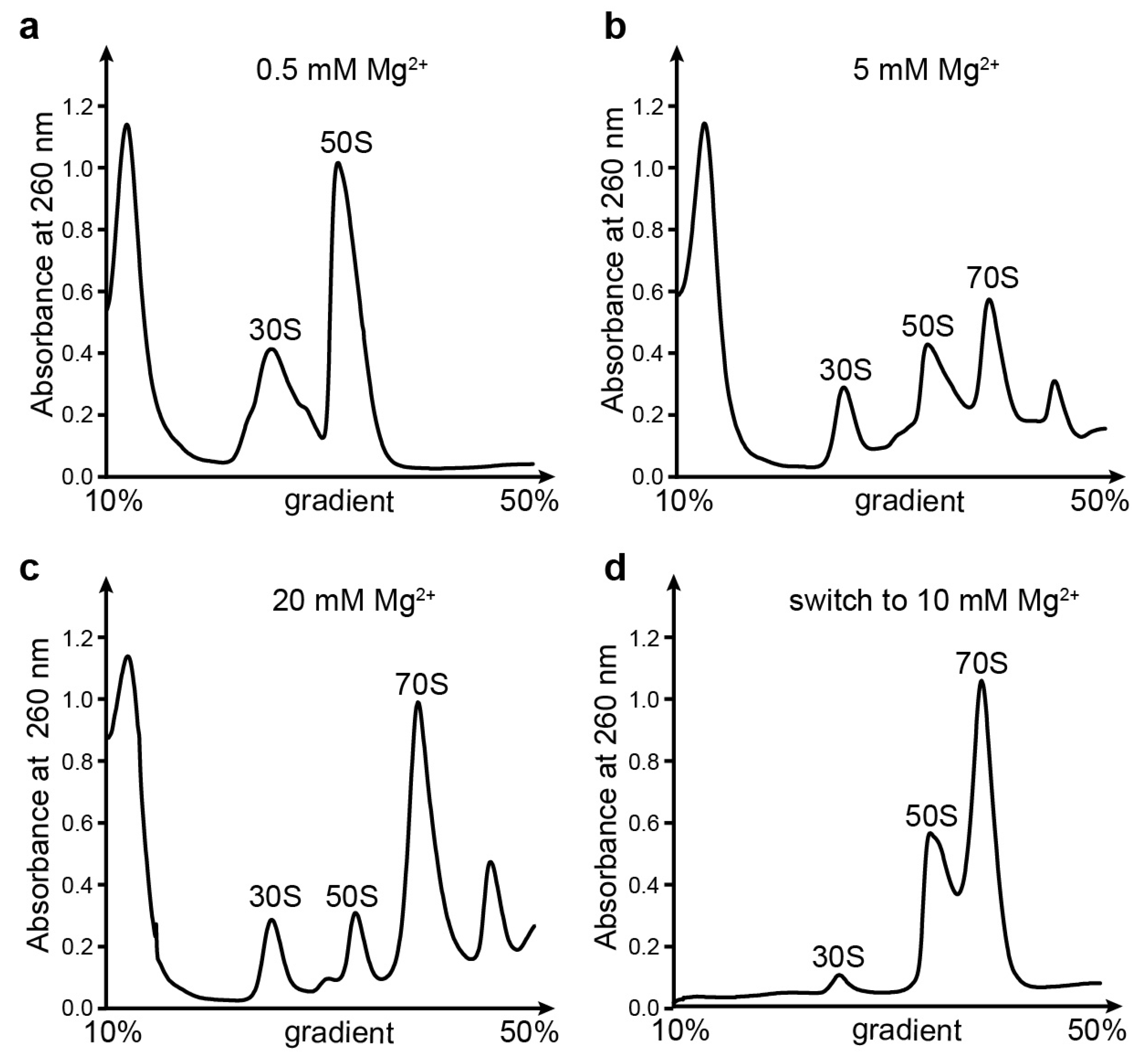
2.2. Sucrose Gradient Centrifugation Analysis
2.3. Cryo-EM Sample Preparation and Data Collection
2.4. Image Processing
2.5. Model Building
2.6. CryoDRGN Analysis
3. Results
3.1. Ribosomal Subunits Are Destroyed by EDTA Treatment
3.2. Structural Distortion of 30S at a Low Magnesium Ion Concentration
3.3. Overall Structures of the 30S Subunits under Different Mg2+ Concentrations
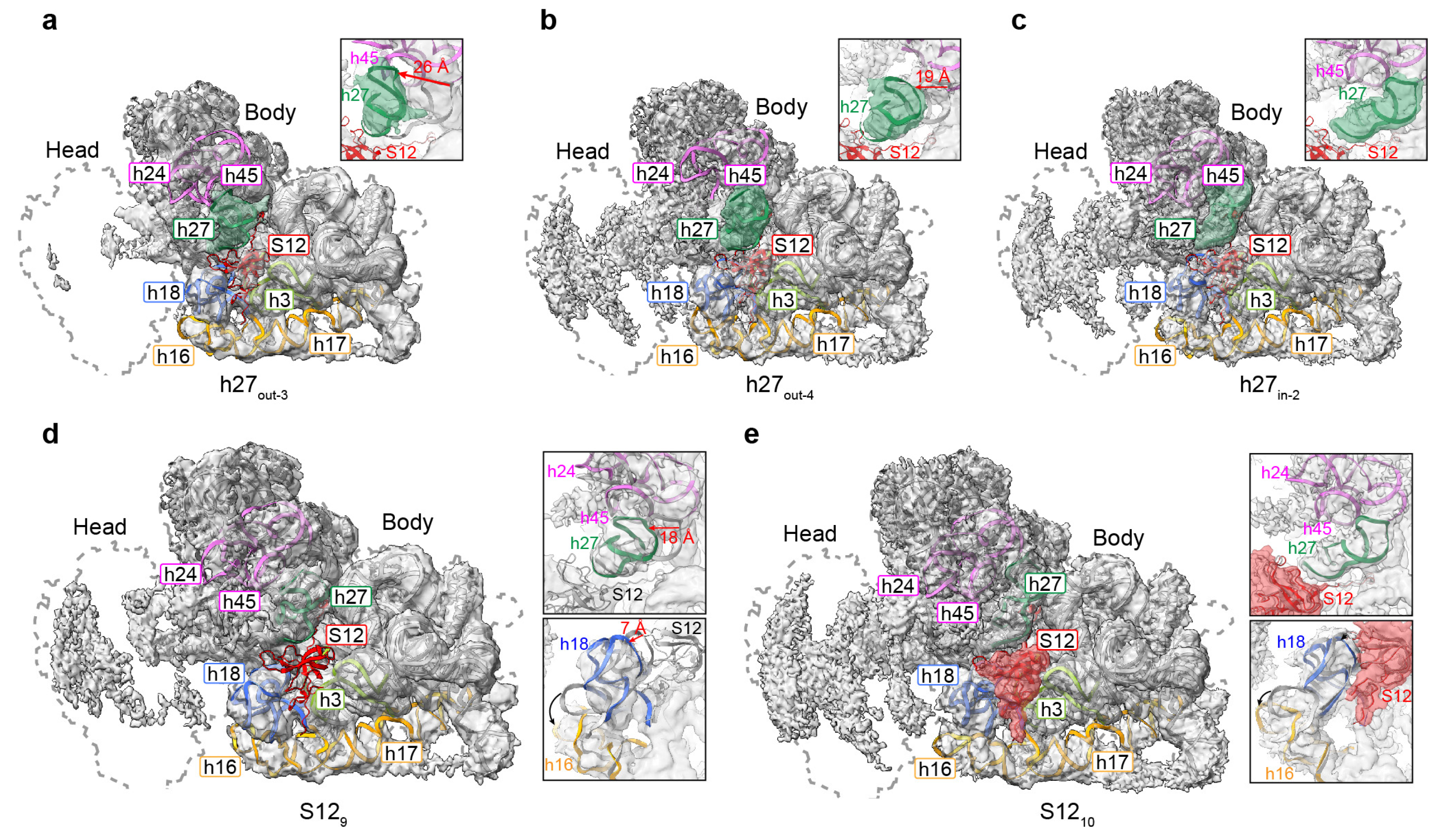
3.4. Movement of Incompact Helices and Loss of S12 in the Decoding Center
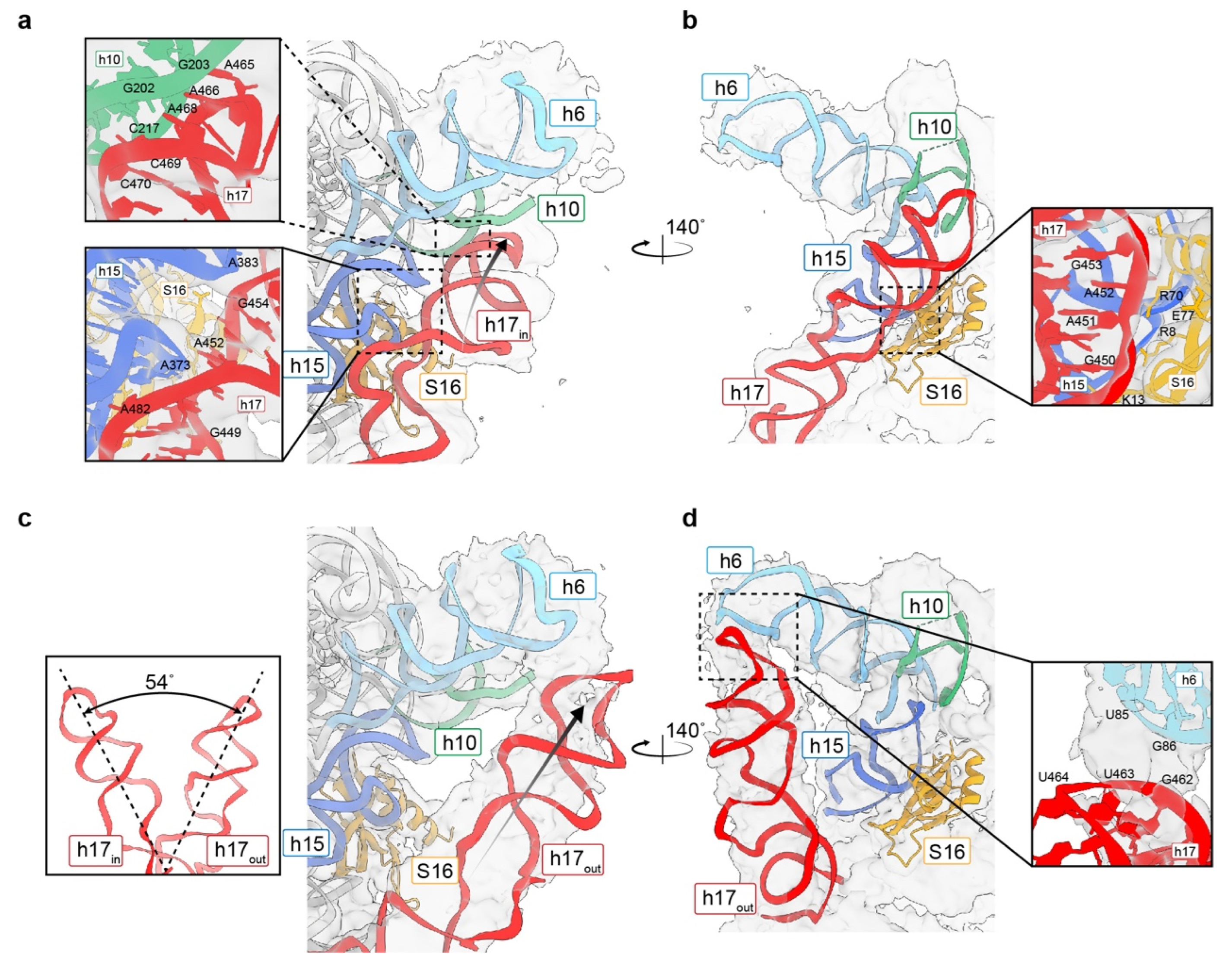
3.5. Movement of h17 towards h6
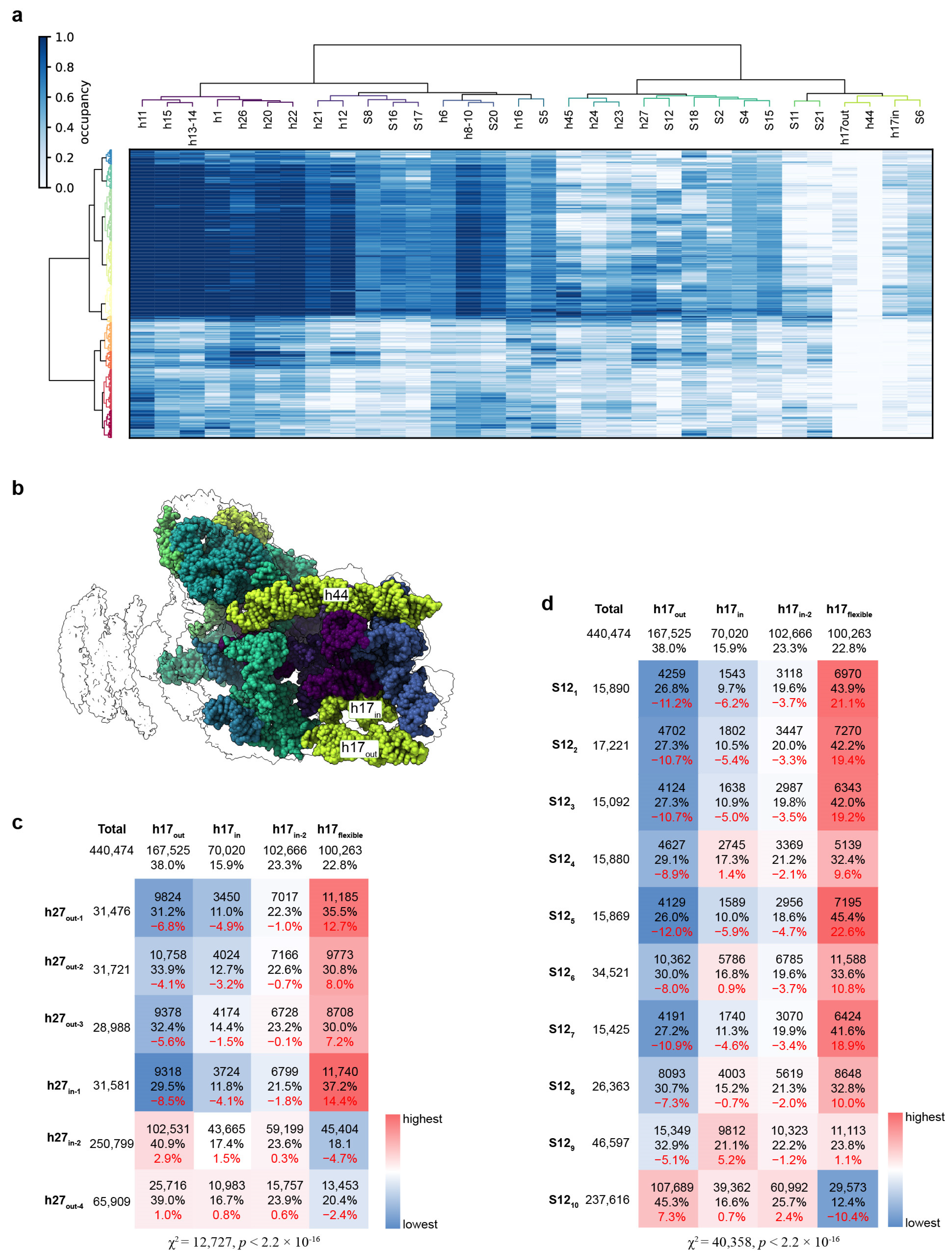
3.6. Correlation of the Structural Distortions between Different Blocks
3.7. Mg2+ Is Essential for Ribosome Structure Stability
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Matsarskaia, O.; Roosen-Runge, F.; Schreiber, F. Multivalent ions and biomolecules: Attempting a comprehensive perspective. Chemphyschem 2020, 21, 1742–1767. [Google Scholar] [CrossRef] [PubMed]
- Gesteland, R.F. Unfolding of Escherichia coli ribosomes by removal of magnesium. J. Mol. Biol. 1966, 18, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.J.; Moore, P.B.; Steitz, T.A. The contribution of metal ions to the structural stability of the large ribosomal subunit. Rna 2004, 10, 1366–1379. [Google Scholar] [CrossRef] [PubMed]
- Watson, Z.L.; Ward, F.R.; Méheust, R.; Ad, O.; Schepartz, A.; Banfield, J.F.; Cate, J.H. Structure of the bacterial ribosome at 2 Å resolution. eLife 2020, 9, e60482. [Google Scholar] [CrossRef]
- Akanuma, G. Diverse relationships between metal ions and the ribosome. Biosci. Biotechnol. Biochem. 2021, 85, 1582–1593. [Google Scholar] [CrossRef] [PubMed]
