Niche Preference of Escherichia coli in a Peri-Urban Pond Ecosystem
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Source
2.2. Isolation of E. coli
2.3. Phylogroup Analysis
2.4. uidA and mutS Sequence Analysis
2.5. Population Genetic Analysis
2.6. Virulence Gene Assays
2.7. Antibiotic Resistance Assays
3. Results
3.1. Bacterial Isolates
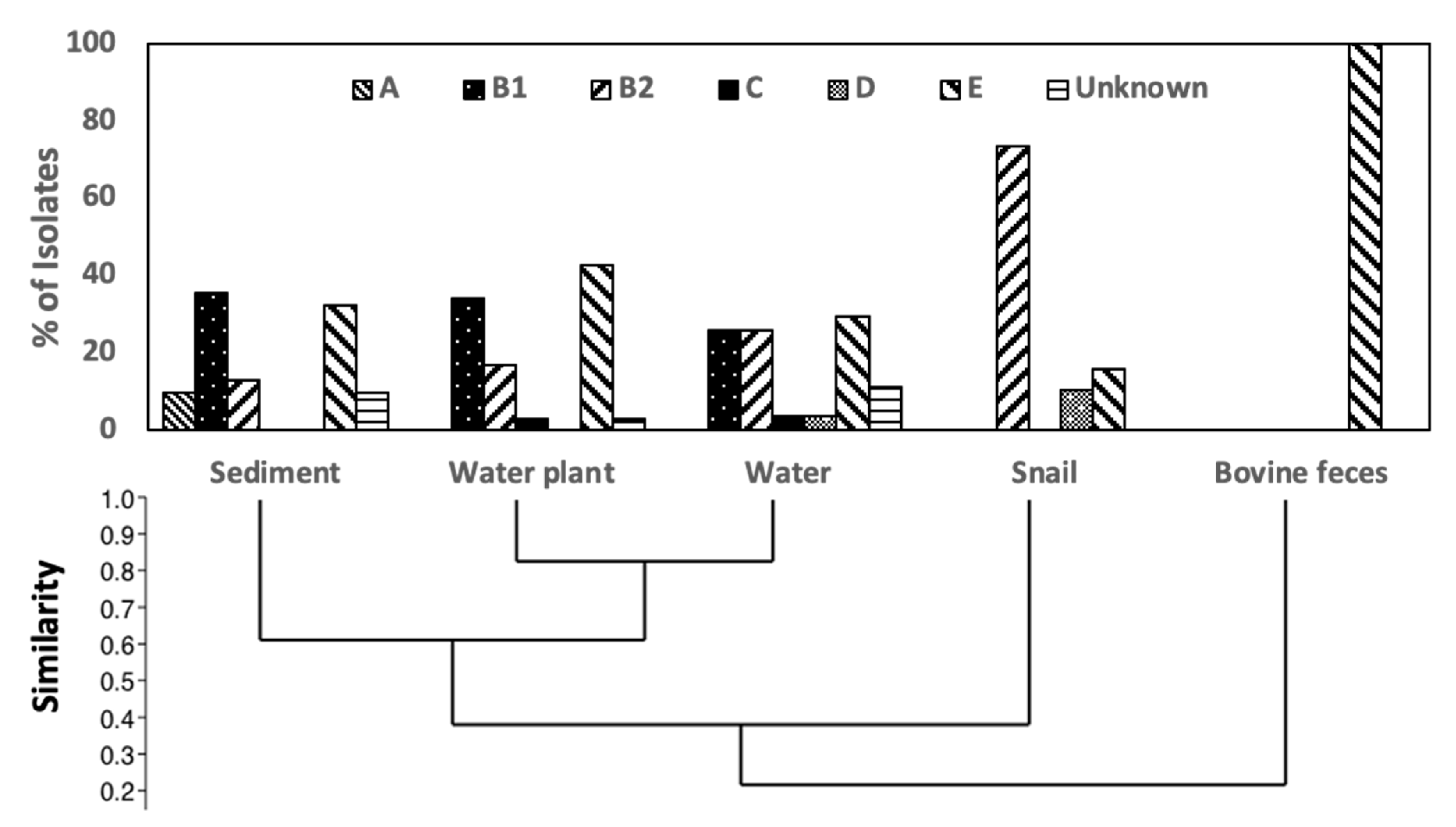
3.2. Phylogroup Distribution
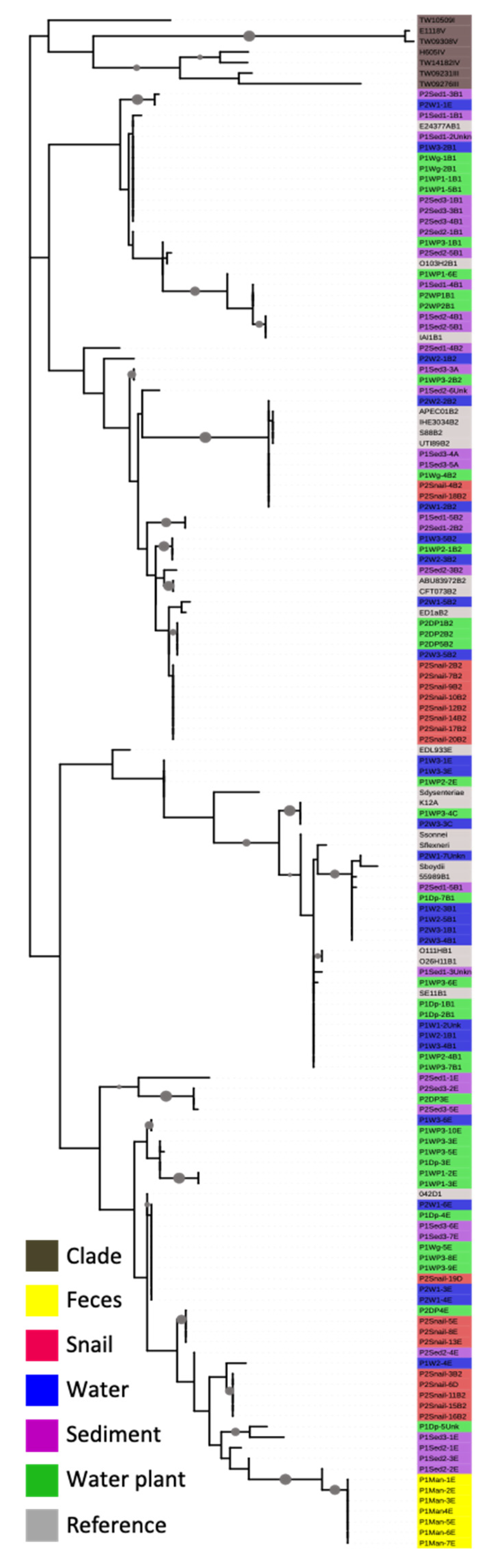
3.3. Phylogenetic Analysis
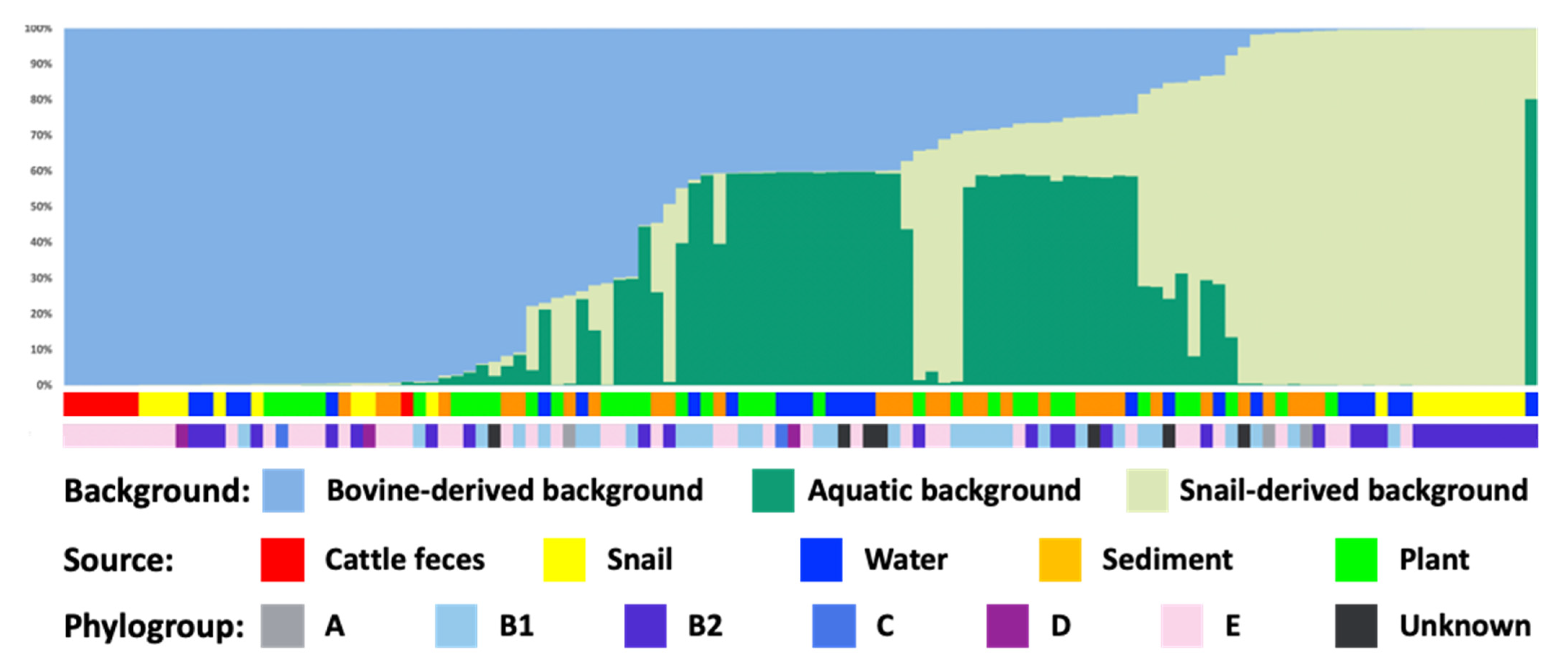
3.4. Population Genetic Analysis
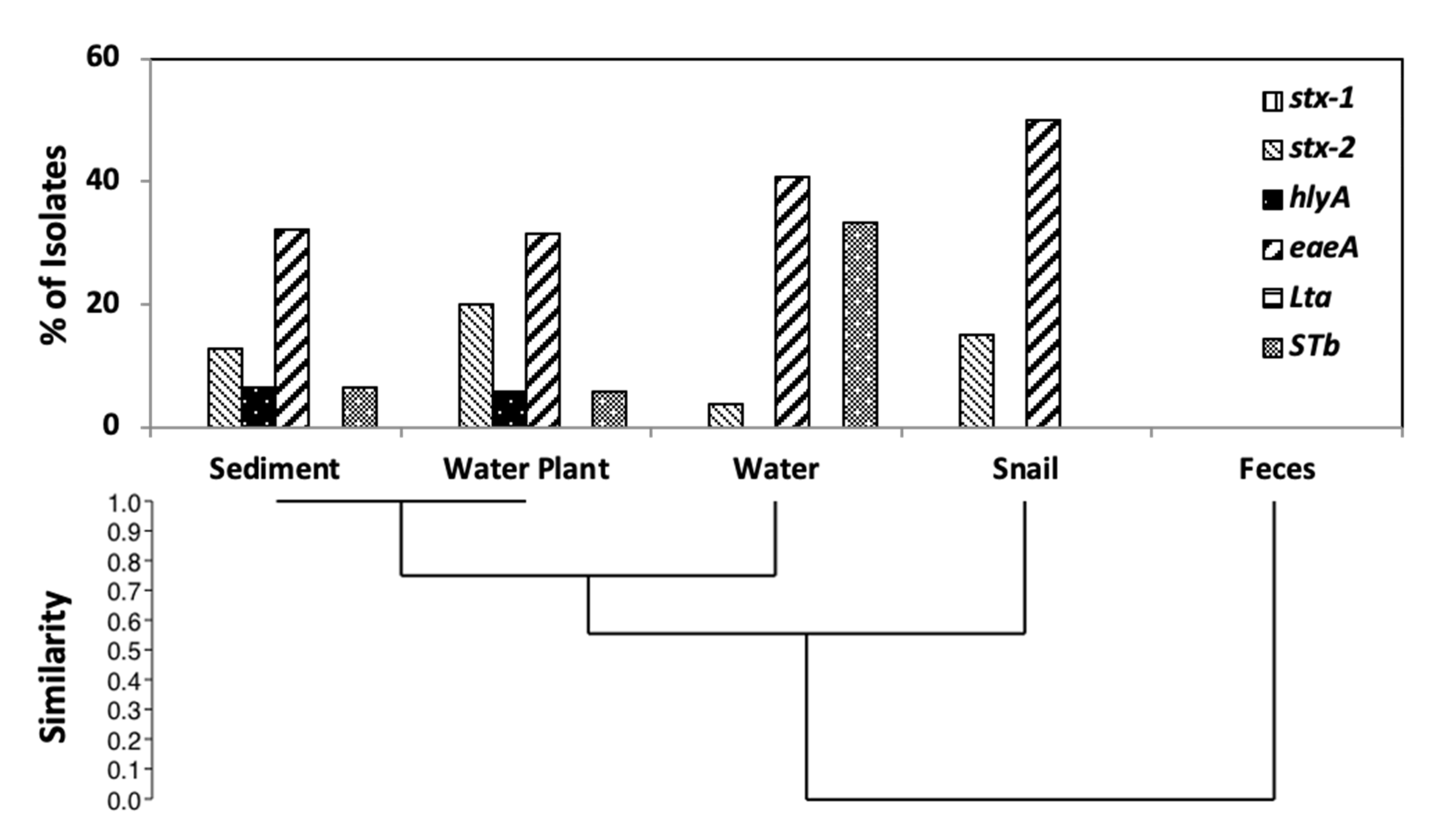
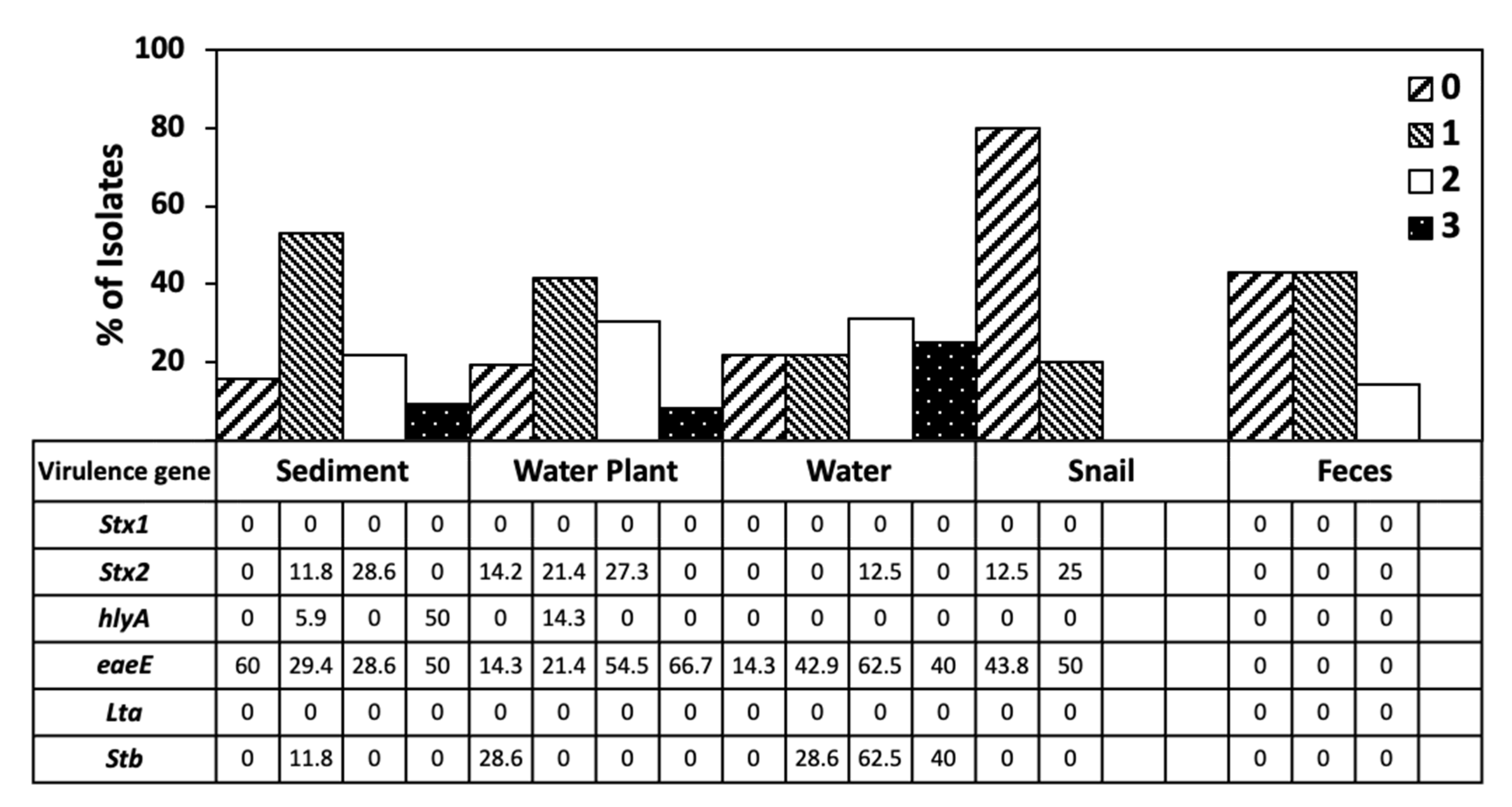
3.5. Virulence Gene Distribution
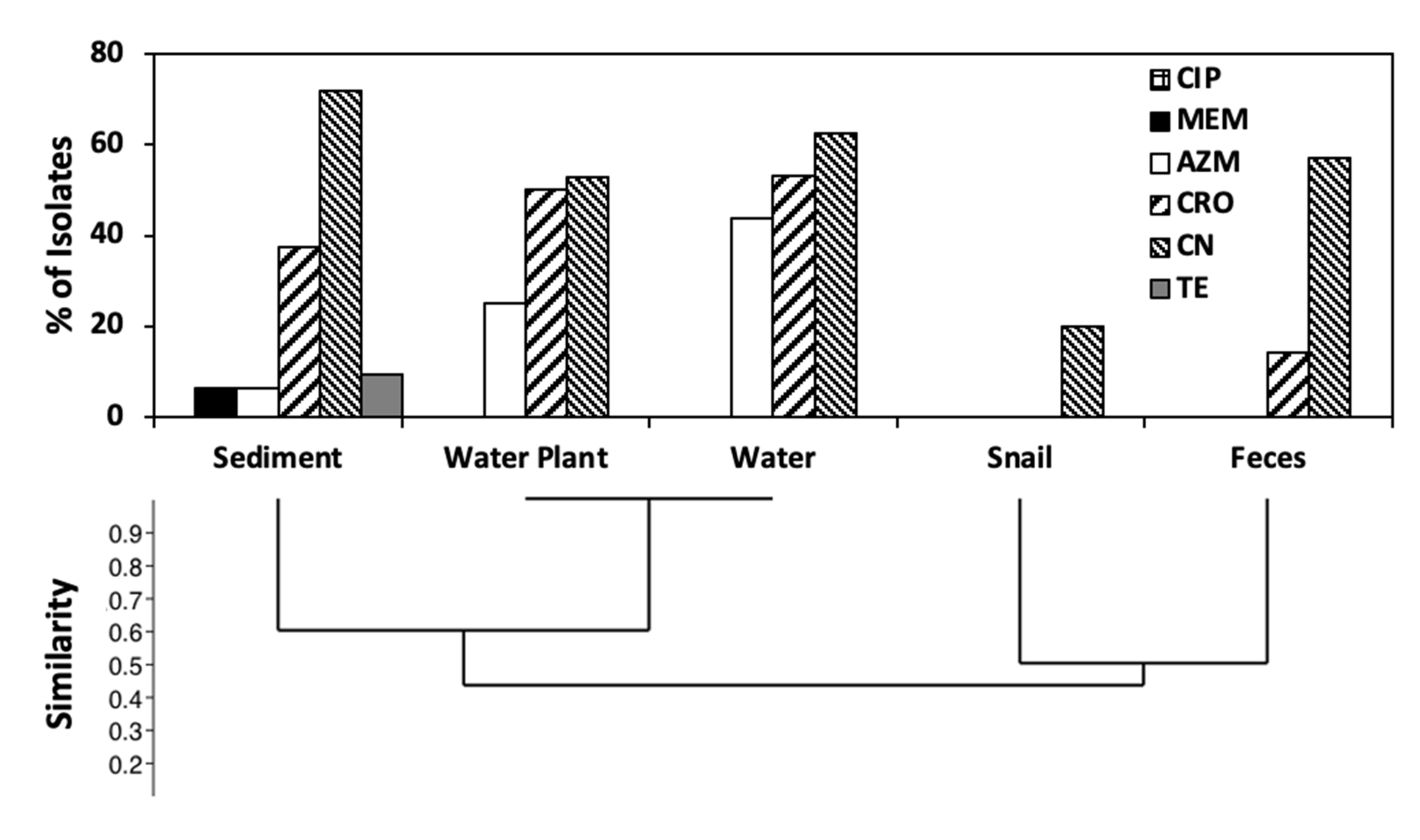
3.6. Antibiotic Resistance Profiling
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jang, J.; Hur, H.G.; Sadowsky, M.J.; Byappanahalli, M.N.; Yan, T.; Ishii, S. Environmental Escherichia coli: Ecology and public health implications—A review. J. Appl. Microbiol. 2017, 123, 570–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blount, Z.D. The unexhausted potential of E. coli. eLife 2015, 4, e05826. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.M. The Influence of Ecological Factors on the Distribution and the Genetic Structure of Escherichia coli. EcoSal Plus 2004, 1. [Google Scholar] [CrossRef]
- Bergthorsson, U.; Ochman, H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol. Biol. Evol. 1998, 15, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Walk, S.T.; Alm, E.W.; Calhoun, L.M.; Mladonicky, J.M.; Whittam, T.S. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 2007, 9, 2274–2288. [Google Scholar] [CrossRef]
