Dynamic Frequency-Decoupled Refinement Network for Polyp Segmentation
Abstract
1. Introduction
- Existing methods struggle to effectively separate high-frequency components (e.g., edges and textures) from low-frequency components (e.g., global shapes and structural coherence). This often results in imprecise boundaries and incomplete segmentations, particularly for polyps with irregular morphology or highly textured surfaces.
- Most segmentation models primarily extract features in the spatial domain, neglecting the complementary insights that frequency-domain representations can provide. The frequency domain is particularly effective in capturing both local details and global patterns, making it crucial for addressing low-contrast and visually ambiguous polyp regions.
- We introduce the DFDR module, consisting of Frequency Adaptive Decoupling (FAD) and Frequency Adaptive Refinement (FAR) submodules. FAD utilizes frequency-domain analysis via Fast Fourier Transform (FFT) to adaptively separate high-frequency (edge-focused) and low-frequency (structure-focused) components, while FAR refines these features through edge enhancement and global consistency modeling. This enables precise boundary delineation and improved segmentation robustness.
- DFDRNet incorporates DFDR modules at multiple levels of the encoder–decoder structure, allowing for progressive refinement of spatial and frequency-domain features across scales. This hierarchical design enhances the network’s ability to capture fine-grained polyp details while preserving global structural coherence.
- We validate DFDRNet on three widely used polyp segmentation datasets, demonstrating superior performance over CNN-based and transformer-based methods. Extensive ablation studies further confirm the effectiveness of DFDR in dynamically optimizing frequency components and improving segmentation accuracy while maintaining computational efficiency.
