Genetic Variants Associated with Suspected Neonatal Hypoxic Ischaemic Encephalopathy: A Study in a South African Context
Abstract
:1. Introduction
2. Results
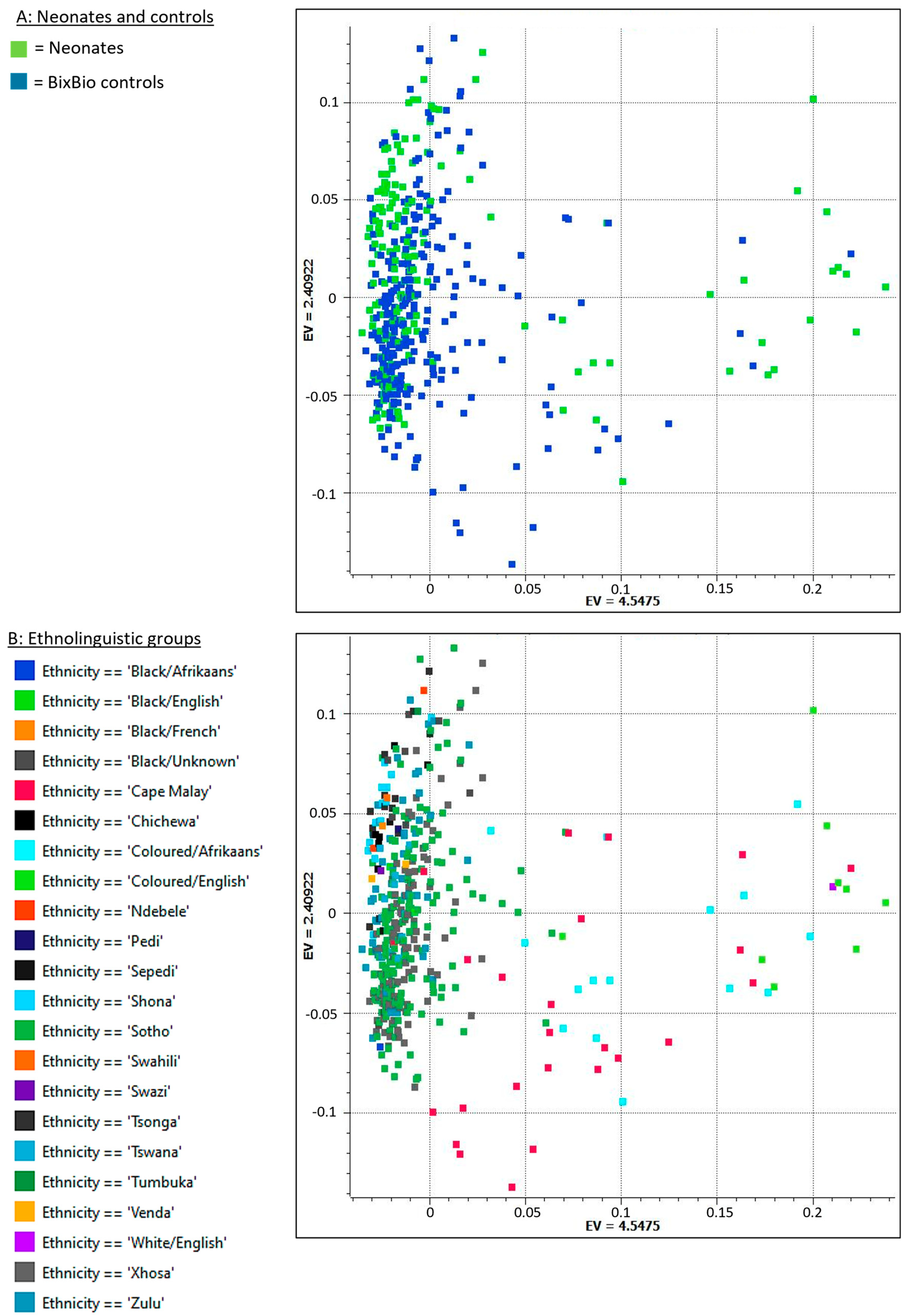
2.1. Data Set and Quality Control
2.2. Variant Data Set
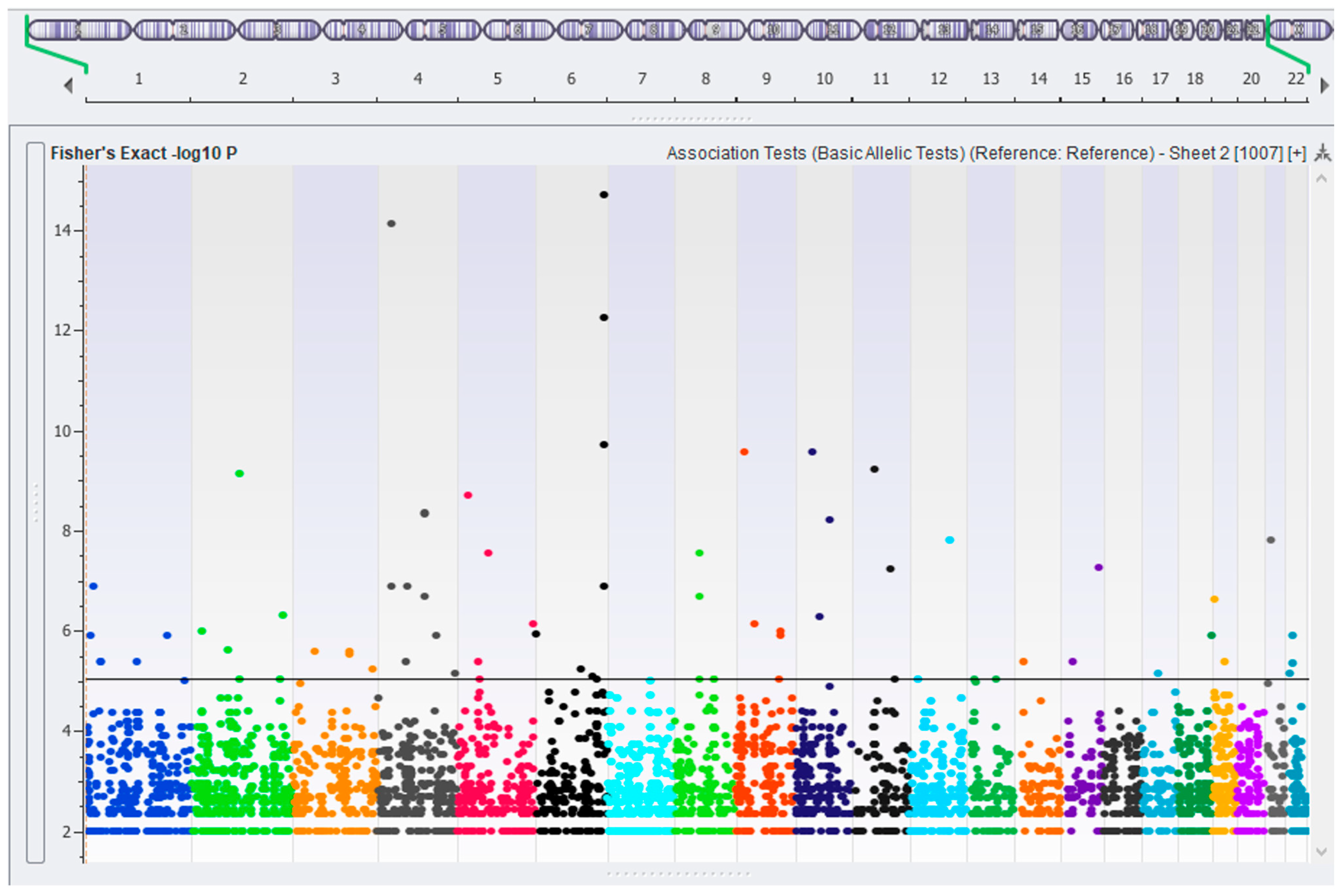
2.3. Association Testing—Case vs. Control
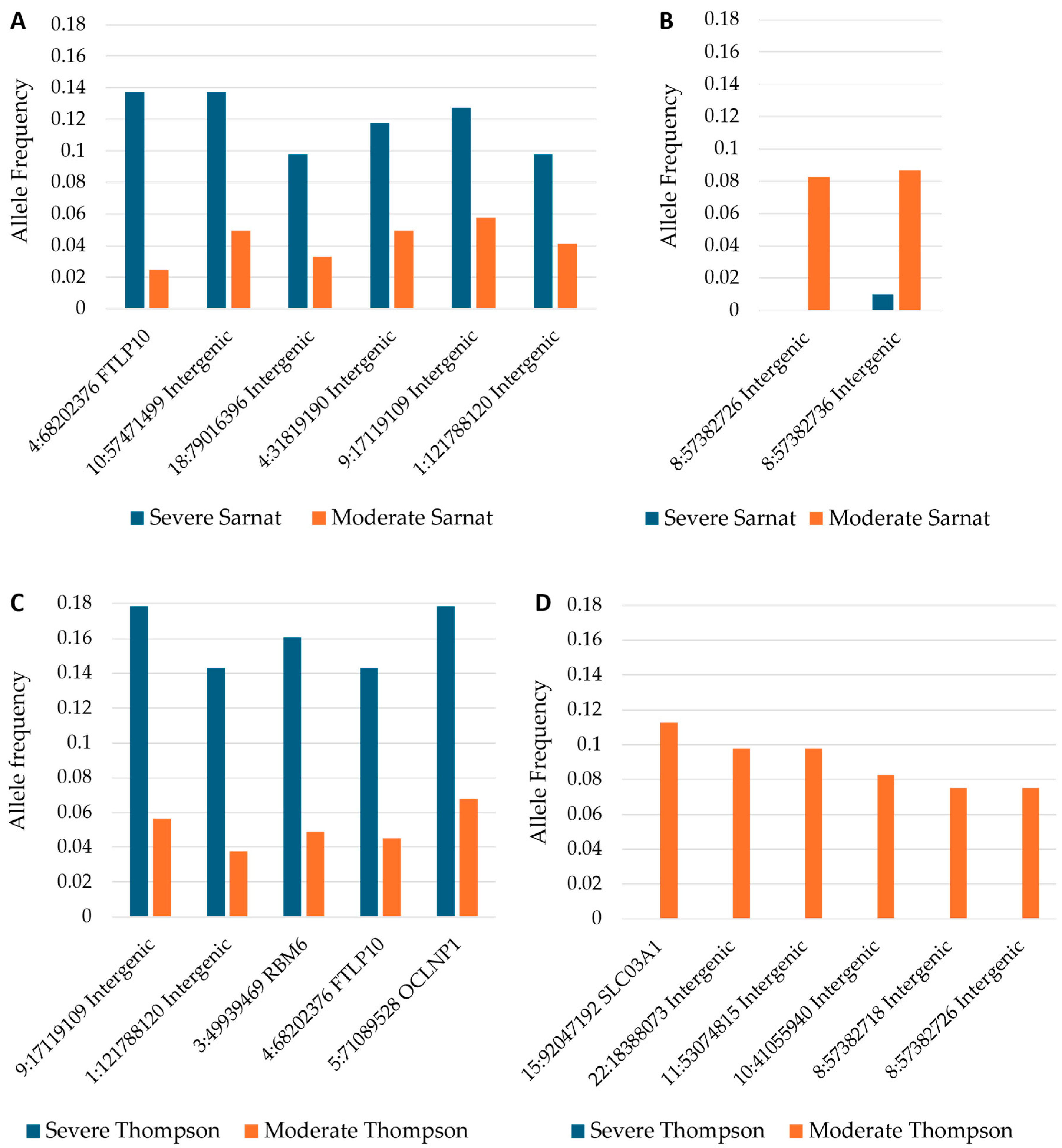
2.4. Allele Frequencies of Variants in Severity and Progression Groups
3. Discussion
Limitations and Future Work
4. Materials and Methods
4.1. Patient Recruitment
4.2. Inclusion and Exclusion Criteria
4.3. Clinical Treatment and Monitoring
4.4. Ancestry-Matched Controls
4.5. Blood Collection and DNA Isolation
4.6. DNA Sequencing and Variant Calling
4.7. Data Quality Control
4.8. Relatedness
4.9. Population Stratification
4.10. Variant Filtering and Prioritization
4.11. Variant Association Testing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- McIntyre, S.; Nelson, K.B.; Mulkey, S.B.; Lechpammer, M.; Molloy, E.; Badawi, N.; Newborn Brain Society, G.; Publications, C. Neonatal encephalopathy: Focus on epidemiology and underexplored aspects of etiology. Semin. Fetal Neonatal Med. 2021, 26, 101265. [Google Scholar] [CrossRef] [PubMed]
- Aslam, S.; Strickland, T.; Molloy, E.J. Neonatal encephalopathy: Need for recognition of multiple etiologies for optimal management. Front. Pediatr. 2019, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Nelson, K.B. Is it HIE? And why that matters. Acta Paediatr. 2007, 96, 1113–1114. [Google Scholar] [CrossRef] [PubMed]
- Douglas-Escobar, M.; Weiss, M.D. Hypoxic-ischemic encephalopathy: A review for the clinician. JAMA Pediatr. 2015, 169, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Molloy, E.J.; Bearer, C. Neonatal encephalopathy versus hypoxic-ischemic encephalopathy. Pediatr. Res. 2018, 84, 574. [Google Scholar] [CrossRef]
- Thompson, C.M.; Puterman, A.S.; Linley, L.L.; Hann, F.M.; van der Elst, C.W.; Molteno, C.D.; Malan, A.F. The value of a scoring system for hypoxic ischaemic encephalopathy in predicting neurodevelopmental outcome. Acta Paediatr. 1997, 86, 757–761. [Google Scholar] [CrossRef]
- Sarnat, H.B.; Sarnat, M.S. Neonatal encephalopathy following fetal distress: A clinical and electroencephalographic study. Arch. Neurol. 1976, 33, 696. [Google Scholar] [CrossRef]
- Bruckmann, E.K.; Velaphi, S. Intrapartum asphyxia and hypoxic ischaemic encephalopathy in a public hospital: Incidence and predictors of poor outcome. SAMJ S. Afr. Med. J. 2015, 105, 298–303. [Google Scholar] [CrossRef]
- Kukka, A.J.; Waheddoost, S.; Brown, N.; Litorp, H.; Wrammert, J.; KC, A. Incidence and outcomes of intrapartum-related neonatal encephalopathy in low-income and middle-income countries: A systematic review and meta-analysis. BMJ Global Health 2022, 7, e010294. [Google Scholar] [CrossRef]
- Greco, P.; Nencini, G.; Piva, I.; Scioscia, M.; Volta, C.A.; Spadaro, S.; Neri, M.; Bonaccorsi, G.; Greco, F.; Cocco, I.; et al. Pathophysiology of hypoxic–ischemic encephalopathy: A review of the past and a view on the future. Acta Neurol. Belg. 2020, 120, 277–288. [Google Scholar] [CrossRef]