- Ratajczak, M.; Laroche, E.; Berthe, T.; Clermont, O.; Pawlak, B.; Denamur, E.; Petit, F. Influence of hydrological conditions on the Escherichia coli population structure in the water of a creek on a rural watershed. BMC Microbiol. 2010, 10, 222. [Google Scholar] [CrossRef] [Green Version]
- NandaKafle, G.; Seale, T.; Flint, T.; Nepal, M.; Venter, S.N.; Brözel, V.S. Distribution of Diverse Escherichia coli between Cattle and Pasture. Microbes Environ. 2017, 32, 226–233. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.R.; Stell, A.L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 2000, 181, 261–272. [Google Scholar] [CrossRef] [Green Version]
- Le Gall, T.; Clermont, O.; Gouriou, S.; Picard, B.; Nassif, X.; Denamur, E.; Tenaillon, O. Extraintestinal virulence is a coincidental by-product of commensalism in B2 phylogenetic group Escherichia coli strains. Mol. Biol. Evol. 2007, 24, 2373–2384. [Google Scholar] [CrossRef] [Green Version]
- Picard, B.; Garcia, J.S.; Gouriou, S.; Duriez, P.; Brahimi, N.; Bingen, E.; Elion, J.; Denamur, E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 1999, 67, 546–553. [Google Scholar] [CrossRef] [Green Version]
- Unno, T.; Han, D.; Jang, J.; Lee, S.N.; Ko, G.; Choi, H.Y.; Kim, J.H.; Sadowsky, M.J.; Hur, H.G. Absence of Escherichia coli phylogenetic group B2 strains in humans and domesticated animals from Jeonnam Province, Republic of Korea. Appl. Environ. Microbiol. 2009, 75, 5659–5666. [Google Scholar] [CrossRef] [Green Version]
