Genotypic and Phenotypic Structure of the Population of Phytophthora infestans in Egypt Revealed the Presence of European Genotypes
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Collection and Isolation of P. infestans
2.2. Determination of Mating Type
2.3. Metalaxyl Sensitivity Assay
2.4. Determination of Virulence Profile
2.5. DNA Extraction and SSR Marker Analysis
2.6. Data Analysis
2.7. Screening of Effector Genes and Detection of Mutation
3. Results
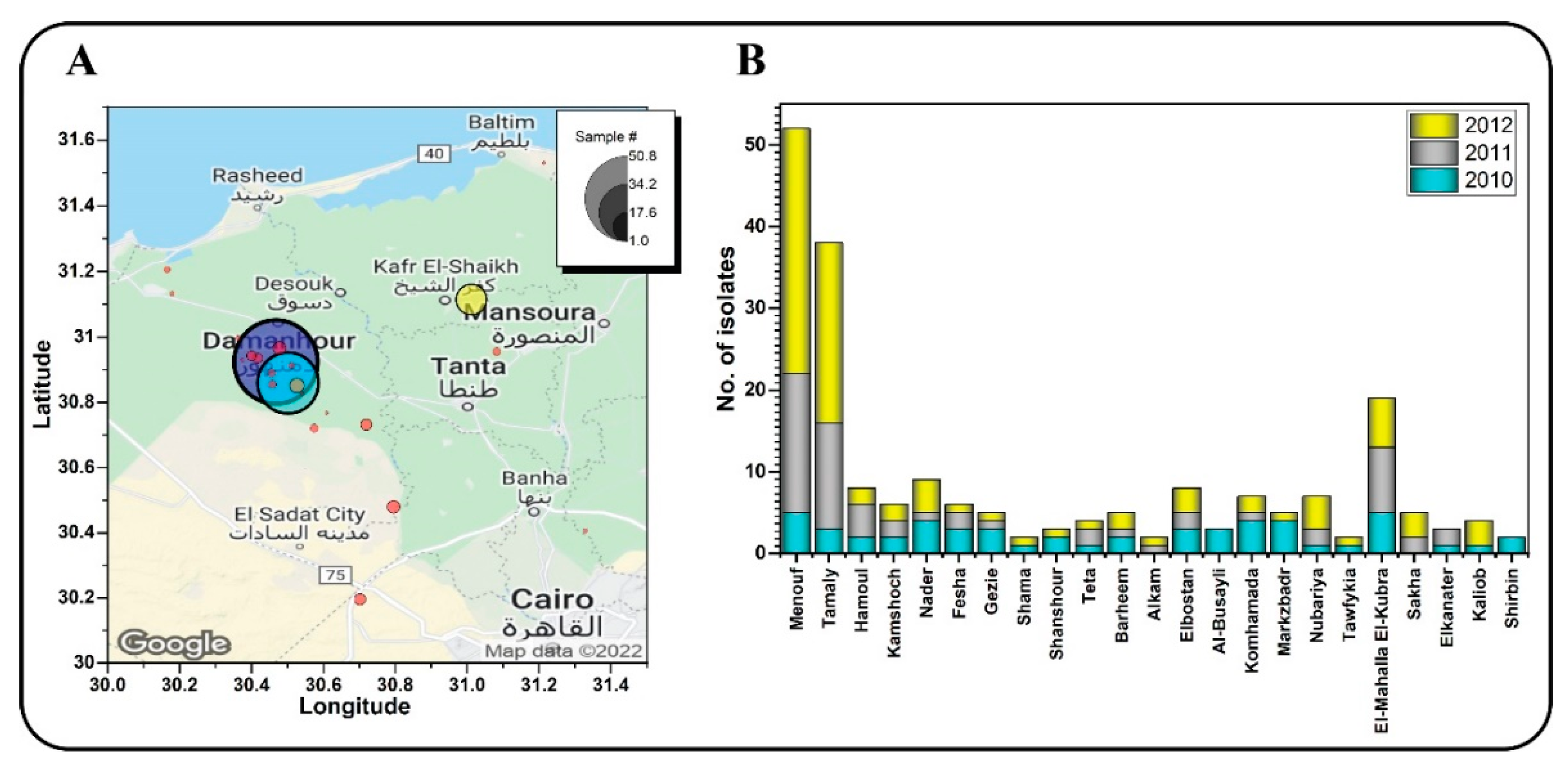
3.1. Samples Collection and Isolation of P. infestans
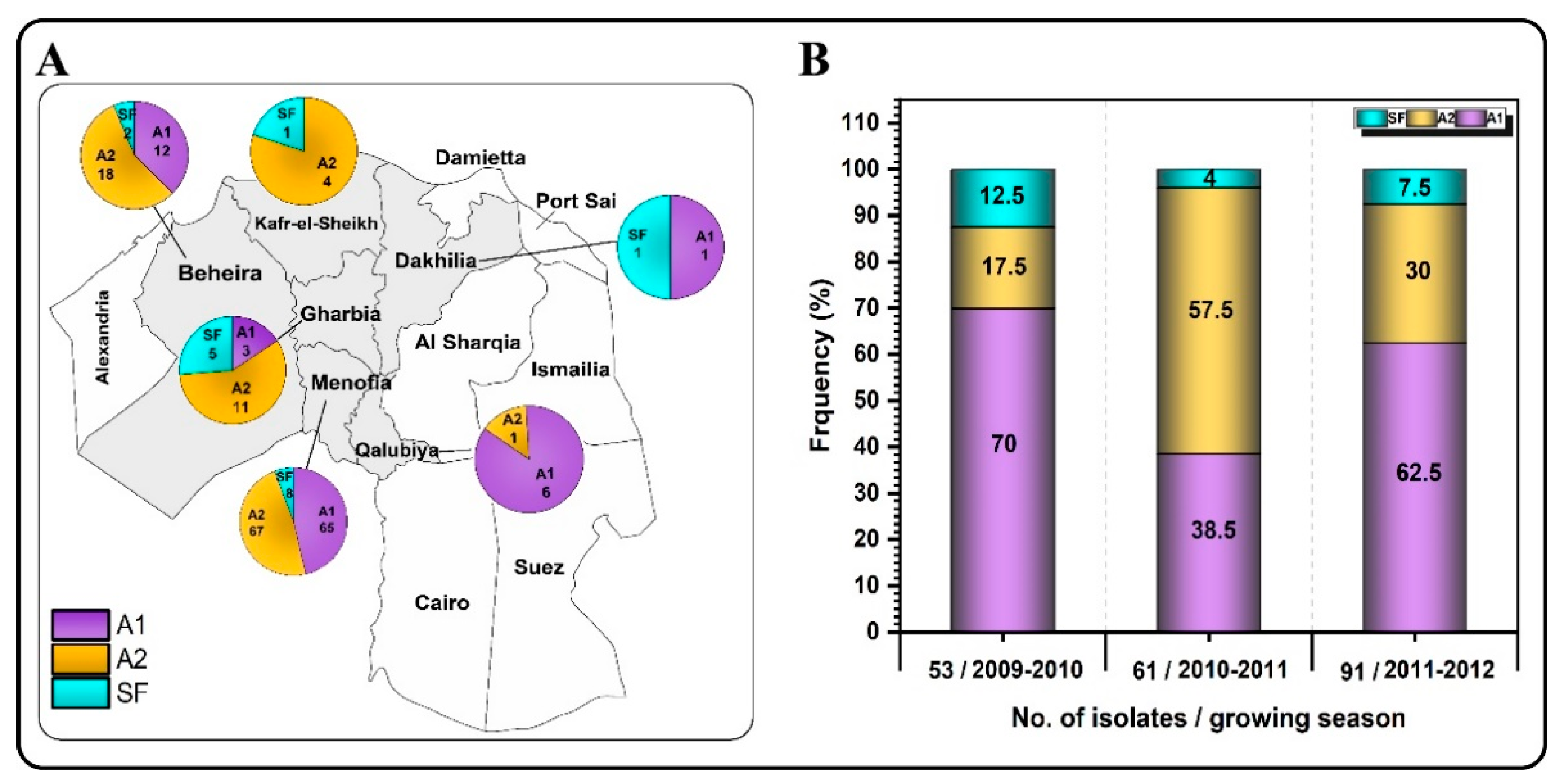
3.2. Determination of Mating Type
3.3. Metalaxyl Sensitivity Assay
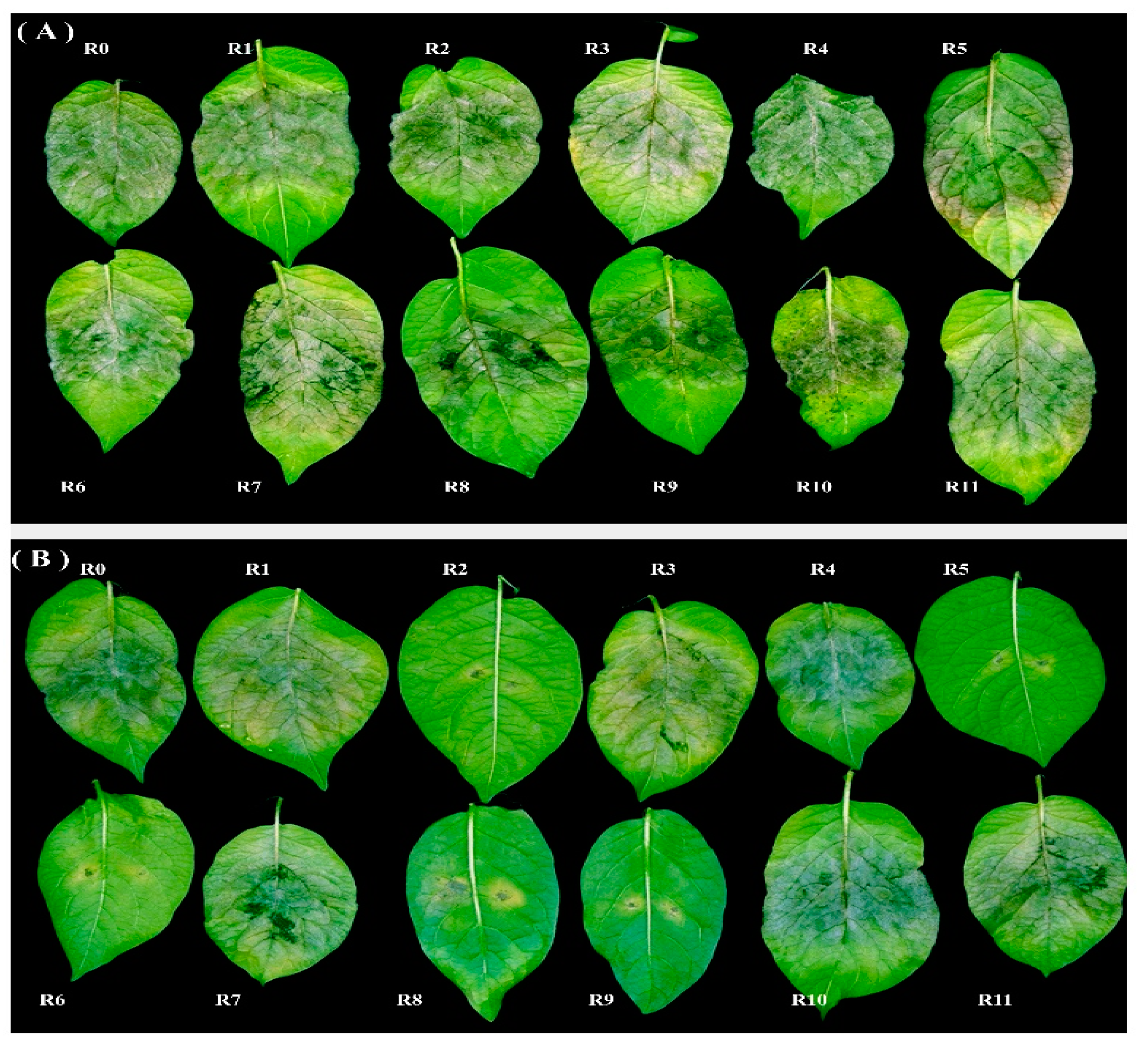
3.4. Determination of Virulence Profile
3.5. Genotyping of the Egyptian Population
3.6. Egyptian Population Structure
3.7. Analysis of Effector Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aguilera-Galvez, C.; Champouret, N.; Rietman, H.; Lin, X.; Wouters, D.; Chu, Z.; Jones, J.; Vossen, J.; Visser, R.; Wolters, P. Two different R gene loci co-evolved with Avr2 of Phytophthora infestans and confer distinct resistance specificities in potato. Stud. Mycol. 