- Akanuma, G.; Yamazaki, K.; Yagishi, Y.; Iizuka, Y.; Ishizuka, M.; Kawamura, F.; Kato-Yamada, Y. Magnesium Suppresses Defects in the Formation of 70S Ribosomes as Well as in Sporulation Caused by Lack of Several Individual Ribosomal Proteins. J. Bacteriol. 2018, 200, e00212-18. [Google Scholar] [CrossRef]
- Gavrilova, L.P.; Ivanov, D.A.; Spirin, A.S. Studies on the structure of ribosomes. 3. Stepwise unfolding of the 50 s particles without loss of ribosomal protein. J. Mol. Biol. 1966, 16, 473–489. [Google Scholar] [CrossRef]
- Hori, N.; Denesyuk, N.A.; Thirumalai, D. Shape changes and cooperativity in the folding of the central domain of the 16S ribosomal RNA. Proc. Natl. Acad. Sci. USA 2021, 118, e2020837118. [Google Scholar] [CrossRef]
- St John, A.C.; Goldberg, A.L. Effects of starvation for potassium and other inorganic ions on protein degradation and ribonucleic acid synthesis in Escherichia coli. J. Bacteriol. 1980, 143, 1223–1233. [Google Scholar] [CrossRef]
- Selmer, M.; Dunham, C.M.; Murphy, F.V.t.; Weixlbaumer, A.; Petry, S.; Kelley, A.C.; Weir, J.R.; Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 2006, 313, 1935–1942. [Google Scholar] [CrossRef]
- Konevega, A.L.; Soboleva, N.G.; Makhno, V.I.; Semenkov, Y.P.; Wintermeyer, W.; Rodnina, M.V.; Katunin, V.I. Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA 2004, 10, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Akanuma, G.; Kobayashi, A.; Suzuki, S.; Kawamura, F.; Shiwa, Y.; Watanabe, S.; Yoshikawa, H.; Hanai, R.; Ishizuka, M. Defect in the formation of 70S ribosomes caused by lack of ribosomal protein L34 can be suppressed by magnesium. J. Bacteriol. 2014, 196, 3820–3830. [Google Scholar] [CrossRef] [PubMed]