- Chandran, A.; Mazumder, A. Occurrence of Diarrheagenic Virulence Genes and Genetic Diversity in Escherichia coli Isolates from Fecal Material of Various Avian Hosts in British Columbia, Canada. Appl. Environ. Microbiol. 2014, 80, 1933–1940. [Google Scholar] [CrossRef] [Green Version]
- Chandran, A.; Mazumder, A. Prevalence of Diarrhea-Associated Virulence Genes and Genetic Diversity in Escherichia coli Isolates from Fecal Material of Various Animal Hosts. Appl. Environ. Microbiol. 2013, 79, 7371–7380. [Google Scholar] [CrossRef] [Green Version]
- McLellan, S.L.; Daniels, A.D.; Salmore, A.K. Genetic characterization of Escherichia coli populations from host sources of fecal pollution by using DNA fingerprinting. Appl. Environ. Microbiol. 2003, 69, 2587–2594. [Google Scholar] [CrossRef] [Green Version]
- Anderson, M.A.; Whitlock, J.E.; Harwood, V.J. Diversity and distribution of Escherichia coli genotypes and antibiotic resistance phenotypes in feces of humans, cattle, and horses. Appl. Environ. Microbiol. 2006, 72, 6914–6922. [Google Scholar] [CrossRef] [Green Version]
- Braz, V.S.; Melchior, K.; Moreira, C.G. Escherichia coli as a Multifaceted Pathogenic and Versatile Bacterium. Front. Cell Infect. Microbiol. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Rasko, D.A.; Rosovitz, M.J.; Myers, G.S.; Mongodin, E.F.; Fricke, W.F.; Gajer, P.; Crabtree, J.; Sebaihia, M.; Thomson, N.R.; Chaudhuri, R. The pangenome structure of Escherichia coli: Comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 2008, 190, 6881–6893. [Google Scholar] [CrossRef] [Green Version]