2. Related Works
2.1. Traditional Methods for Polyp Segmentation
2.2. Deep Learning Methods for Polyp Segmentation
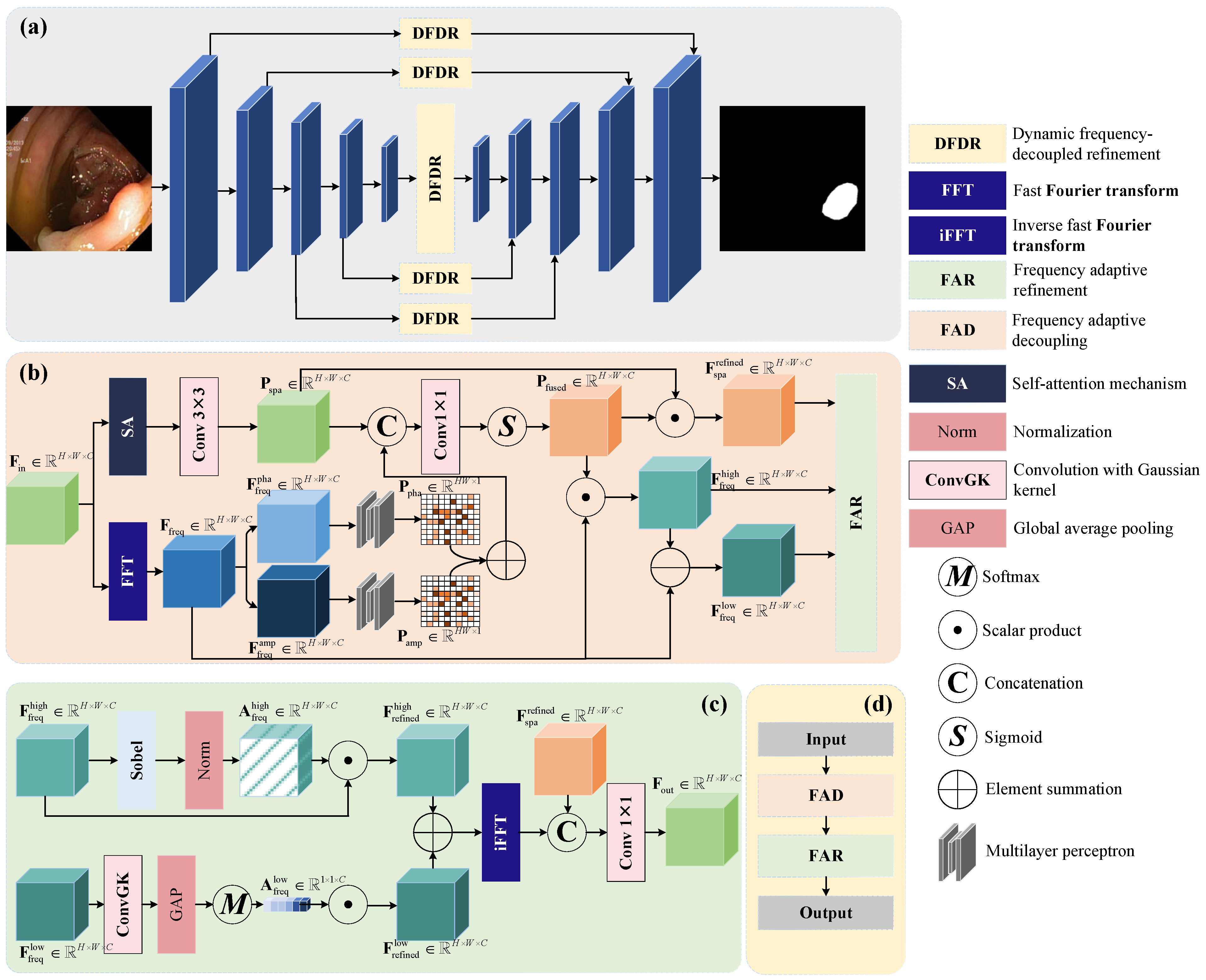
3. Method
3.1. Overview
3.2. Details of DFDR Module
3.2.1. Frequency Adaptive Decoupling (FAD)
3.2.2. Frequency Adaptive Refinement (FAR)
4. Experiments
4.1. Datasets
4.1.1. Kvasir-SEG
4.1.2. CVC-ClinicDB
4.1.3. CVC-ColonDB
4.2. Implementation Details
4.3. Compared with Sota Methods
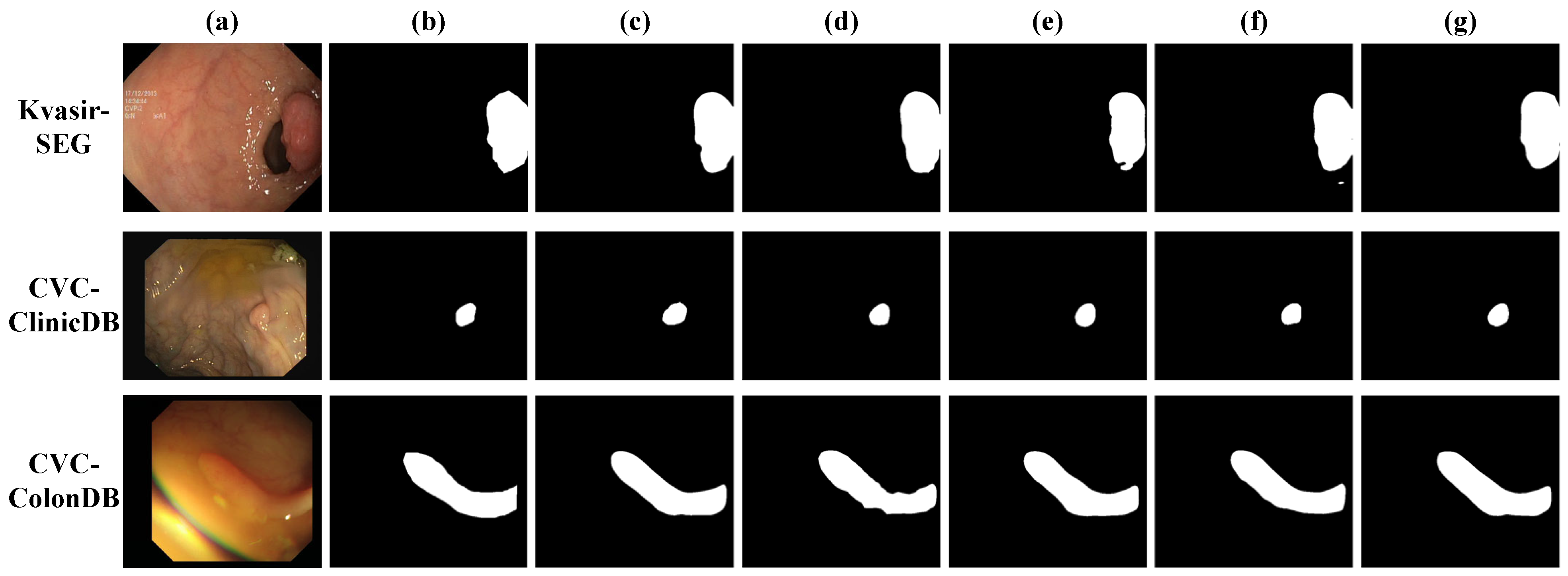
4.3.1. Results on Kvasir-SEG
4.3.2. Results on CVC-ClinicDB
4.3.3. Results on CVC-ColonDB
4.3.4. Effects of DFDR Module
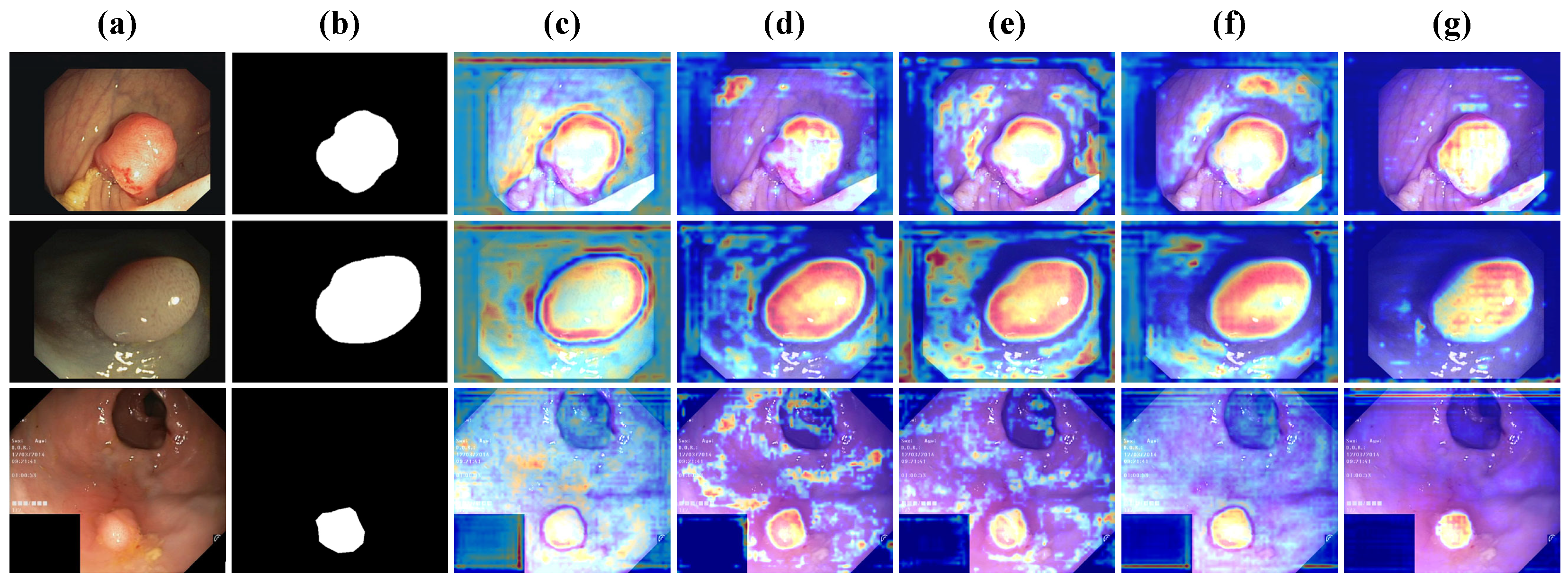
4.4. Results of Different Stages
4.5. Efficiency Analysis
4.5.1. Comparisons with Variants
4.5.2. Efficiency Analysis of Using Different GPUs
4.6. Ablation Study of Incorporating Different Frequency Components
- DFDRNet (Ours): The full model utilizing both high-frequency (HF) and low-frequency (LF) components;
- DFDRNet (No HF): A variant where high-frequency components are removed from the frequency processing pipeline, eliminating edge enhancement and fine-texture preservation;
- DFDRNet (No LF): A variant where low-frequency components are discarded, removing global structural consistency modeling;
- DFDRNet (No Frequency Processing): A baseline model without any frequency decomposition, relying solely on spatial domain information.
- Effect of Removing High-Frequency (HF) Components: Without HF information, the Dice score decreases by 1.3% on average across datasets. This indicates that high-frequency components are critical for capturing fine-grained details, particularly at object boundaries.