- Failmezger, J.; Nitschel, R.; Sánchez-Kopper, A.; Kraml, M.; Siemann-Herzberg, M. Site-Specific Cleavage of Ribosomal RNA in Escherichia coli-Based Cell-Free Protein Synthesis Systems. PLoS ONE 2016, 11, e0168764. [Google Scholar] [CrossRef]
- Görisch, H.; Goss, D.J.; Parkhurst, L.J. Kinetics of ribosome dissociation and subunit association studied in a light-scattering stopped-flow apparatus. Biochemistry 1976, 15, 5743–5753. [Google Scholar] [CrossRef]
- Nierhaus, K.H. Mg2+, K+, and the ribosome. J. Bacteriol. 2014, 196, 3817–3819. [Google Scholar] [CrossRef] [PubMed]
- Alatossava, T.; Jütte, H.; Kuhn, A.; Kellenberger, E. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J. Bacteriol. 1985, 162, 413–419. [Google Scholar] [CrossRef]
- Johansson, M.; Zhang, J.; Ehrenberg, M. Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA selection. Proc. Natl. Acad. Sci. USA 2012, 109, 131–136. [Google Scholar] [CrossRef]
- Zhang, J.; Ieong, K.W.; Johansson, M.; Ehrenberg, M. Accuracy of initial codon selection by aminoacyl-tRNAs on the mRNA-programmed bacterial ribosome. Proc. Natl. Acad. Sci. USA 2015, 112, 9602–9607. [Google Scholar] [CrossRef]
- Pape, T.; Wintermeyer, W.; Rodnina, M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999, 18, 3800–3807. [Google Scholar] [CrossRef]
- Schluenzen, F.; Tocilj, A.; Zarivach, R.; Harms, J.; Gluehmann, M.; Janell, D.; Bashan, A.; Bartels, H.; Agmon, I.; Franceschi, F.; et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 2000, 102, 615–623. [Google Scholar] [CrossRef]
- Talkington, M.W.; Siuzdak, G.; Williamson, J.R. An assembly landscape for the 30S ribosomal subunit. Nature 2005, 438, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Adilakshmi, T.; Bellur, D.L.; Woodson, S.A. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature 2008, 455, 1268–1272. [Google Scholar] [CrossRef] [PubMed]