- Horn, A.R.; Swingler, G.H.; Myer, L.; Harrison, M.C.; Linley, L.L.; Nelson, C.; Tooke, L.; Rhoda, N.R.; Robertson, N.J. Defining hypoxic ischemic encephalopathy in newborn infants: Benchmarking in a South African population. J. Perinat. Med. 2013, 41, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Chopra, M.; Gable, D.L.; Love-Nichols, J.; Tsao, A.; Rockowitz, S.; Sliz, P.; Barkoudah, E.; Bastianelli, L.; Coulter, D.; Davidson, E.; et al. Mendelian etiologies identified with whole exome sequencing in cerebral palsy. Ann. Clin. Transl. Neurol. 2022, 9, 193–205. [Google Scholar] [CrossRef] [PubMed]
- McMichael, G.; Bainbridge, M.N.; Haan, E.; Corbett, M.; Gardner, A.; Thompson, S.; van Bon, B.W.M.; van Eyk, C.L.; Broadbent, J.; Reynolds, C.; et al. Whole-exome sequencing points to considerable genetic heterogeneity of cerebral palsy. Mol. Psychiatry 2015, 20, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Takezawa, Y.; Kikuchi, A.; Haginoya, K.; Niihori, T.; Numata-Uematsu, Y.; Inui, T.; Yamamura-Suzuki, S.; Miyabayashi, T.; Anzai, M.; Suzuki-Muromoto, S.; et al. Genomic analysis identifies masqueraders of full-term cerebral palsy. Ann. Clin. Transl. Neurol. 2018, 5, 538–551. [Google Scholar] [CrossRef]
- Moreno-De-Luca, A.; Millan, F.; Pesacreta, D.R.; Elloumi, H.Z.; Oetjens, M.T.; Teigen, C.; Wain, K.E.; Scuffins, J.; Myers, S.M.; Torene, R.I.; et al. Molecular diagnostic yield of exome sequencing in patients with cerebral palsy. JAMA 2021, 325, 467–475. [Google Scholar] [CrossRef]
- May, H.J.; Lippa, N.; Fasheun, J.A.; Bain, J.M.; Baugh, E.H.; Bier, L.E.; Revah-Politi, A.; Roye, D.P., Jr.; Goldstein, D.B.; Carmel, J.B.; et al. Genetic testing in individuals with cerebral palsy. Dev. Med. Child. Neurol. 2021, 63, 1448–1455. [Google Scholar] [CrossRef]
- Yechieli, M.; Gulsuner, S.; Ben-Pazi, H.; Fattal, A.; Aran, A.; Kuzminsky, A.; Sagi, L.; Guttman, D.; Schneebaum Sender, N.; Gross-Tsur, V.; et al. Diagnostic yield of chromosomal microarray and trio whole exome sequencing in cryptogenic cerebral palsy. J. Med. Genet. 2022, 59, 759–767. [Google Scholar] [CrossRef]
- Jackson, M.; Marks, L.; May, G.H.W.; Wilson, J.B. The genetic basis of disease. Essays Biochem. 2020, 62, 643–723. [Google Scholar] [CrossRef]
- Ciesielski, T.H.; Sirugo, G.; Iyengar, S.K.; Williams, S.M. Characterizing the pathogenicity of genetic variants: The consequences of context. NPJ Genom. Med. 2024, 9, 3. [Google Scholar] [CrossRef]
- Cano-Gamez, E.; Trynka, G. From GWAS to function: Using functional genomics to identify the mechanisms underlying complex diseases. Front. Genet. 2020, 11, 424. [Google Scholar] [CrossRef]
- Visscher, P.M.; Yengo, L.; Cox, N.J.; Wray, N.R. Discovery and implications of polygenicity of common diseases. Science 2021, 373, 1468–1473. [Google Scholar] [CrossRef] [PubMed]