- Effect of Removing Low-Frequency (LF) Components: The absence of LF components leads to a 1.0% reduction in Dice and a 1.2% drop in IoU, emphasizing the role of global structural coherence in polyp segmentation. Low-frequency signals provide crucial contextual information that ensures consistent segmentation across larger regions.
- Effect of Removing All Frequency Processing: The most substantial performance drop occurs when both HF and LF components are removed. Compared with the full DFDRNet model, Dice decreases by 2.6%, while IoU and Sensitivity also decline. This confirms that frequency-aware feature refinement is essential for DFDRNet’s success.
- Comparison across Datasets: The impact of frequency removal is most pronounced on CVC-ColonDB, where structural variations are more complex. The Dice score for DFDRNet (No Frequency Processing) is 3.0% lower than that for the full model, highlighting that frequency refinement is especially crucial in challenging segmentation scenarios.
| Metrics | DFDRNet (Ours) | DFDRNet (No HF) | DFDRNet (No LF) | DFDRNet (No Frequency Processing) |
|---|---|---|---|---|
| Kvasir-SEG | ||||
| Dice | 0.9285 | 0.9155 | 0.9190 | 0.9025 |
| IoU | 0.8750 | 0.8605 | 0.8655 | 0.8470 |
| Sensitivity | 0.9920 | 0.9855 | 0.9890 | 0.9805 |
| Specificity | 0.9400 | 0.9305 | 0.9350 | 0.9215 |
| CVC-ClinicDB | ||||
| Dice | 0.9405 | 0.9260 | 0.9315 | 0.9185 |
| IoU | 0.8915 | 0.8755 | 0.8820 | 0.8690 |
| Sensitivity | 0.9930 | 0.9870 | 0.9900 | 0.9830 |
| Specificity | 0.9455 | 0.9335 | 0.9390 | 0.9270 |
| CVC-ColonDB | ||||
| Dice | 0.8205 | 0.8050 | 0.8105 | 0.7905 |
| IoU | 0.7805 | 0.7655 | 0.7700 | 0.7515 |
| Sensitivity | 0.9020 | 0.8905 | 0.8950 | 0.8800 |
| Specificity | 0.8720 | 0.8600 | 0.8650 | 0.8505 |
5. Discussions
5.1. Theoretical Justification for Frequency Processing
- Frequency Adaptive Decoupling (FAD): Decomposes input features into separate HF and LF components via Fast Fourier Transform (FFT) to enhance discriminative feature extraction.