- Dutca, L.M.; Culver, G.M. Assembly of the 5′ and 3′ minor domains of 16S ribosomal RNA as monitored by tethered probing from ribosomal protein S20. J. Mol. Biol. 2008, 376, 92–108. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mulder, A.M.; Yoshioka, C.; Beck, A.H.; Bunner, A.E.; Milligan, R.A.; Potter, C.S.; Carragher, B.; Williamson, J.R. Visualizing ribosome biogenesis: Parallel assembly pathways for the 30S subunit. Science 2010, 330, 673–677. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Palovcak, E.; Armache, J.P.; Verba, K.A.; Cheng, Y.; Agard, D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef]
- Rohou, A.; Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015, 192, 216–221. [Google Scholar] [CrossRef]
- Zivanov, J.; Nakane, T.; Forsberg, B.O.; Kimanius, D.; Hagen, W.J.; Lindahl, E.; Scheres, S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 2018, 7, e42166. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Cryst. D Biol. Cryst. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Kinman, L.F.; Powell, B.M.; Zhong, E.D.; Berger, B.; Davis, J.H. Uncovering structural ensembles from single-particle cryo-EM data using cryoDRGN. Nat. Protoc. 2023, 18, 319–339. [Google Scholar] [CrossRef]
- King, T.C.; Rucinsky, T.; Schlessinger, D.; Milanovich, F. Escherichia coli ribosome unfolding in low Mg2+ solutions observed by laser Raman spectroscopy and electron microscopy. Nucleic Acids Res. 1981, 9, 647–661. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Spahn, C.M.; Gomez-Lorenzo, M.G.; Grassucci, R.A.; Jørgensen, R.; Andersen, G.R.; Beckmann, R.; Penczek, P.A.; Ballesta, J.P.; Frank, J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004, 23, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Mohan, S.; Donohue, J.P.; Noller, H.F. Molecular mechanics of 30S subunit head rotation. Proc. Natl. Acad. Sci. USA 2014, 111, 13325–13330. [Google Scholar] [CrossRef]