2018, 89, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Mazumdar, P.; Singh, P.; Kethiravan, D.; Ramathani, I.; Ramakrishnan, N. Late blight in tomato: Insights into the pathogenesis of the aggressive pathogen Phytophthora infestans and future research priorities. Planta 2021, 253, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Chowdappa, P.; Kumar, N.B.; Madhura, S.; Kumar, M.S.; Myers, K.L.; Fry, W.E.; Squires, J.N.; Cooke, D.E. Emergence of 13_ A 2 Blue lineage of Phytophthora infestans was responsible for severe outbreaks of late blight on tomato in South-West India. J. Phytopath. 2013, 161, 49–58. [Google Scholar] [CrossRef]
- Fry, W.E.; Grünwald, N.J.; Cooke, D.E.; McLeod, A.; Forbes, G.A.; Cao, K. Population genetics and population diversity of Phytophthora infestans. In Oomycete Genetics and Genomics: Diversity, Interactions, and Research Tools; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009; pp. 139–164. [Google Scholar]
- Kamoun, S.; Furzer, O.; Jones, J.D.; Judelson, H.S.; Ali, G.S.; Dalio, R.J.; Roy, S.G.; Schena, L.; Zambounis, A.; Panabières, F. The top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 2015, 16, 413–434. [Google Scholar] [CrossRef]
- Martin, M.D.; Ho, S.Y.; Wales, N.; Ristaino, J.B.; Gilbert, M.T.P. Persistence of the mitochondrial lineage responsible for the Irish potato famine in extant new world Phytophthora infestans. Mol. Biol. Evol. 2014, 31, 1414–1420. [Google Scholar] [CrossRef]