- Touchon, M.; Hoede, C.; Tenaillon, O.; Barbe, V.; Baeriswyl, S.; Bidet, P.; Bingen, E.; Bonacorsi, S.; Bouchier, C.; Bouvet, O.; et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009, 5, e1000344. [Google Scholar] [CrossRef] [Green Version]
- Robins-Browne, R.M.; Holt, K.E.; Ingle, D.J.; Hocking, D.M.; Yang, J.; Tauschek, M. Are Escherichia coli Pathotypes Still Relevant in the Era of Whole-Genome Sequencing? Front. Cell Infect. Microbiol. 2016, 6, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukjancenko, O.; Wassenaar, T.M.; Ussery, D.W. Comparison of 61 sequenced Escherichia coli genomes. Microb. Ecol. 2010, 60, 708–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNally, A.; Oren, Y.; Kelly, D.; Pascoe, B.; Dunn, S.; Sreecharan, T.; Vehkala, M.; Valimaki, N.; Prentice, M.B.; Ashour, A.; et al. Combined Analysis of Variation in Core, Accessory and Regulatory Genome Regions Provides a Super-Resolution View into the Evolution of Bacterial Populations. PLoS Genet. 2016, 12, e1006280. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Ksoll, W.B.; Hicks, R.E.; Sadowsky, M.J. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 2006, 72, 612–621. [Google Scholar] [CrossRef] [Green Version]
- Brennan, F.P.; Abram, F.; Chinalia, F.A.; Richards, K.G.; O’Flaherty, V. Characterization of environmentally persistent Escherichia coli isolates leached from an Irish soil. Appl. Environ. Microbiol. 2010, 76, 2175–2180. [Google Scholar] [CrossRef] [Green Version]