- Boyle, E.A.; Li, Y.I.; Pritchard, J.K. An expanded view of complex traits: From polygenic to omnigenic. Cell 2017, 169, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.S.; Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 2015, 372, 793–795. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z. Practicing precision medicine with intelligently integrative clinical and multi-omics data analysis. Hum. Genom. 2020, 14, 35. [Google Scholar] [CrossRef] [PubMed]
- Zeggini, E.; Gloyn, A.L.; Barton, A.C.; Wain, L.V. Translational genomics and precision medicine: Moving from the lab to the clinic. Science 2019, 365, 1409–1413. [Google Scholar] [CrossRef]
- van Eyk, C.; MacLennan, S.C.; MacLennan, A.H. All patients with a cerebral palsy diagnosis merit genomic sequencing. JAMA Pediatr. 2023, 177, 455–456. [Google Scholar] [CrossRef]
- Sobotka, S.A.; Ross, L.F. Newborn screening for neurodevelopmental disorders may exacerbate health disparities. Pediatrics 2023, 152, e2023061727. [Google Scholar] [CrossRef]
- Harris, H.K.; Sideridis, G.D.; Barbaresi, W.J.; Harstad, E. Pathogenic yield of genetic testing in autism spectrum disorder. Pediatrics 2020, 146, e20193211. [Google Scholar] [CrossRef]
- Trevino, V.; Falciani, F.; Barrera-Saldaña, H.A. DNA microarrays: A powerful genomic tool for biomedical and clinical research. Mol. Med. 2007, 13, 527–541. [Google Scholar] [CrossRef]
- Marian, A.J. Sequencing your genome: What does it mean? Methodist. Debakey Cardiovasc. J. 2014, 10, 3–6. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, H.; Kugathasan, S.; Annese, V.; Bradfield, J.P.; Russell, R.K.; Sleiman, P.M.; Imielinski, M.; Glessner, J.; Hou, C.; et al. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn disease. Am. J. Hum. Genet. 2009, 84, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Potrony, M.; Puig-Butille, J.A.; Farnham, J.M.; Giménez-Xavier, P.; Badenas, C.; Tell-Martí, G.; Aguilera, P.; Carrera, C.; Malvehy, J.; Teerlink, C.C.; et al. Genome-wide linkage analysis in Spanish melanoma-prone families identifies a new familial melanoma susceptibility locus at 11q. Eur. J. Hum. Genet. 2018, 26, 1188–1193. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; De Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Primers 2021, 1, 59. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- MacArthur, D.G.; Manolio, T.A.; Dimmock, D.P.; Rehm, H.L.; Shendure, J.; Abecasis, G.R.; Adams, D.R.; Altman, R.B.; Antonarakis, S.E.; Ashley, E.A.; et al. Guidelines for investigating causality of sequence variants in human disease. Nature 2014, 508, 469–476. [Google Scholar] [CrossRef]
- Choudhury, A.; Matovu, E.; Aron, S.; Botigué, L.R.; Sengupta, D.; Botha, G.; Bensellak, T.; Wells, G.; Kumuthini, J.; Shriner, D.; et al. High-depth African genomes inform human migration and health. Nature 2020, 586, 741–748. [Google Scholar] [CrossRef]
- Lander, E.; Kruglyak, L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat. Genet. 1995, 11, 241–247. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Holborn, M.A.; Ford, G.; Turner, S.; Mellet, J.; van Rensburg, J.; Joubert, F.; Pepper, M.S. The NESHIE and CP Genetics Resource (NCGR): A database of genes and variants reported in neonatal encephalopathy with suspected hypoxic ischemic encephalopathy (NESHIE) and consequential cerebral palsy (CP). Genomics 2022, 114, 110508. [Google Scholar] [CrossRef] [PubMed]