- Frequency Adaptive Refinement (FAR): Dynamically adjusts and fuses these components, reinforcing structural consistency while preserving detailed boundaries.
- Multi-Level Integration: The DFDR module is embedded at multiple stages of the network, ensuring progressive refinement across feature hierarchies.
5.2. Empirical Evidence Supporting Frequency-Based Refinement
- Improved Boundary Accuracy: DFDRNet achieves the highest Dice and IoU scores, surpassing both CNN-based (U-Net, ResUNet) and transformer-based (Swin, Polyp-LVT) approaches by a significant margin.
- Robustness in Low-Contrast Conditions: Frequency-based refinement enables DFDRNet to segment polyps with vague boundaries more effectively, as demonstrated in CVC-ColonDB (Table 3).
- Better Balance of Sensitivity and Specificity: While CNN-based models tend to miss polyps (low Sensitivity) and transformer-based models over-segment (low Specificity), DFDRNet optimally balances both.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Selvaraj, J.; Umapathy, S.; Rajesh, N.A. Artificial intelligence based real time colorectal cancer screening study: Polyp segmentation and classification using multi-house database. Biomed. Signal Process. Control 2025, 99, 106928. [Google Scholar] [CrossRef]
- Jafar, A.; Abidin, Z.U.; Naqvi, R.A.; Lee, S.W. Unmasking colorectal cancer: A high-performance semantic network for polyp and surgical instrument segmentation. Eng. Appl. Artif. Intell. 2024, 138, 109292. [Google Scholar] [CrossRef]
- Li, W.; Xiong, X.; Li, S.; Fan, F. HybridVPS: Hybrid-Supervised Video Polyp Segmentation Under Low-Cost Labels. IEEE Signal Process. Lett. 2024, 31, 111–115. [Google Scholar] [CrossRef]
- Yue, G.; Li, Y.; Jiang, W.; Zhou, W.; Zhou, T. Boundary Refinement Network for Colorectal Polyp Segmentation in Colonoscopy Images. IEEE Signal Process. Lett. 2024, 31, 954–958. [Google Scholar] [CrossRef]
- Ming, Q.; Xiao, X. Towards Accurate Medical Image Segmentation With Gradient-Optimized Dice Loss. IEEE Signal Process. Lett. 2024, 31, 191–195. [Google Scholar] [CrossRef]
- Li, X.; Xu, F.; Liu, F.; Tong, Y.; Lyu, X.; Zhou, J. Semantic Segmentation of Remote Sensing Images by Interactive Representation Refinement and Geometric Prior-Guided Inference. IEEE Trans. Geosci. Remote Sens. 2024, 62, 1–18. [Google Scholar] [CrossRef]
- Yang, B.; Zhang, X.; Li, S.; Higashita, R.; Liu, J. HA-Net: Hierarchical Attention Network Based on Multi-Task Learning for Ciliary Muscle Segmentation in AS-OCT. IEEE Signal Process. Lett. 2023, 30, 1342–1346. [Google Scholar] [CrossRef]
- Li, Z.; Yang, Z.; Wu, W.; Guo, Z.; Zhu, D. Boundary Refinement Network for Polyp Segmentation With Deformable Attention. IEEE Signal Process. Lett. 2025, 32, 121–125. [Google Scholar] [CrossRef]
- Li, X.; Xu, F.; Yu, A.; Lyu, X.; Gao, H.; Zhou, J. A Frequency Decoupling Network for Semantic Segmentation of Remote Sensing Images. IEEE Trans. Geosci. Remote Sens. 2025, 63, 1–21. [Google Scholar] [CrossRef]
- Rajasekar, D.; Theja, G.; Prusty, M.R.; Chinara, S. Efficient colorectal polyp segmentation using wavelet transformation and AdaptUNet: A hybrid U-Net. Heliyon 2024, 10, e33655. [Google Scholar] [CrossRef]
- Selvaraj, J.; Umapathy, S.; Rajesh, N.A. Advancing colorectal polyp detection: An automated segmentation approach with colrectseg-unet. Biomed. Eng. Appl. Basis Commun. 2024, 36, 2450031. [Google Scholar] [CrossRef]
- Li, X.; Xu, F.; Tao, F.; Tong, Y.; Gao, H.; Liu, F.; Chen, Z.; Lyu, X. A Cross-Domain Coupling Network for Semantic Segmentation of Remote Sensing Images. IEEE Geosci. Remote Sens. Lett. 2024, 21, 1–5. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging 2019, 39, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, Q.; Wang, Y. Road extraction by deep residual u-net. IEEE Geosci. Remote Sens. Lett. 2018, 15, 749–753. [Google Scholar] [CrossRef]
- Fan, D.P.; Ji, G.P.; Zhou, T.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Pranet: Parallel reverse attention network for polyp segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 263–273. [Google Scholar]