- Guo, Z.; Noller, H.F. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc. Natl. Acad. Sci. USA 2012, 109, 20391–20394. [Google Scholar] [CrossRef]
- Maksimova, E.; Kravchenko, O.; Korepanov, A.; Stolboushkina, E. Protein Assistants of Small Ribosomal Subunit Biogenesis in Bacteria. Microorganisms 2022, 10, 747. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sato, N.S.; Watanabe, K.; Suzuki, T. Functional genetic selection of the decoding center in E. coli 16S rRNA. Nucleic Acids Res Suppl. 2003, 3, 319–320. [Google Scholar] [CrossRef]
- Smith, T.F.; Lee, J.C.; Gutell, R.R.; Hartman, H. The origin and evolution of the ribosome. Biol. Direct 2008, 3, 16. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shem, A.; Jenner, L.; Yusupova, G.; Yusupov, M. Crystal structure of the eukaryotic ribosome. Science 2010, 330, 1203–1209. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, D.; Gregory, S.T.; O’Connor, M. Error-prone and error-restrictive mutations affecting ribosomal protein S12. J. Mol. Biol. 2011, 410, 1–9. [Google Scholar] [CrossRef]
- Schuwirth, B.S.; Borovinskaya, M.A.; Hau, C.W.; Zhang, W.; Vila-Sanjurjo, A.; Holton, J.M.; Cate, J.H. Structures of the bacterial ribosome at 3.5 A resolution. Science 2005, 310, 827–834. [Google Scholar] [CrossRef]
- Drygin, D.; Zimmermann, R.A. Magnesium ions mediate contacts between phosphoryl oxygens at positions 2122 and 2176 of the 23S rRNA and ribosomal protein L1. Rna 2000, 6, 1714–1726. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maguire, M.E.; Cowan, J.A. Magnesium chemistry and biochemistry. Biometals 2002, 15, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Shayan, R.; Rinaldi, D.; Larburu, N.; Plassart, L.; Balor, S.; Bouyssié, D.; Lebaron, S.; Marcoux, J.; Gleizes, P.E.; Plisson-Chastang, C. Good Vibrations: Structural Remodeling of Maturing Yeast Pre-40S Ribosomal Particles Followed by Cryo-Electron Microscopy. Molecules 2020, 25, 1125. [Google Scholar] [CrossRef] [PubMed]
- Barandun, J.; Hunziker, M.; Klinge, S. Assembly and structure of the SSU processome-a nucleolar precursor of the small ribosomal subunit. Curr. Opin. Struct. Biol. 2018, 49, 85–93. [Google Scholar] [CrossRef]
- Schedlbauer, A.; Iturrioz, I.; Ochoa-Lizarralde, B.; Diercks, T.; Lopez-Alonso, J.P.; Lavin, J.L.; Kaminishi, T.; Capuni, R.; Dhimole, N.; de Astigarraga, E.; et al. A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit. Sci. Adv. 2021, 7, eabf7547. [Google Scholar] [CrossRef]
- Gabashvili, I.S.; Agrawal, R.K.; Spahn, C.M.; Grassucci, R.A.; Svergun, D.I.; Frank, J.; Penczek, P. Solution structure of the E. coli 70S ribosome at 11.5 A resolution. Cell 2000, 100, 537–549. [Google Scholar] [CrossRef]
- Yusupov, M.M.; Yusupova, G.Z.; Baucom, A.; Lieberman, K.; Earnest, T.N.; Cate, J.H.; Noller, H.F. Crystal structure of the ribosome at 5.5 A resolution. Science 2001, 292, 883–896. [Google Scholar] [CrossRef]
- Fourmy, D.; Yoshizawa, S.; Puglisi, J.D. Paromomycin binding induces a local conformational change in the A-site of 16S rRNA. J. Mol. Biol. 1998, 277, 333–345. [Google Scholar] [CrossRef]
- Culver, G.M.; Kirthi, N. Assembly of the 30S Ribosomal Subunit. EcoSal Plus 2008, 3. [Google Scholar] [CrossRef]
- Mizushima, S.; Nomura, M. Assembly mapping of 30S ribosomal proteins from E. coli. Nature 1970, 226, 1214. [Google Scholar] [CrossRef]