- Bartonicek, N.; Clark, M.B.; Quek, X.C.; Torpy, J.R.; Pritchard, A.L.; Maag, J.L.V.; Gloss, B.S.; Crawford, J.; Taft, R.J.; Hayward, N.K.; et al. Intergenic disease-associated regions are abundant in novel transcripts. Genome Biol. 2017, 18, 241. [Google Scholar] [CrossRef] [PubMed]
- Short, P.J.; McRae, J.F.; Gallone, G.; Sifrim, A.; Won, H.; Geschwind, D.H.; Wright, C.F.; Firth, H.V.; FitzPatrick, D.R.; Barrett, J.C.; et al. De novo mutations in regulatory elements in neurodevelopmental disorders. Nature 2018, 555, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Kleinjan, D.A.; van Heyningen, V. Long-range control of gene expression: Emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 2005, 76, 8–32. [Google Scholar] [CrossRef]
- Barrett, L.W.; Fletcher, S.; Wilton, S.D. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol. Life Sci. 2012, 69, 3613–3634. [Google Scholar] [CrossRef]
- Gordon, C.T.; Lyonnet, S. Enhancer mutations and phenotype modularity. Nat. Genet. 2014, 46, 3–4. [Google Scholar] [CrossRef]
- Mirabella, A.C.; Foster, B.M.; Bartke, T. Chromatin deregulation in disease. Chromosoma 2016, 125, 75–93. [Google Scholar] [CrossRef]
- Zou, H.; Wu, L.X.; Tan, L.; Shang, F.F.; Zhou, H.H. Significance of single-nucleotide variants in long intergenic non-protein coding RNAs. Front. Cell Dev. Biol. 2020, 8, 347. [Google Scholar] [CrossRef]
- Wei, S.C.; Tan, Y.Y.; Weng, M.T.; Lai, L.C.; Hsiao, J.H.; Chuang, E.Y.; Shun, C.T.; Wu, D.C.; Kao, A.W.; Chuang, C.S.; et al. SLCO3A1, A novel Crohn’s disease-associated gene, regulates nf-κB activity and associates with intestinal perforation. PLoS ONE 2014, 9, e100515. [Google Scholar] [CrossRef]
- Haliburton, G.D.; McKinsey, G.L.; Pollard, K.S. Disruptions in a cluster of computationally identified enhancers near FOXC1 and GMDS may influence brain development. Neurogenetics 2016, 17, 1–9. [Google Scholar] [CrossRef]
- Chatterjee, M.; Schild, D.; Teunissen, C.E. Contactins in the central nervous system: Role in health and disease. Neural Regen. Res. 2019, 14, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Micol, J.-B.; Pastore, A.; Inoue, D.; Duployez, N.; Kim, E.; Lee, S.C.-W.; Durham, B.H.; Chung, Y.R.; Cho, H.; Zhang, X.J.; et al. ASXL2 is essential for haematopoiesis and acts as a haploinsufficient tumour suppressor in leukemia. Nat. Commun. 2017, 8, 15429. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.H.; Raftrey, B.C.; D’Amato, G.; Surya, V.N.; Poduri, A.; Chen, H.I.; Goldstone, A.B.; Woo, J.; Fuller, G.G.; Dunn, A.R.; et al. DACH1 stimulates shear stress-guided endothelial cell migration and coronary artery growth through the CXCL12-CXCR4 signaling axis. Genes. Dev. 2017, 31, 1308–1324. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lyu, Y.; Tu, K.; Xu, Q.; Yang, Y.; Salman, S.; Le, N.; Lu, H.; Chen, C.; Zhu, Y.; et al. Histone citrullination by PADI4 is required for HIF-dependent transcriptional responses to hypoxia and tumor vascularization. Sci. Adv. 2021, 7, eabe3771. [Google Scholar] [CrossRef]