- Zhang, R.; Lai, P.; Wan, X.; Fan, D.J.; Gao, F.; Wu, X.J.; Li, G. Lesion-aware dynamic kernel for polyp segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 99–109. [Google Scholar]
- Li, X.; Xu, F.; Lyu, X.; Gao, H.; Tong, Y.; Cai, S.; Li, S.; Liu, D. Dual attention deep fusion semantic segmentation networks of large-scale satellite remote-sensing images. Int. J. Remote Sens. 2021, 42, 3583–3610. [Google Scholar] [CrossRef]
- Li, X.; Xu, F.; Liu, F.; Lyu, X.; Tong, Y.; Xu, Z.; Zhou, J. A Synergistical Attention Model for Semantic Segmentation of Remote Sensing Images. IEEE Trans. Geosci. Remote Sens. 2023, 61, 3243954. [Google Scholar] [CrossRef]
- Strudel, R.; Garcia, R.; Laptev, I.; Schmid, C. Segmenter: Transformer for semantic segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 7262–7272. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Wang, W.; Xie, E.; Li, X.; Fan, D.P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. Pyramid vision transformer: A versatile backbone for dense prediction without convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 568–578. [Google Scholar]
- Li, X.; Xu, F.; Li, L.; Xu, N.; Liu, F.; Yuan, C.; Chen, Z.; Lyu, X. AAFormer: Attention-Attended Transformer for Semantic Segmentation of Remote Sensing Images. IEEE Geosci. Remote Sens. Lett. 2024, 21, 1–5. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Zhang, Y.; Liu, H.; Hu, Q. Transfuse: Fusing transformers and cnns for medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part I 24. Springer: Berlin/Heidelberg, Germany, 2021; pp. 14–24. [Google Scholar]
- Wang, J.; Huang, Q.; Tang, F.; Meng, J.; Su, J.; Song, S. Stepwise feature fusion: Local guides global. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 110–120. [Google Scholar]
- Duc, N.T.; Oanh, N.T.; Thuy, N.T.; Triet, T.M.; Dinh, V.S. Colonformer: An efficient transformer based method for colon polyp segmentation. IEEE Access 2022, 10, 80575–80586. [Google Scholar] [CrossRef]
- Dong, B.; Wang, W.; Fan, D.P.; Li, J.; Fu, H.; Shao, L. Polyp-PVT: Polyp Segmentation with Pyramid Vision Transformers. CAAI Artif. Intell. Res. 2023, 2. [Google Scholar] [CrossRef]
- Xia, Y.; Yun, H.; Liu, Y.; Luan, J.; Li, M. MGCBFormer: The multiscale grid-prior and class-inter boundary-aware transformer for polyp segmentation. Comput. Biol. Med. 2023, 167, 107600. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhao, Y.; Li, F.; Wang, L. MIA-Net: Multi-information aggregation network combining transformers and convolutional feature learning for polyp segmentation. Knowl.-Based Syst. 2022, 247, 108824. [Google Scholar] [CrossRef]
- Wu, Z.; Lv, F.; Chen, C.; Hao, A.; Li, S. Colorectal Polyp Segmentation in the Deep Learning Era: A Comprehensive Survey. arXiv 2024, arXiv:2401.11734. [Google Scholar]
- Salpea, N.; Tzouveli, P.; Kollias, D. Medical image segmentation: A review of modern architectures. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 691–708. [Google Scholar]
- Xia, S.; Krishnan, S.M.; Tjoa, M.P.; Goh, P.M. A novel methodology for extracting colon’s lumen from colonoscopic images. J. Syst. Cybern. Informatics 2003, 1, 7–12. [Google Scholar]
- Wang, Z.; Li, L.; Anderson, J.; Harrington, D.P.; Liang, Z. Computer-aided detection and diagnosis of colon polyps with morphological and texture features. In Proceedings of the Medical Imaging 2004: Image Processing, San Diego, CA, USA, 14–19 February 2004; SPIE: Bellingham, WA, USA, 2004; Volume 5370, pp. 972–979. [Google Scholar]
- Jerebko, A.; Lakare, S.; Cathier, P.; Periaswamy, S.; Bogoni, L. Symmetric curvature patterns for colonic polyp detection. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Copenhagen, Denmark, 1–6 October 2006; Springer: Berlin/Heidelberg, Germany, 2006; pp. 169–176. [Google Scholar]
- Eriyanti, N.A.; Sigit, R.; Harsono, T. Classification of colon polyp on endoscopic image using support vector machine. In Proceedings of the 2021 International Electronics Symposium (IES), Surabaya, East Java, Indonesia, 29–30 September 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 244–250. [Google Scholar]