- Wang, S.; Boevink, P.C.; Welsh, L.; Zhang, R.; Whisson, S.C.; Birch, P.R. Delivery of cytoplasmic and apoplastic effectors from Phytophthora infestans haustoria by distinct secretion pathways. New Phytol. 2017, 216, 205–215. [Google Scholar] [CrossRef]
- Cooke, D.; Lees, A. Markers, old and new, for examining Phytophthora infestans diversity. Plant Pathol. 2004, 53, 692–704. [Google Scholar] [CrossRef]
- Li, Y.; Cooke, D.E.; Jacobsen, E.; van der Lee, T. Efficient multiplex simple sequence repeat genotyping of the oomycete plant pathogen Phytophthora infestans. J. Microbiol. Methods 2013, 92, 316–322. [Google Scholar] [CrossRef]
- Saville, A.C.; Martin, M.D.; Ristaino, J.B. Historic late blight outbreaks caused by a widespread dominant lineage of Phytophthora infestans (Mont.) de Bary. PLoS ONE 2016, 11, e0168381. [Google Scholar] [CrossRef]
- Mariette, N.; Androdias, A.; Mabon, R.; Corbiere, R.; Marquer, B.; Montarry, J.; Andrivon, D. Local adaptation to temperature in populations and clonal lineages of the Irish potato famine pathogen Phytophthora infestans. Ecol. Evol. 2016, 6, 6320–6331. [Google Scholar] [CrossRef]
- Cooke, D.E.; Cano, L.M.; Raffaele, S.; Bain, R.A.; Cooke, L.R.; Etherington, G.J.; Deahl, K.L.; Farrer, R.A.; Gilroy, E.M.; Goss, E.M. Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLoS Pathog. 2012, 8, e1002940. [Google Scholar] [CrossRef] [PubMed]