- Lyautey, E.; Lu, Z.; Lapen, D.R.; Wilkes, G.; Scott, A.; Berkers, T.; Edge, T.A.; Topp, E. Distribution and diversity of Escherichia coli populations in the south nation river drainage basin, eastern Ontario, Canada. Appl. Environ. Microbiol. 2010, 76, 1486–1496. [Google Scholar] [CrossRef] [Green Version]
- Van Elsas, J.D.; Hill, P.; Chronakova, A.; Grekova, M.; Topalova, Y.; Elhottova, D.; Kristufek, V. Survival of genetically marked Escherichia coli O157:H7 in soil as affected by soil microbial community shifts. ISME J. 2007, 1, 204–214. [Google Scholar] [CrossRef] [Green Version]
- Byappanahalli, M.N.; Richard, L.W.; Shivelya, D.A.; John, F.; Ishii, S.; Sadowsky, M.J. Population structure of cladophora-borne Escherichia coli in nearshore water of lake Michigan. Water Res. 2007, 41, 3649–3654. [Google Scholar] [CrossRef]
- Casarez, E.A.; Pillai, S.D.; Di Giouanni, G.D. Genotype diversity of Escherichia coli isolates in natural waters determined by PFGE and ERIC-PCR. Water Res. 2007, 41, 3643–3648. [Google Scholar] [CrossRef]
- McLellan, S.L. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl. Environ. Microbiol. 2004, 70, 4658–4665. [Google Scholar] [CrossRef] [Green Version]
- Beversdorf, L.J.; Bornstein-Forst, S.M.; McLellan, S.L. The potential for beach sand to serve as a reservoir for Escherichia coli and the physical influences on cell die-off. J. Appl. Microbiol. 2007, 102, 1372–1381. [Google Scholar] [CrossRef]