- Sasmal, P.; Bhuyan, M.K.; Dutta, S.; Iwahori, Y. An unsupervised approach of colonic polyp segmentation using adaptive markov random fields. Pattern Recognit. Lett. 2022, 154, 7–15. [Google Scholar] [CrossRef]
- Condessa, F.; Bioucas-Dias, J. Segmentation and detection of colorectal polyps using local polynomial approximation. In Proceedings of the Image Analysis and Recognition: 9th International Conference, ICIAR 2012, Aveiro, Portugal, 25–27 June 2012; Proceedings, Part II 9. Springer: Berlin/Heidelberg, Germany, 2012; pp. 188–197. [Google Scholar]
- Sadagopan, R.; Ravi, S.; Adithya, S.V.; Vivekanandhan, S. PolyEffNetV1: A CNN based colorectal polyp detection in colonoscopy images. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2023, 237, 406–418. [Google Scholar] [CrossRef] [PubMed]
- Malik, J.; Kiranyaz, S.; Kunhoth, S.; Ince, T.; Al-Maadeed, S.; Hamila, R.; Gabbouj, M. Colorectal cancer diagnosis from histology images: A comparative study. arXiv 2019, arXiv:1903.11210. [Google Scholar]
- Ma, M.; Wang, H.; Song, B.; Hu, Y.; Gu, X.; Liang, Z. Random forest based computer-aided detection of polyps in CT colonography. In Proceedings of the 2014 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC), Seattle, WA, USA, 8–15 November 2014; IEEE: Piscataway, NJ, USA, 2014; pp. 1–4. [Google Scholar]
- Varghese, A.; Jain, S.; Jawahar, M.; Prince, A.A. Auto-pore segmentation of digital microscopic leather images for species identification. Eng. Appl. Artif. Intell. 2023, 126, 107049. [Google Scholar] [CrossRef]
- Si, C.; Rahim, M.S.M.; Mianzhou, Y.; Li, N.; Hongyu, C. UNet-Based Polyp Segmentation: A Survey. In Proceedings of the 2023 IEEE International Conference on Computing (ICOCO), Kedah, Malaysia, 9–12 October 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 154–159. [Google Scholar]
- Gupta, M.; Mishra, A. A systematic review of deep learning based image segmentation to detect polyp. Artif. Intell. Rev. 2024, 57, 7. [Google Scholar] [CrossRef]
- Mei, J.; Zhou, T.; Huang, K.; Zhang, Y.; Zhou, Y.; Wu, Y.; Fu, H. A survey on deep learning for polyp segmentation: Techniques, challenges and future trends. Vis. Intell. 2025, 3, 1. [Google Scholar] [CrossRef]
- Sanchez-Peralta, L.F.; Bote-Curiel, L.; Picon, A.; Sanchez-Margallo, F.M.; Pagador, J.B. Deep learning to find colorectal polyps in colonoscopy: A systematic literature review. Artif. Intell. Med. 2020, 108, 101923. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Zhao, H.; Tong, Y.; Mu, R.; Wang, Y.; Zhang, S.; Zhao, Y.; Wang, W.; Zhang, M.; Liu, Y.; et al. A frequency attention-embedded network for polyp segmentation. Sci. Rep. 2025, 15, 4961. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Yao, S.; Liu, D.; Chang, B.; Chen, Z.; Wang, J.; Wei, J. CAFE-Net: Cross-attention and feature exploration network for polyp segmentation. Expert Syst. Appl. 2024, 238, 121754. [Google Scholar] [CrossRef]
- Xiao, B.; Hu, J.; Li, W.; Pun, C.M.; Bi, X. CTNet: Contrastive Transformer Network for Polyp Segmentation. IEEE Trans. Cybern. 2024, 54, 5040–5053. [Google Scholar] [CrossRef] [PubMed]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Park, K.B.; Lee, J.Y. SwinE-Net: Hybrid deep learning approach to novel polyp segmentation using convolutional neural network and Swin Transformer. J. Comput. Des. Eng. 2022, 9, 616–632. [Google Scholar] [CrossRef]
- Xu, Z.; Tang, F.; Chen, Z.; Zhou, Z.; Wu, W.; Yang, Y.; Liang, Y.; Jiang, J.; Cai, X.; Su, J. Polyp-mamba: Polyp segmentation with visual mamba. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024; Springer: Berlin/Heidelberg, Germany, 2024; pp. 510–521. [Google Scholar]
- Li, J.; Wang, J.; Lin, F.; Heidari, A.A.; Chen, Y.; Chen, H.; Wu, W. PRCNet: A parallel reverse convolutional attention network for colorectal polyp segmentation. Biomed. Signal Process. Control 2024, 95, 106336. [Google Scholar] [CrossRef]