- Chmielarz, M.; Sobkowiak, S.; Dębski, K.; Cooke, D.; Brurberg, M.; Śliwka, J. Diversity of Phytophthora infestans from Poland. Plant Pathol. 2014, 63, 203–211. [Google Scholar] [CrossRef]
- Rekad, F.Z.; Cooke, D.E.L.; Puglisi, I.; Randall, E.; Guenaoui, Y.; Bouznad, Z.; Evoli, M.; Pane, A.; Schena, L.; di San Lio, G.M. Characterization of Phytophthora infestans populations in northwestern Algeria during 2008–2014. Fungal Biol. 2017, 121, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Njoroge, A.W.; Andersson, B.; Lees, A.K.; Mutai, C.; Forbes, G.A.; Yuen, J.E.; Pelle, R. Genotyping of Phytophthora infestans in Eastern Africa reveals a dominating invasive European lineage. Phytopathology 2019, 109, 670–680. [Google Scholar] [CrossRef]
- Van Loon, K.D. The seed potato market. In Potato Biology and Biotechnology; Oxford: Oxford, UK, 2007; pp. 45–51. [Google Scholar]
- Njoroge, A.; Tusiime, G.; Forbes, G.; Yuen, J. Displacement of US-1 clonal lineage by a new lineage of Phytophthora infestans on potato in Kenya and Uganda. Plant Pathol. 2016, 65, 587–592. [Google Scholar] [CrossRef]
- Workman, D. Trade Map, International Trade Centre. 2022. Available online: https://www.worldstopexports.com/potatoes-exports-by-country, (accessed on 25 March 2022).