- Byappanahalli, M.N.; Fujioka, R.S. Evidence that tropical soil environment can support the growth of Escherichia coli. Water Sci. Technol. 1998, 38, 171–174. [Google Scholar] [CrossRef]
- Byappanahalli, M.N.; Whitman, R.L.; Shively, D.A.; Sadowsky, M.J.; Ishii, S. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 2006, 8, 504–513. [Google Scholar] [CrossRef]
- Petit, F.; Clermont, O.; Delannoy, S.; Servais, P.; Gourmelon, M.; Fach, P.; Oberle, K.; Fournier, M.; Denamur, E.; Berthe, T. Change in the Structure of Escherichia coli Population and the Pattern of Virulence Genes along a Rural Aquatic Continuum. Front. Microbiol. 2017, 8, 609. [Google Scholar] [CrossRef]
- Blount, Z.D.; Barrick, J.E.; Davidson, C.J.; Lenski, R.E. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 2012, 489, 513–518. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Paleontol. Electron. 2001, 4, 1–9. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Hothorn, T. Comprehensive R Archive Network; Party, v.1.0-17; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Walk, S.T.; Alm, E.W.; Gordon, D.M.; Ram, J.L.; Toranzos, G.A.; Tiedje, J.M.; Whittam, T.S. Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 2009, 75, 6534–6544. [Google Scholar] [CrossRef] [Green Version]
- Ferenci, T. What is driving the acquisition of mutS and rpoS polymorphisms in Escherichia coli? Trends Microbiol. 2003, 11, 457–461. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A. SE-AL v. 2.0a11: Sequence Alignment Program. 2002. Available online: http://tree.bio.ed.ac.uk/software/seal/ (accessed on 17 September 2021).