- Brannvoll, A.; Xue, X.; Kwon, Y.; Kompocholi, S.; Simonsen, A.K.W.; Viswalingam, K.S.; Gonzalez, L.; Hickson, I.D.; Oestergaard, V.H.; Mankouri, H.W.; et al. The ZGRF1 helicase promotes recombinational repair of replication-blocking DNA damage in human cells. Cell Rep. 2020, 32, 107849. [Google Scholar] [CrossRef]
- Schmidt, M.F.; Gan, Z.Y.; Komander, D.; Dewson, G. Ubiquitin signalling in neurodegeneration: Mechanisms and therapeutic opportunities. Cell Death Differ. 2021, 28, 570–590. [Google Scholar] [CrossRef]
- Benson, D.L.; Huntley, G.W. Are we listening to everything the PARK genes are telling us? J. Comp. Neurol. 2019, 527, 1527–1540. [Google Scholar] [CrossRef]
- Tarocco, A.; Morciano, G.; Perrone, M.; Cafolla, C.; Ferrè, C.; Vacca, T.; Pistocchi, G.; Meneghin, F.; Cocchi, I.; Lista, G.; et al. Increase of Parkin and ATG5 plasmatic levels following perinatal hypoxic-ischemic encephalopathy. Sci. Rep. 2022, 12, 7795. [Google Scholar] [CrossRef]
- Leith, W.M.; Zeegers, M.P.; Freeman, M.D. A predictive model for perinatal hypoxic ischemic encephalopathy using linked maternal and neonatal hospital data. Ann. Epidemiol. 2024, 89, 29–36. [Google Scholar] [CrossRef]
- Li, Y.; Gonzalez, P.; Zhang, L. Fetal stress and programming of hypoxic/ischemic-sensitive phenotype in the neonatal brain: Mechanisms and possible interventions. Prog. Neurobiol. 2012, 98, 145–165. [Google Scholar] [CrossRef]
- Machour, F.E.; Abu-Zhayia, E.R.; Awwad, S.W.; Bidany-Mizrahi, T.; Meinke, S.; Bishara, L.A.; Heyd, F.; Aqeilan, R.I.; Ayoub, N. RBM6 splicing factor promotes homologous recombination repair of double-strand breaks and modulates sensitivity to chemotherapeutic drugs. Nucleic Acids Res. 2021, 49, 11708–11727. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Chen, Z.; Yu, J. Pseudogenes and their potential functions in hematopoiesis. Exp. Hematol. 2021, 103, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Aguet, F.; Anand, S.; Ardlie, K.G.; Gabriel, S.; Getz, G.A.; Graubert, A.; Hadley, K.; Handsaker, R.E.; Huang, K.H.; Kashin, S.; et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Lin, Y.-H.; Yeh, C.-T.; Chen, C.-Y.; Lin, K.-H. Pseudogene: Relevant or Irrelevant? Biomed. J. 2024, 100790. [Google Scholar] [CrossRef]
- Krishnan, V.; Kumar, V.; Shankaran, S.; Thayyil, S. Rise and Fall of Therapeutic Hypothermia in Low-Resource Settings: Lessons from the HELIX Trial. Indian J. Pediatr. 2021. [Google Scholar] [CrossRef]
- Thayyil, S.; Pant, S.; Montaldo, P.; Shukla, D.; Oliveira, V.; Ivain, P.; Bassett, P.; Swamy, R.; Mendoza, J.; Moreno-Morales, M.; et al. Hypothermia for moderate or severe neonatal encephalopathy in low-income and middle-income countries (HELIX): A randomised controlled trial in India, Sri Lanka, and Bangladesh. Lancet Glob. Health 2021, 9, e1273–e1285. [Google Scholar] [CrossRef]