- Saad, N. Epiphytology of Potato Blight in Arab Republic of Egypt. Ph.D. Thesis, Cairo University, Egypt, Cairo, 1978. [Google Scholar]
- El-Korany, A.E. Occurrence of the self-fertile phenotype of Phytophthora infestans in El-Behera governorate. Egypt. J. Agric. Sci 2002, 27, 7855–7863. [Google Scholar]
- El-Korany, A.E. Occurrence of oospores of Phytophthora infestans in the field and under controlled conditions. J. Agric. Environ. Sci. Alex. Univ. 2008, 7, 31–52. [Google Scholar]
- El-Sheikh, M.A.; El-Farnawany, M.; EI-Korany, A. Characterization of the Egyptian population of tomato isolates of Phytophthora infestans. J. Agric. Environ. Sci. 2005, 37–55, 37–55. [Google Scholar]
- El-Korany, A. Pathological Studies on the Late Blight of Potato Caused by Phytophthora infestans; Suez Canal University: Ismailia, Egypt, 1994. [Google Scholar]
- El-Komy, M. The Role of Certain Resistance Factors Affecting the Development of Late Blight Disease of Potato. Ph.D. Thesis, Faculty of Agriculture, Alexandria University, Alexandria, Egypt, 2007. [Google Scholar]
- El-Ganainy, S.; Ahmed, Y.; Soliman, M.; Ismail, A.; Tohamy, A.; Randall, E.; Cooke, D. A Shift in the Population of Phytophthora infestans on Egyptian Potato Crops. In Phytopathol; American Phytopathological Society: St. Paul, MN, USA, 2016; p. 140. [Google Scholar]
- Barton, N.H.; Charlesworth, B. Why sex and recombination? Science 1998, 281, 1986–1990. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.D.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef]
- Tyler, B.M.; Tripathy, S.; Zhang, X.; Dehal, P.; Jiang, R.H.; Aerts, A.; Arredondo, F.D.; Baxter, L.; Bensasson, D.; Beynon, J.L. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 2006, 313, 1261–1266. [Google Scholar] [CrossRef] [PubMed]
- Caten, C.; Jinks, J. Spontaneous variability of single isolates of Phytophthora infestans. I. Cultural variation. Can. J. Bot. 1968, 46, 329–348. [Google Scholar] [CrossRef]
- Martin, F.N.; Abad, Z.G.; Balci, Y.; Ivors, K. Identification and detection of Phytophthora: Reviewing our progress, identifying our needs. Plant Dis. 2012, 96, 1080–1103. [Google Scholar] [CrossRef] [PubMed]
- Cooke, D.; Young, V.; Birch, P.; Toth, R.; Gourlay, F.; Day, J.; Carnegie, S.; Duncan, J. Phenotypic and genotypic diversity of Phytophthora infestans populations in Scotland (1995–1997). Plant Pathol. 2003, 52, 181–192. [Google Scholar] [CrossRef]
- Goodwin, S.B.; Spielman, L.J.; Matuszak, J.; Bergeron, S.; Fry, W. Clonal diversity and genetic differentiation of Phytophthora infestans populations in northern and central Mexico. Phytopathology 1992, 82, 955–961. [Google Scholar] [CrossRef]
- Forbes, G. Manual for laboratory work on Phytophthora infestans. In CIP’s Training Manual; International Potato Center: Lima, Peru, 1997. [Google Scholar]
- Daggett, S.; Götz, E.; Therrien, C. Phenotypic changes in populations of Phytophthora infestans from eastern Germany. Phytopathology 1993, 83, 319. [Google Scholar] [CrossRef]
- Therrien, C.; Tooley, P.; Spielman, L.; Fry, W.; Ritch, D.; Shelly, S. Nuclear DNA content, allozyme phenotypes and metalaxyl sensitivity of Phytophthora infestans from Japan. Mycol. Res. 1993, 97, 945–950. [Google Scholar] [CrossRef]
- Malcolmson, J.F.; Black, W. New R genes in Solanum demissum Lindl. and their complementary races of Phytophthora infestans (Mont.) de Bary. Euphytica 1966, 15, 199–203. [Google Scholar] [CrossRef]
- Flier, W.; Turkensteen, L. Foliar aggressiveness of Phytophthora infestans in three potato growing regions in the Netherlands. Eur. J. Plant Pathol. 1999, 105, 381–388. [Google Scholar] [CrossRef]
- Wang, H.; Qi, M.; Cutler, A.J. A simple method of preparing plant samples for PCR. Nucleic Acids Res. 1993, 21, 4153. [Google Scholar] [CrossRef]