- Yue, G.; Zhuo, G.; Yan, W.; Zhou, T.; Tang, C.; Yang, P.; Wang, T. Boundary uncertainty aware network for automated polyp segmentation. Neural Netw. 2024, 170, 390–404. [Google Scholar] [CrossRef]
- Lin, L.; Lv, G.; Wang, B.; Xu, C.; Liu, J. Polyp-LVT: Polyp segmentation with lightweight vision transformers. Knowl.-Based Syst. 2024, 300, 112181. [Google Scholar] [CrossRef]
- Wang, J.; Chen, F.; Ma, Y.; Wang, L.; Fei, Z.; Shuai, J.; Tang, X.; Zhou, Q.; Qin, J. Xbound-former: Toward cross-scale boundary modeling in transformers. IEEE Trans. Med. Imaging 2023, 42, 1735–1745. [Google Scholar] [CrossRef]
- Goceri, E. Polyp segmentation using a hybrid vision transformer and a hybrid loss function. J. Imaging Informatics Med. 2024, 37, 851–863. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Zhang, P.; Wu, F.; Li, X. Fcanet: Frequency channel attention networks. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 783–792. [Google Scholar]
- Dai, T.; Wang, J.; Guo, H.; Li, J.; Wang, J.; Zhu, Z. FreqFormer: Frequency-aware transformer for lightweight image super-resolution. In Proceedings of the International Joint Conference on Artificial Intelligence, Jeju, Republic of Korea, 3–9 August 2024; pp. 731–739. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; De Lange, T.; Johansen, D.; Johansen, H.D. Kvasir-seg: A segmented polyp dataset. In Proceedings of the MultiMedia Modeling: 26th International Conference, MMM 2020, Daejeon, Republic of Korea, 5–8 January 2020; Proceedings, Part II 26. Springer: Berlin/Heidelberg, Germany, 2020; pp. 451–462. [Google Scholar]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 2015, 43, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, N.; Gurudu, S.R.; Liang, J. Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans. Med. Imaging 2015, 35, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I. Deep Learning; MIT Press: Cambridge, UK, 2016. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Johansen, D.; De Lange, T.; Halvorsen, P.; Johansen, H.D. Resunet++: An advanced architecture for medical image segmentation. In Proceedings of the 2019 IEEE International Symposium on Multimedia (ISM), San Diego, CA, USA, 9–11 December 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 225–2255. [Google Scholar]
- Zhou, Y.; Huang, J.; Wang, C.; Song, L.; Yang, G. Xnet: Wavelet-based low and high frequency fusion networks for fully-and semi-supervised semantic segmentation of biomedical images. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 1–6 October 2023; pp. 21085–21096. [Google Scholar]
- Tong, Y.; Chen, Z.; Zhou, Z.; Hu, Y.; Li, X.; Qiao, X. An Edge-Enhanced Network for Polyp Segmentation. Bioengineering 2024, 11, 959. [Google Scholar] [CrossRef]
- Zhang, N.; Yu, L.; Zhang, D.; Wu, W.; Tian, S.; Kang, X. APT-Net: Adaptive encoding and parallel decoding transformer for medical image segmentation. Comput. Biol. Med. 2022, 151, 106292. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Fu, J.; Liu, J.; Tian, H.; Li, Y.; Bao, Y.; Fang, Z.; Lu, H. Dual Attention Network for Scene Segmentation. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–17 June 2019; pp. 3141–3149. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 5999–6009. [Google Scholar]
| Methods | Dice | IoU | Sensitivity | Specificity |
|---|---|---|---|---|
| U-Net [13] | 0.8120 | 0.7405 | 0.9430 | 0.8507 |
| UNet++ [14] | 0.8109 | 0.7349 | 0.9739 | 0.7971 |
| ResUNet [15] | 0.8179 | 0.7459 | 0.9499 | 0.8569 |
| ResUNet++ [64] | 0.8245 | 0.7734 | 0.8937 | 0.8299 |
| PraNet [16] | 0.8876 | 0.8303 | 0.9667 | 0.9015 |
| XNet [65] | 0.8583 | 0.8076 | 0.9239 | 0.8686 |
| EENet [66] | 0.9208 | 0.8664 | 0.9912 | 0.9319 |
| Segmenter [20] | 0.9023 | 0.8451 | 0.9781 | 0.9102 |
| Swin [21] | 0.8998 | 0.8425 | 0.9764 | 0.9077 |
| Polyp-LVT [55] | 0.9091 | 0.8513 | 0.9799 | 0.9116 |
| APT-Net [67] | 0.9175 | 0.8627 | 0.9904 | 0.9298 |
| XBound-Former [56] | 0.9225 | 0.8703 | 0.9914 | 0.9345 |
| FAENet [47] | 0.9174 | 0.8632 | 0.9881 | 0.9280 |
| DFDRNet (ours) | 0.9285 | 0.8750 | 0.9920 | 0.9400 |