- Leimbach, A.; Hacker, J.; Dobrindt, U. E. coli as an all-rounder: The thin line between commensalism and pathogenicity. Curr. Top. Microbiol. Immunol. 2013, 358, 3–32. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011, 39, W475–W478. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Fagan, P.K.; Hornitzky, M.A.; Bettelheim, K.A.; Djordjevic, S.P. Detection of Shiga-liketoxin (stx(1) and stx(2)), Intimin (eaeA), and enterohemorrhagic Escherichia coli (EHEC) Hemolysin (EHEC hlyA) genes in animal feces by multiplex PCR. Appl. Environ. Microbiol. 1999, 65, 868–872. [Google Scholar] [CrossRef] [Green Version]
- Osek, J. Multiplex polymerase chain reaction assay for identification of enterotoxigenic Escherichia coli strains. J. Vet. Diagn. Invest. 2001, 13, 308–311. [Google Scholar] [CrossRef] [Green Version]
- Wayne, P.A.; CLSI. Twenty-Fifth Informational Performance Standards for Antimicrobial Susceptibility Testing Supplement. In CLSI Document M100-S25; Clinical and Laboratory Standards Institute: New York, NY, USA, 2015. [Google Scholar]
- Chang, G.W.; Brill, J.; Lum, R. Proportion of beta-D-glucuronidase-negative Escherichia coli in human fecal samples. Appl. Environ. Microbiol. 1989, 55, 335–339. [Google Scholar] [CrossRef] [Green Version]
- Mora, A.; Lopez, C.; Dhabi, G.; Lopez-Beceiro, A.M.; Fidalgo, L.E.; Diaz, E.A.; Martinez-Carrasco, C.; Mamani, R.; Herrera, A.; Blanco, J.E.; et al. Seropathotypes, Phylogroups, Stx Subtypes, and Intimin Types of Wildlife-Carried, Shiga Toxin-Producing Escherichia coli Strains with the Same Characteristics as Human-Pathogenic Isolates. Appl. Environ. Microb. 2012, 78, 2578–2585. [Google Scholar] [CrossRef] [Green Version]
- Dhaka, P.; Vijay, D.; Vergis, J.; Negi, M.; Kumar, M.; Mohan, V.; Doijad, S.; Poharkar, K.V.; Malik, S.S.; Barbuddhe, S.B.; et al. Genetic diversity and antibiogram profile of diarrhoeagenic Escherichia coli pathotypes isolated from human, animal, foods and associated environmental sources. Infect. Ecol. Epidemiol. 2016, 6, 31055. [Google Scholar] [CrossRef]
- Schwidder, M.; Heinisch, L.; Schmidt, H. Genetics, Toxicity, and Distribution of Enterohemorrhagic Escherichia coli Hemolysin. Toxins 2019, 11, 502. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Sha, Q.; Lacher, D.W.; Del Valle, J.; Mosci, R.E.; Moore, J.A.; Scribner, K.T.; Manning, S.D. Characterization of enteropathogenic and Shiga toxin-producing Escherichia coli in cattle and deer in a shared agroecosystem. Front. Cell Infect. Microbiol. 2015, 5, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francis, D.H.; Willgohs, J.A. Evaluation of a live avirulent Escherichia coli vaccine for K88+, LT+ enterotoxigenic colibacillosis in weaned pigs. Am. J. Vet. Res. 1991, 52, 1051–1055. [Google Scholar]
- Hansen, D.L.; Ishii, S.; Sadowsky, M.J.; Hicks, R.E. Escherichia coli Populations in Great Lakes Waterfowl Exhibit Spatial Stability and Temporal Shifting. Appl. Environ. Microb. 2009, 75, 1546–1551. [Google Scholar] [CrossRef] [Green Version]
- White, A.P.; Sibley, K.A.; Sibley, C.D.; Wasmuth, J.D.; Schaefer, R.; Surette, M.G.; Edge, T.A.; Neumann, N.F. Intergenic sequence comparison of Escherichia coli isolates reveals lifestyle adaptations but not host specificity. Appl. Environ. Microbiol. 2011, 77, 7620–7632. [Google Scholar] [CrossRef] [Green Version]
- Meric, G.; Kemsley, E.K.; Falush, D.; Saggers, E.J.; Lucchini, S. Phylogenetic distribution of traits associated with plant colonization in Escherichia coli. Environ. Microbiol. 2013, 15, 487–501. [Google Scholar] [CrossRef]
- Jang, J.; Di, D.Y.W.; Lee, A.; Unno, T.; Sadowsky, M.J.; Hur, H.G. Seasonal and Genotypic Changes in Escherichia coli Phylogenetic Groups in the Yeongsan River Basin of South Korea. PLoS ONE 2014, 9, e100585. [Google Scholar] [CrossRef]
- Dusek, N.; Hewitt, A.J.; Schmidt, K.N.; Bergholz, P.W. Landscape-Scale Factors Affecting the Prevalence of Escherichia coli in Surface Soil Include Land Cover Type, Edge Interactions, and Soil pH. Appl. Environ. Microbiol. 2018, 84, e02714–e02717. [Google Scholar] [CrossRef] [Green Version]
- Kimura, M. The Neutral Theory of Molecular Evolution; Cambridge University Press: Cambridge, UK, 1983. [Google Scholar]