- Bruvo, R.; Michiels, N.K.; D’Souza, T.G.; Schulenburg, H. A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Mol. Ecol. 2004, 13, 2101–2106. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.V.; Jasieniuk, M. POLYSAT: An R package for polyploid microsatellite analysis. Mol. Ecol. Res. 2011, 11, 562–566. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.A.; Churchill, G.; Autrique, J.; Tanksley, S.; Sorrells, M. Optimizing parental selection for genetic linkage maps. Genome 1993, 36, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Gilroy, E.M.; Breen, S.; Whisson, S.C.; Squires, J.; Hein, I.; Kaczmarek, M.; Turnbull, D.; Boevink, P.C.; Lokossou, A.; Cano, L.M. Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 2011, 191, 763–776. [Google Scholar] [CrossRef]
- Armstrong, M.R.; Whisson, S.C.; Pritchard, L.; Bos, J.I.; Venter, E.; Avrova, A.O.; Rehmany, A.P.; Böhme, U.; Brooks, K.; Cherevach, I. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. USA 2005, 102, 7766–7771. [Google Scholar] [CrossRef]
- Van Poppel, P.M.; Guo, J.; van de Vondervoort, P.J.; Jung, M.W.; Birch, P.R.; Whisson, S.C.; Govers, F. The Phytophthora infestans avirulence gene Avr4 encodes an RXLR-dEER effector. Mol. Plant-Microbe Interact. 2008, 21, 1460–1470. [Google Scholar] [CrossRef]
- Arafa, R.A.; Soliman, N.E.K.; Moussa, O.M.; Kamel, S.M.; Shirasawa, K. Characterization of Egyptian Phytophthora infestans population using simple sequence repeat markers. J. Gen. Plant Pathol. 2018, 84, 104–107. [Google Scholar] [CrossRef]
- Arafa, R.A.; Kamel, S.M.; Rakha, M.T.; Soliman, N.E.K.; Moussa, O.M.; Shirasawa, K. Analysis of the lineage of Phytophthora infestans isolates using mating type assay, traditional markers, and next generation sequencing technologies. PLoS ONE 2020, 15, e0221604. [Google Scholar] [CrossRef]
- Niederhauser, J.S. Phytophthora infestans: The Mexican Connection; Cambridge University Press: Cambridge, UK, 1991. [Google Scholar]
- Drenth, A.; Janssen, E.M.; Govers, F. Formation and survival of oospores of Phytophthora infestans under natural conditions. Plant Pathol. 1995, 44, 86–94. [Google Scholar] [CrossRef]
- Smirnov, A.; Elansky, S. Oospore formation in the field populations of Phytophthora infestans in Moscow region. Mikol. Fitopatol. 1999, 33, 421–425. [Google Scholar]
- Fernández-Pavía, S.; Grünwald, N.; Diaz-Valasis, M.; Cadena-Hinojosa, M.; Fry, W. Soilborne oospores of Phytophthora infestans in central Mexico survive winter fallow and infect potato plants in the field. Plant Dis. 2004, 88, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Erwin, D.C.; Ribeiro, O.K. Phytophthora: Diseases Worldwide; APS Press: St. Paul, MN, USA, 1996. [Google Scholar]
- Clinton, G.P. Oospores of potato blight. Science 1911, 33, 744–747. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mosa, A.A.; Kato, M.; Sato, N.; Kobayashi, K.; Ogoshi, A. Occurrence of the A2 mating type of Phytophthora infestans on potato in Japan. Jpn. J. Phytopathol. 1989, 55, 615–620. [Google Scholar] [CrossRef]
- Fyfe, A.; Shaw, D. An analysis of self-fertility in field isolates of Phytophthora infestans. Mycol. Res. 1992, 96, 390–394. [Google Scholar] [CrossRef]
- Lees, A.; Stewart, J.; Lynott, J.; Carnegie, S.; Campbell, H.; Roberts, A. The effect of a dominant Phytophthora infestans genotype (13_A2) in Great Britain on host resistance to foliar late blight in commercial potato cultivars. Potato Res. 2012, 55, 125–134. [Google Scholar] [CrossRef]
- Li, Y. Multiplex SSR Analysis of Phytophthora infestans in Different Countries and the Importance for Potato Breeding; Wageningen University and Research: Wageningen, The Netherlands, 2012. [Google Scholar]
- Göre, M.E.; Altın, N.; Myers, K.; Cooke, D.E.L.; Fry, W.E.; Özer, G. Population structure of Phytophthora infestans in Turkey reveals expansion and spread of dominant clonal lineages and virulence. Plant Pathol. 2021, 70, 898–911. [Google Scholar] [CrossRef]