| Methods | Dice | IoU | Sensitivity | Specificity |
|---|---|---|---|---|
| U-Net [13] | 0.7618 | 0.6988 | 0.8766 | 0.7729 |
| UNet++ [14] | 0.7940 | 0.7290 | 0.9270 | 0.7950 |
| ResUNet [15] | 0.7957 | 0.7299 | 0.9155 | 0.8073 |
| ResUNet++ [64] | 0.8590 | 0.7881 | 0.9885 | 0.8716 |
| PraNet [16] | 0.8990 | 0.8490 | 0.9901 | 0.9110 |
| XNet [65] | 0.8943 | 0.8204 | 0.9910 | 0.9073 |
| EENet [66] | 0.9316 | 0.8817 | 0.9915 | 0.9586 |
| Segmenter [20] | 0.9105 | 0.8605 | 0.9803 | 0.8250 |
| Swin [21] | 0.9080 | 0.8572 | 0.9785 | 0.9221 |
| Polyp-LVT [55] | 0.9251 | 0.8694 | 0.9911 | 0.9304 |
| APT-Net [67] | 0.9210 | 0.8770 | 0.9900 | 0.9380 |
| XBound-Former [56] | 0.9340 | 0.8840 | 0.9914 | 0.9405 |
| FAENet [47] | 0.9330 | 0.8832 | 0.9930 | 0.9601 |
| DFDRNet (ours) | 0.9405 | 0.8915 | 0.9930 | 0.9455 |
| Methods | Dice | IoU | Sensitivity | Specificity |
|---|---|---|---|---|
| U-Net [13] | 0.6283 | 0.5764 | 0.7889 | 0.6956 |
| UNet++ [14] | 0.6549 | 0.6013 | 0.8343 | 0.7155 |
| ResUNet [15] | 0.6563 | 0.6021 | 0.8239 | 0.7265 |
| ResUNet++ [64] | 0.7086 | 0.6500 | 0.8896 | 0.7844 |
| PraNet [16] | 0.7415 | 0.7003 | 0.8910 | 0.8199 |
| XNet [65] | 0.7564 | 0.6939 | 0.8919 | 0.8165 |
| EENet [66] | 0.8042 | 0.7611 | 0.8923 | 0.8627 |
| Segmenter [20] | 0.7705 | 0.7400 | 0.8850 | 0.8250 |
| Swin [21] | 0.7682 | 0.7078 | 0.8831 | 0.7227 |
| Polyp-LVT [55] | 0.7825 | 0.7353 | 0.8919 | 0.8373 |
| APT-Net [67] | 0.7995 | 0.7504 | 0.8900 | 0.8552 |
| XBound-Former [56] | 0.8103 | 0.7670 | 0.8935 | 0.8685 |
| FAENet [47] | 0.8022 | 0.7635 | 0.8802 | 0.8399 |
| DFDRNet (ours) | 0.8205 | 0.7805 | 0.9020 | 0.8720 |
| Metrics | DFDRNet (Ours) | DFDRNet-SE [68] | DFDRNet-CBAM [69] | DFDRNet-DAM [70] | DFDRNet-MHSA [71] |
|---|---|---|---|---|---|
| Kvasir-SEG | |||||
| Dice | 0.9285 | 0.9210 | 0.9230 | 0.9245 | 0.9260 |
| IoU | 0.8750 | 0.8700 | 0.8725 | 0.8735 | 0.8745 |
| Sensitivity | 0.9920 | 0.9905 | 0.9910 | 0.9915 | 0.9918 |
| Specificity | 0.9400 | 0.9360 | 0.9375 | 0.9380 | 0.9390 |
| CVC-ClinicDB | |||||
| Dice | 0.9405 | 0.9350 | 0.9370 | 0.9385 | 0.9395 |
| IoU | 0.8915 | 0.8850 | 0.8865 | 0.8890 | 0.8905 |
| Sensitivity | 0.9930 | 0.9910 | 0.9920 | 0.9925 | 0.9930 |
| Specificity | 0.9455 | 0.9420 | 0.9435 | 0.9440 | 0.9450 |
| CVC-ColonDB | |||||
| Dice | 0.8205 | 0.8120 | 0.8150 | 0.8175 | 0.8190 |
| IoU | 0.7805 | 0.7700 | 0.7740 | 0.7765 | 0.7780 |
| Sensitivity | 0.9020 | 0.8970 | 0.9005 | 0.9010 | 0.9020 |
| Specificity | 0.8720 | 0.8600 | 0.8655 | 0.8670 | 0.8700 |
| Methods | FLOPs (GMac) | Params (M) | Inference Time (ms) |
|---|---|---|---|
| DFDRNet-SE [68] | 18.2 | 23.1 | 21.5 |
| DFDRNet-CBAM [69] | 19.4 | 24.8 | 23.2 |
| DFDRNet-DAM [70] | 20.1 | 25.3 | 24.5 |
| DFDRNet-MHSA [71] | 21.5 | 26.7 | 26.0 |
| DFDRNet (ours) | 22.8 | 28.5 | 27.3 |
| GPU | Memory (GB) | CUDA Cores | Inference Time (ms) |
|---|---|---|---|
| NVIDIA A40 | 48GB | 10752 | 21.6 |
| NVIDIA RTX 3090 | 24GB | 10496 | 22.1 |
| NVIDIA Tesla V100 | 16GB | 5120 | 25.8 |
| NVIDIA RTX 2080 Ti | 11GB | 4352 | 27.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tong, Y.; Chai, J.; Chen, Z.; Zhou, Z.; Hu, Y.; Li, X.; Qiao, X.; Hu, K. Dynamic Frequency-Decoupled Refinement Network for Polyp Segmentation. Bioengineering 2025, 12, 277. https://doi.org/10.3390/bioengineering12030277
Tong Y, Chai J, Chen Z, Zhou Z, Hu Y, Li X, Qiao X, Hu K. Dynamic Frequency-Decoupled Refinement Network for Polyp Segmentation. Bioengineering. 2025; 12(3):277. https://doi.org/10.3390/bioengineering12030277
Chicago/Turabian StyleTong, Yao, Jingxian Chai, Ziqi Chen, Zuojian Zhou, Yun Hu, Xin Li, Xuebin Qiao, and Kongfa Hu. 2025. "Dynamic Frequency-Decoupled Refinement Network for Polyp Segmentation" Bioengineering 12, no. 3: 277. https://doi.org/10.3390/bioengineering12030277
APA StyleTong, Y., Chai, J., Chen, Z., Zhou, Z., Hu, Y., Li, X., Qiao, X., & Hu, K. (2025). Dynamic Frequency-Decoupled Refinement Network for Polyp Segmentation. Bioengineering, 12(3), 277. https://doi.org/10.3390/bioengineering12030277