- Janssen, L.; Dupont, L.; Bekhouche, M.; Noel, A.; Leduc, C.; Voz, M.; Peers, B.; Cataldo, D.; Apte, S.S.; Dubail, J.; et al. ADAMTS3 activity is mandatory for embryonic lymphangiogenesis and regulates placental angiogenesis. Angiogenesis 2016, 19, 53–65. [Google Scholar] [CrossRef]
- Altuntaş, C.; Alper, M.; Keleş, Y.; Sav, F.N.; Köçkar, F. Hypoxic regulation of ADAMTS-2 and -3 (a disintegrin and matrix metalloproteinase with thrombospondin motifs 2 and 3) procollagen N proteinases by HIF-1α in endothelial cells. Mol. Cell Biochem. 2023, 478, 1151–1160. [Google Scholar] [CrossRef]
- Horn, A.R.; Swingler, G.H.; Myer, L.; Linley, L.L.; Raban, M.S.; Joolay, Y.; Harrison, M.C.; Chandrasekaran, M.; Rhoda, N.R.; Robertson, N.J. Early clinical signs in neonates with hypoxic ischemic encephalopathy predict an abnormal amplitude-integrated electroencephalogram at age 6 hours. BMC Pediatr. 2013, 13, 52. [Google Scholar] [CrossRef]
- Sherman, R.M.; Forman, J.; Antonescu, V.; Puiu, D.; Daya, M.; Rafaels, N.; Boorgula, M.P.; Chavan, S.; Vergara, C.; Ortega, V.E.; et al. Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat. Genet. 2019, 51, 30–35. [Google Scholar] [CrossRef]
- Mitchell, B.D.; Fornage, M.; McArdle, P.F.; Cheng, Y.-C.; Pulit, S.L.; Wong, Q.; Dave, T.; Williams, S.R.; Corriveau, R.; Gwinn, K.; et al. Using previously genotyped controls in genome-wide association studies (GWAS): Application to the Stroke Genetics Network (SiGN). Front. Genet. 2014, 5, 95. [Google Scholar] [CrossRef]
- Mukherjee, S.; Simon, J.; Bayuga, S.; Ludwig, E.; Yoo, S.; Orlow, I.; Viale, A.; Offit, K.; Kurtz, R.C.; Olson, S.H.; et al. Including additional controls from public databases improves the power of a genome-wide association study. Hum. Hered. 2011, 72, 21–34. [Google Scholar] [CrossRef]
- Little, J.; Higgins, J.P.; Ioannidis, J.P.; Moher, D.; Gagnon, F.; von Elm, E.; Khoury, M.J.; Cohen, B.; Davey-Smith, G.; Grimshaw, J.; et al. STrengthening the REporting of Genetic Association Studies (STREGA)-an extension of the STROBE statement. Genet. Epidemiol. 2009, 33, 581–598. [Google Scholar] [CrossRef]
- Bozeman, M.; Golden Helix, Inc. SNP & Variation Suite™ (Version 8.9.1) [Software]. Available online: http://www.goldenhelix.com (accessed on 1 September 2023).
- Halko, N.; Martinsson, P.-G.; Shkolnisky, Y.; Tygert, M. An algorithm for the principal component analysis of large data sets. SIAM J. Sci. Comput. 2011, 33, 2580–2594. [Google Scholar] [CrossRef]
- Jalali Sefid Dashti, M.; Gamieldien, J. A practical guide to filtering and prioritizing genetic variants. BioTechniques 2017, 62, 18–30. [Google Scholar] [CrossRef]
- Chen, S.; Francioli, L.C.; Goodrich, J.K.; Collins, R.L.; Kanai, M.; Wang, Q.; Alföldi, J.; Watts, N.A.; Vittal, C.; Gauthier, L.D.; et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature 2024, 625, 92–100. [Google Scholar] [CrossRef]