- Kanetis, L.I.; Pittas, L.; Nikoloudakis, N.; Cooke, D.E.L.; Ioannou, N. Characterization of Phytophthora infestans populations in Cyprus, the Southernmost potato-producing European country. Plant Dis. 2021, 105, 3407–3417. [Google Scholar] [CrossRef]
- Harbaoui, K.; van der Lee, T.; Vleeshouwers, V.G.; Khammassy, N.; Harrabi, M.; Hamada, W. Characterisation of Phytophthora infestans isolates collected from potato and tomato crops in Tunisia during 2006–2008. Potato Res. 2013, 56, 11–29. [Google Scholar] [CrossRef]
- Dey, T.; Saville, A.; Myers, K.; Tewari, S.; Cooke, D.E.; Tripathy, S.; Fry, W.E.; Ristaino, J.B.; Guha Roy, S. Large sub-clonal variation in Phytophthora infestans from recent severe late blight epidemics in India. Sci. Rep. 2018, 8, 1–12. [Google Scholar]
- Saville, A.C.; La Spada, F.; Faedda, R.; Migheli, Q.; Scanu, B.; Ermacora, P.; Gilardi, G.; Fedele, G.; Rossi, V.; Lenzi, N. Population structure of Phytophthora infestans collected on potato and tomato in Italy. Plant Pathol. 2021, 70, 2165–2178. [Google Scholar] [CrossRef]
- Harbaoui, K.; Hamada, W.; Li, Y.; Vleeshouwers, V.G.; van der Lee, T. Increased difficulties to control late blight in Tunisia are caused by a genetically diverse Phytophthora infestans population next to the clonal lineage NA-01. Plant Dis. 2014, 98, 898–908. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haas, B.J.; Kamoun, S.; Zody, M.C.; Jiang, R.H.; Handsaker, R.E.; Cano, L.M.; Grabherr, M.; Kodira, C.D.; Raffaele, S.; Torto-Alalibo, T. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 2009, 461, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 2006, 44, 41–60. [Google Scholar] [CrossRef] [PubMed]
- Vleeshouwers, V.G.; Rietman, H.; Krenek, P.; Champouret, N.; Young, C.; Oh, S.-K.; Wang, M.; Bouwmeester, K.; Vosman, B.; Visser, R.G. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE 2008, 3, e2875. [Google Scholar] [CrossRef]
- Du, Y.; Weide, R.; Zhao, Z.; Msimuko, P.; Govers, F.; Bouwmeester, K. RXLR effector diversity in Phytophthora infestans isolates determines recognition by potato resistance proteins; the case study AVR1 and R1. Stud. Mycol. 2018, 89, 85–93. [Google Scholar] [CrossRef]


| Primer | Sequence (5′-3′) | References |
|---|---|---|
| AVR1F1 | TTGCCCTGTTGTGTATGCAT | [12] |
| AVR1R1 | GGCCATCAATTCCTCAGGT | [12] |
| Pex147F | CCATGCGTCTGGCAATTATGCT | [43] |
| Pex147R | CTGAAAACTAATATCCAGTGA | [43] |
| PiAvr4F | ATGCGTTCGCTTCACATTTTGCTGG | [44] |
| PiAvr4R | CTAAGATATGGGCCGTCTAGCTTGGAG | [44] |
| PiAVR2_F2 | GACCAAACGGCGTACTTCAT | [42] |
| PiAVR2_R2 | CGCCGAGCTCTTAACTCCT | [42] |
| Piavr2_F7 | ACGCTTCTATCCGACAACGA | [42] |
| Piavr2_R7 | ATTGGTGGTAATGCCTGCG | [42] |
| NitRedF | GGACCGCTGGGCCACTTCAC | [42] |
| NitRedR | CGCTGGCTTGCAGGCGTACT | [42] |
| Sampling Year | Mating Type | Metalaxyl Sensitivity | ||||
|---|---|---|---|---|---|---|
| A1 | A2 | SF | R | S | IR | |
| 2010 | 37 | 9 | 7 | 25 | 25 | 3 |
| 2011 | 24 | 35 | 2 | 32 | 22 | 7 |
| 2012 | 57 | 27 | 7 | 35 | 41 | 15 |
| Total | 118 | 71 | 16 | 92 | 88 | 25 |
| SSR locus | Fragments | Alleles Size |
|---|---|---|
| Pi02 | 4 | 266, 268, 270, 272 |
| Pi4B | 3 | 205, 213, 217 |
| G11 | 9 | 142, 154, 156, 160, 164, 204, 206, 208, 210 |
| Pi04 | 2 | 166, 170 |
| Pi63 | 3 | 270, 273, 279 |
| Pi70 | 1 | 192 |
| D13 | 8 | 118, 134, 140, 142, 154, 156, 210, 214 |
| SSR11 | 2 | 331, 341 |
| SSR2 | 2 | 173, 175 |
| SSR4 | 3 | 284, 288, 294 |
| SSR6 | 3 | 240, 242, 244 |
| SSR8 | 2 | 260, 266 |
| Isolate Name | Genotype | Avr1 | Avr2 | Avr2-Like | Avr3a | Avr4 |
|---|---|---|---|---|---|---|
| EG_02 | 13_A2_5 | - | - | + | + | + |
| EG_05 | 13_A2_5 | - | - | + | + | + |
| EG_15 | 13_A2_43 | - | - | + | n/d | n/d |