- Lynch, M.; Walsh, B. The Origins of Genome Architecture; Sinauer Associates Sunderland: Sunderland, MA, USA, 2007; Volume 98. [Google Scholar]
- Maddamsetti, R. Gene flow in microbial communities could explain unexpected patterns of synonymous variation in the Escherichia coli core genome. Mob. Genet. Elem. 2016, 6, e1137380. [Google Scholar] [CrossRef] [PubMed]
- Retchless, A.C.; Lawrence, J.G. Temporal Fragmentation of Speciation in Bacteria. Science 2007, 317, 1093–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheppard, S.K.; McCarthy, N.D.; Falush, D.; Maiden, M.C. Convergence of Campylobacter species: Implications for bacterial evolution. Science 2008, 320, 237–239. [Google Scholar] [CrossRef]
- Luo, C.; Walk, S.T.; Gordon, D.M.; Feldgarden, M.; Tiedje, J.M.; Konstantinidis, K.T. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc. Natl. Acad. Sci. USA 2011, 108, 7200–7205. [Google Scholar] [CrossRef] [Green Version]
- Karberg, K.A.; Olsen, G.J.; Davis, J.J. Similarity of genes horizontally acquired by Escherichia coli and Salmonella enterica is evidence of a supraspecies pangenome. Proc. Natl. Acad. Sci. USA 2011, 108, 20154–20159. [Google Scholar] [CrossRef] [Green Version]
- Goudeau, D.M.; Parker, C.T.; Zhou, Y.G.; Sela, S.; Kroupitski, Y.; Brandl, M.T. The Salmonella Transcriptome in Lettuce and Cilantro Soft Rot Reveals a Niche Overlap with the Animal Host Intestine. Appl. Environ. Microbiol. 2013, 79, 250–262. [Google Scholar] [CrossRef] [Green Version]
- Byappanahalli, M.N.; Nevers, M.B.; Whitman, R.L.; Ishii, S. Application of a Microfluidic Quantitative Polymerase Chain Reaction Technique to Monitor Bacterial Pathogens in Beach Water and Complex Environmental Matrices. Environ. Sci. Technol. Lett. 2015, 2, 347–351. [Google Scholar] [CrossRef]
- Zhang, Q.; Eichmiller, J.J.; Staley, C.; Sadowsky, M.J.; Ishii, S. Correlations between pathogen concentration and fecal indicator marker genes in beach environments. Sci. Total Environ. 2016, 573, 826–830. [Google Scholar] [CrossRef]
- Hamilton, M.J.; Hadi, A.Z.; Griffith, J.F.; Ishii, S.; Sadowsky, M.J. Large scale analysis of virulence genes in Escherichia coli strains isolated from Avalon Bay, CA. Water Res. 2010, 44, 5463–5473. [Google Scholar] [CrossRef] [Green Version]
- Chandran, A.; Mazumder, A. Pathogenic Potential, Genetic Diversity, and Population Structure of Escherichia coli Strains Isolated from a Forest-Dominated Watershed (Comox Lake) in British Columbia, Canada. Appl. Environ. Microbiol. 2015, 81, 1779–1789. [Google Scholar] [CrossRef] [PubMed]
- Amaya, E.; Reyes, D.; Paniagua, M.; Calderon, S.; Rashid, M.U.; Colque, P.; Kuhn, I.; Mollby, R.; Weintraub, A.; Nord, C.E. Antibiotic resistance patterns of Escherichia coli isolates from different aquatic environmental sources in Leon, Nicaragua. Clin. Microbiol. Infect. 2012, 18, E347–E354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayah, R.S.; Kaneene, J.B.; Johnson, Y.; Miller, R. Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl. Environ. Microbiol. 2005, 71, 1394–1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibekwe, A.M.; Murinda, S.E.; Graves, A.K. Genetic Diversity and Antimicrobial Resistance of Escherichia coli from Human and Animal Sources Uncovers Multiple Resistances from Human Sources. PLoS ONE 2011, 6, e20819. [Google Scholar] [CrossRef] [Green Version]
- Vaz-Moreira, I.; Nunes, O.C.; Manaia, C.M. Bacterial diversity and antibiotic resistance in water habitats: Searching the links with the human microbiome. Fems Microbiol. Rev. 2014, 38, 761–778. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
NandaKafle, G.; Huegen, T.; Potgieter, S.C.; Steenkamp, E.; Venter, S.N.; Brözel, V.S. Niche Preference of Escherichia coli in a Peri-Urban Pond Ecosystem. Life 2021, 11, 1020. https://doi.org/10.3390/life11101020
NandaKafle G, Huegen T, Potgieter SC, Steenkamp E, Venter SN, Brözel VS. Niche Preference of Escherichia coli in a Peri-Urban Pond Ecosystem. Life. 2021; 11(10):1020. https://doi.org/10.3390/life11101020
Chicago/Turabian StyleNandaKafle, Gitanjali, Taylor Huegen, Sarah C. Potgieter, Emma Steenkamp, Stephanus N. Venter, and Volker S. Brözel. 2021. "Niche Preference of Escherichia coli in a Peri-Urban Pond Ecosystem" Life 11, no. 10: 1020. https://doi.org/10.3390/life11101020