- Wells, A.; Heckerman, D.; Torkamani, A.; Yin, L.; Sebat, J.; Ren, B.; Telenti, A.; di Iulio, J. Ranking of non-coding pathogenic variants and putative essential regions of the human genome. Nat. Commun. 2019, 10, 5241. [Google Scholar] [CrossRef]

| Severity Categories of Neonates (N = 172) | Number (%) Normal | Number (%) Mild | Number (%) Moderate | Number (%) Severe | Number (%) Not Measured a |
|---|---|---|---|---|---|
| Sarnat baseline | 0 (0) | 0 (0) | 121 (70) | 51 (30) | 0 (0) |
| Thompson baseline | 0 (0) | 11 (6) | 133 (77) | 28 (16) | 0 (0) |
| Thompson day 4/5 b | 7 (4) | 81 (47) | 71 (41) | 10 (6) | 3 (2) |
| Progression Categories of Neonates (N = 172) | Number (%) Improved c | Number (%) Not Improved | Number (%) Not Measured a | ||
| Thompson progression baseline to day 4/5 b | 104 (60) | 65 (38) | 3 (2) | ||
| Number of Variants in the Gene | Gene Symbol | Gene Name | Known or Suspected Gene/Gene Product Function a |
|---|---|---|---|
| 1 | ADAMTS3 | ADAM metallopeptidase with thrombospondin type 1 motif 3 | Protease, a role in the processing of type II fibrillar collagen in articular cartilage |
| 1 | ANAPC1P4 | ANAPC1 pseudogene 4 | Pseudogene |
| 1 | ASXL2 | ASXL transcriptional regulator 2 | Epigenetic regulator, binds histone-modifying enzymes and involved in the assembly of transcription factors |
| 1 | CDC73 | Cell division cycle 73 | RNA polymerase II core binding |
| 1 | CNTN5 | Contactin 5 | Glycosylphosphatidylinositol (GPI)-anchored neuronal membrane protein that functions as a cell adhesion molecule |
| 2 | CPNE4 | Copine 4 | Calcium-dependent, phospholipid-binding protein, may be involved in membrane trafficking, mitogenesis, and development |
| 1 | DACH1 | Dachshund family transcription factor 1 | Chromatin-associated protein, associates with other DNA-binding transcription factors to regulate gene expression and cell fate determination during development |
| 1 | FGD6 | FYVE, RhoGEF and PH domain containing 6 | Guanyl-nucleotide exchange factor activity, small GTPase binding |
| 1 | FTLP10 | Ferritin light chain pseudogene 10 | Pseudogene |
| 1 | GMDS | GDP-mannose 4,6-dehydratase | Conversion of GDP-mannose to GDP-4-keto-6-deoxymannose, using NADP+ as a cofactor |
| 1 | INVS | Inversin | Interacts with nephrocystin and infers a connection between primary cilia function and left–right axis determination |
| 1 | LINC02679 | Long intergenic non-protein coding RNA 2679 | lncRNA |
| 2 | LOC102724710 | Uncharacterized LOC102724710 | ncRNA |
| 1 | NAT16 | N-acetyltransferase 16 (putative) | Predicted to enable acyltransferase activity, transferring groups other than amino-acyl groups |
| 1 | OCLNP1 | Occludin pseudogene | Pseudogene |
| 1 | PADI4 | Peptidyl arginine deiminase 4 | Enzyme responsible for the conversion of arginine residues to citrulline residues |
| 1 | PDSS2 | Decaprenyl diphosphate synthase subunit 2 | Enzyme that synthesizes the prenyl side chain of coenzyme Q, one of the key elements in the respiratory chain |
| 4 | PRKN | Parkin RBR E3 ubiquitin protein ligase | A component of a multiprotein E3 ubiquitin ligase complex that mediates the targeting of substrate proteins for proteasomal degradation |