- Demirci, H.; Wang, L.; Murphy, F.V.t.; Murphy, E.L.; Carr, J.F.; Blanchard, S.C.; Jogl, G.; Dahlberg, A.E.; Gregory, S.T. The central role of protein S12 in organizing the structure of the decoding site of the ribosome. RNA 2013, 19, 1791–1801. [Google Scholar] [CrossRef] [PubMed]
- Ogle, J.M.; Murphy, F.V.; Tarry, M.J.; Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 2002, 111, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Moncany, M.L.; Kellenberger, E. High magnesium content of Escherichia coli B. Experientia 1981, 37, 846–847. [Google Scholar] [CrossRef]
- Gall, A.R.; Datsenko, K.A.; Figueroa-Bossi, N.; Bossi, L.; Masuda, I.; Hou, Y.M.; Csonka, L.N. Mg2+ regulates transcription of mgtA in Salmonella Typhimurium via translation of proline codons during synthesis of the MgtL peptide. Proc. Natl. Acad. Sci. USA 2016, 113, 15096–15101. [Google Scholar] [CrossRef]
- Rozov, A.; Khusainov, I.; El Omari, K.; Duman, R.; Mykhaylyk, V.; Yusupov, M.; Westhof, E.; Wagner, A.; Yusupova, G. Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction. Nat. Commun. 2019, 10, 2519. [Google Scholar] [CrossRef]
- Hury, J.; Nagaswamy, U.; Larios-Sanz, M.; Fox, G.E. Ribosome origins: The relative age of 23S rRNA Domains. Orig. Life Evol. Biosph. 2006, 36, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, C.; Mohan, S.; Kalahar, B.K.; Williams, L.D. Peeling the onion: Ribosomes are ancient molecular fossils. Mol. Biol. Evol. 2009, 26, 2415–2425. [Google Scholar] [CrossRef] [PubMed]
- Agmon, I. The dimeric proto-ribosome: Structural details and possible implications on the origin of life. Int. J. Mol. Sci. 2009, 10, 2921–2934. [Google Scholar] [CrossRef]
- Davidovich, C.; Belousoff, M.; Wekselman, I.; Shapira, T.; Krupkin, M.; Zimmerman, E.; Bashan, A.; Yonath, A. The Proto-Ribosome: An ancient nano-machine for peptide bond formation. Isr. J. Chem. 2010, 50, 29–35. [Google Scholar] [CrossRef]