| EG_18 | 13_A2_84 | - | - | + | n/d | n/d |
| EG_90 | 13_A2_84 | - | - | + | n/d | n/d |
| EG_91 | 13_A2_84 | - | - | + | n/d | n/d |
| EG_95 | 13_A2_84 | - | - | + | + | + |
| EG_96 | 13_A2_84 | - | - | + | + | + |
| EG_11 | 23_A1 | + | + | + | + | + |
| EG_39 | 23_A1 | n/d | + | + | n/d | n/d |
| EG_40 | 23_A1 | + | + | + | + | + |
| EG_75 | 23_A1 | + | + | + | n/d | n/d |
| EG_86 | 23_A1 | n/d | + | + | n/d | n/d |
| EG_87 | 23_A1 | n/d | + | + | n/d | n/d |
| EG_03 | 23_A1 | - | n/d | n/d | + | + |
| EG_07 | 23_A1 | - | n/d | n/d | n/d | n/d |
| EG_79 | 23_A1 | + | n/d | n/d | n/d | n/d |
| EG_06 | misc | - | + | + | + | + |
| T30-4 | A1 | + | + | - | + | + |
| No | Effector Gene | Isolate Name | Genotype | Accession Number |
|---|---|---|---|---|
| 1 | avr1 | EG_40 | EU_23_A1_17 | MG976587 |
| 2 | avr1 | EG_75 | EU_23_A1_18 | MG976588 |
| 3 | avr1 | EG_79 | EU_23_A1_15 | MG976589 |
| 4 | avr2 | EG_06 | misc | MG976590 |
| 5 | Avr2 | EG_11 | EU_23_A1_15 | MG976591 |
| 6 | Avr2 | EG_39 | EU_23_A1_15 | MG976592 |
| 7 | Avr2 | EG_40 | EU_23_A1_17 | MG976593 |
| 8 | Avr2 | EG_75 | EU_23_A1_18 | MG976594 |
| 9 | Avr2* | EG_96 | EU_13_A2_84 | MG976595 |
| 10 | avr2-like | EG_05 | EU_13_A2_5 | MG976596 |
| 11 | avr2-like | EG_06 | misc | MG976597 |
| 12 | avr2-like | EG_11 | EU_23_A1_15 | MG976598 |
| 13 | avr2-like | EG_18 | EU_13_A2_84 | MG976599 |
| 14 | avr2-like | EG_40 | EU_23_A1_17 | MG976600 |
| 15 | avr2-like | EG_96 | EU_13_A2_84 | MG976601 |
| 16 | avr3a | EG_05 | EU_13_A2_5 | MG976602 |
| 17 | avr3a | EG_06 | misc | MG976603 |
| 18 | avr3a | EG_11 | EU_23_A1_15 | MG976604 |
| 19 | avr3a | EG_40 | EU_23_A1_17 | MG976605 |
| 20 | avr4 | EG_05 | EU_13_A2_5 | MG976606 |
| 21 | avr4 | EG_06 | misc | MG976607 |
| 22 | avr4 | EG_11 | EU_23_A1_15 | MG976608 |
| 23 | avr4 | EG_40 | EU_23_A1_17 | MG976609 |
| 24 | avr4 | EG_96_ | EU_13_A2_84 | MG976610 |
| 25 | Avr1 | T30-4 | A1 | XM_002896847 |
| 26 | Avr2 | T30-4 | A1 | XM_002902939 |
| 27 | avr3a | T30-4 | A1 | XM_002898796 |
| 28 | Avr4 | T30-4 | A1 | XM_002904373 |
| Effector | Product Size bp | Total SNPs | Syn-SNPs | 13_A2 | 23_A1 | No of AA | Source of Allelic Variance |
|---|---|---|---|---|---|---|---|
| avr1 | 550 | 3 | 1 | No @ | Yes * | 208 | Mut or Del |
| Avr2 | 500 | 0 | 0 | No | Yes | 116 | Del |
| avr2-Like | 480 | 2 | 0 | Yes | Yes | 116 | Mut |
| avr3a | 450 | 2 | 0 | Yes | Yes | 147 | Mut |
| avr4 | 950 | 13 | 4 | Yes | Yes | 287 | Mut or Tru |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Ganainy, S.M.; Iqbal, Z.; Awad, H.M.; Sattar, M.N.; Tohamy, A.M.; Abbas, A.O.; Squires, J.; Cooke, D.E.L. Genotypic and Phenotypic Structure of the Population of Phytophthora infestans in Egypt Revealed the Presence of European Genotypes. J. Fungi 2022, 8, 468. https://doi.org/10.3390/jof8050468
El-Ganainy SM, Iqbal Z, Awad HM, Sattar MN, Tohamy AM, Abbas AO, Squires J, Cooke DEL. Genotypic and Phenotypic Structure of the Population of Phytophthora infestans in Egypt Revealed the Presence of European Genotypes. Journal of Fungi. 2022; 8(5):468. https://doi.org/10.3390/jof8050468
Chicago/Turabian StyleEl-Ganainy, Sherif Mohamed, Zafar Iqbal, Hossam Mohamed Awad, Muhammad Naeem Sattar, Abdel Mohsen Tohamy, Ahmed O. Abbas, Julie Squires, and David E. L. Cooke. 2022. "Genotypic and Phenotypic Structure of the Population of Phytophthora infestans in Egypt Revealed the Presence of European Genotypes" Journal of Fungi 8, no. 5: 468. https://doi.org/10.3390/jof8050468
APA StyleEl-Ganainy, S. M., Iqbal, Z., Awad, H. M., Sattar, M. N., Tohamy, A. M., Abbas, A. O., Squires, J., & Cooke, D. E. L. (2022). Genotypic and Phenotypic Structure of the Population of Phytophthora infestans in Egypt Revealed the Presence of European Genotypes. Journal of Fungi, 8(5), 468. https://doi.org/10.3390/jof8050468