- Rivas, M.; Fox, G.E. Further Characterization of the Pseudo-Symmetrical Ribosomal Region. Life 2020, 10, 201. [Google Scholar] [CrossRef]
- Wang, W.; Li, W.; Ge, X.; Yan, K.; Mandava, C.S.; Sanyal, S.; Gao, N. Loss of a single methylation in 23S rRNA delays 50S assembly at multiple late stages and impairs translation initiation and elongation. Proc. Natl. Acad. Sci. USA 2020, 117, 15609–15619. [Google Scholar] [CrossRef] [PubMed]
- Nikolay, R.; Hilal, T.; Schmidt, S.; Qin, B.; Schwefel, D.; Vieira-Vieira, C.H.; Mielke, T.; Bürger, J.; Loerke, J.; Amikura, K.; et al. Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization. Mol. Cell 2021, 81, 1200–1215.e1209. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, T.; Jiang, J.; Yu, Q.; Li, X.; Zeng, F. Structural Insights into the Distortion of the Ribosomal Small Subunit at Different Magnesium Concentrations. Biomolecules 2023, 13, 566. https://doi.org/10.3390/biom13030566
Yu T, Jiang J, Yu Q, Li X, Zeng F. Structural Insights into the Distortion of the Ribosomal Small Subunit at Different Magnesium Concentrations. Biomolecules. 2023; 13(3):566. https://doi.org/10.3390/biom13030566
Chicago/Turabian StyleYu, Ting, Junyi Jiang, Qianxi Yu, Xin Li, and Fuxing Zeng. 2023. "Structural Insights into the Distortion of the Ribosomal Small Subunit at Different Magnesium Concentrations" Biomolecules 13, no. 3: 566. https://doi.org/10.3390/biom13030566
APA StyleYu, T., Jiang, J., Yu, Q., Li, X., & Zeng, F. (2023). Structural Insights into the Distortion of the Ribosomal Small Subunit at Different Magnesium Concentrations. Biomolecules, 13(3), 566. https://doi.org/10.3390/biom13030566