| 1 | RBM6 | Rap associating with DIL domain | Enables GTPase binding activity |
| 1 | SLCO3A1 | Solute carrier organic anion transporter family member 3A1 | Sodium-independent organic anion transmembrane transporter activity |
| 2 | THRAP3 | Thyroid hormone receptor-associated protein 3 | Enables phosphoprotein binding, thyroid hormone receptor binding, and transcription coactivator activity |
| 1 | TJP3 | Tight junction protein 3 | Role in linkage between the actin cytoskeleton and tight junctions, also sequesters cyclin D1 at tight junctions during mitosis |
| 1 | ULK4P3 | ULK4 pseudogene 3 | Pseudogene |
| 3 | ZGRF1 | Zinc finger GRF-type containing 1 | GRF zinc fingers are found in a number of DNA-binding proteins |
| Variant | Gene Region (Gene If Applicable) | Allele Frequencies for Severity, Severe (n = 51) vs. Moderate (n = 121) Sarnat at Baseline, p < 0.05 | Allele Frequencies for Severity, Severe (n = 28) vs. Moderate (133) Thompson at Baseline, p < 0.05. Missing n = 11 a | Allele Frequencies for Severity, Severe or Moderate (n = 81) vs. Mild or Normal (n = 88) Thompson Day 4 or 5, p < 0.05. Missing n = 2 b,c | Did Not Improve (n = 65) vs. Improved (n = 104) Thompson from Baseline to Worst Grade Day 4 or 5. Missing n = 3 b | Associated with More Severe and/or Not Improving NESHIE | Associated with Milder and/or Improving NESHIE | Associated with More Severe NESHIE at Baseline but Improvement over Time |
|---|---|---|---|---|---|---|---|---|
| NC_000001.11:g.121788120C>T | Intergenic | 0.098 vs. 0.041 (p = 0.047) | 0.143 vs. 0.038 (p = 0.006) | x | ||||
| NC_000002.12:g.114110393C>T | Intergenic | 0.086 vs. 0.028 (p = 0.031) | 0.092 vs. 0.034 (p = 0.029) | x | ||||
| NC_000004.11:g.31819190G>A | Intergenic | 0.118 vs. 0.050 (p = 0.035) | x | |||||
| NC_000004.12:g.68202376A>C | Intron (FTLP10) | 0.137 vs. 0.025 (p = 1.44 × 10−4) | 0.143 vs. 0.045 (p = 0.012) | x | ||||
| NC_000004.12:g.72456033C>T | Intron (ADAMTS3) | 0.099 vs. 0.034 (p = 0.025) | 0.108 vs. 0.039 (p = 0.021) | x | ||||
| NC_000005.10:g.71089528A>G | Intron (OCLNP1) | 0.179 vs. 0.068 (p = 0.016) | x | |||||
| NC_000010.11:g.57471499C>T | Intergenic | 0.137 vs. 0.050 (p = 0.007) | x | |||||
| NC_000010.11:g.79642025T>A | Intron (LINC02679) | 0.148 vs. 0.057 (p = 0.006) | x | |||||
| NC_000018.10:g.79016396C>T | Downstream intergenic | 0.098 vs. 0.033 (p = 0.030) | x | |||||
| NC_000019.10:g.30075028_30075029del | Intergenic | 0.108 vs. 0.029 (p = 0.004) | x | |||||
| NC_000002.12:g.87718514G>A | Intron (ANAPC1P4) | 0.025 vs. 0.085 (p = 0.018) | 0.015 vs. 0.082 (p = 0.013) | x | ||||
| NC_000006.12:g.162011042_162011043del | Intron (PRKN) | 0.031 vs. 0.096 (p = 0.028) | x | |||||
| NC_000008.11:g.57382718G>A | Intergenic | 0 vs. 0.075 (p = 0.031) | 0.025 vs. 0.085 (p = 0.018) | x | ||||
| NC_000008.11:g.57382726del | Intergenic | 0 vs. 0.083 (p = 0.002) | 0 vs. 0.075 (p = 0.031) | 0.025 vs. 0.085 (p = 0.018) | x | |||
| NC_000008.11:g.57382736T>A | Intergenic | 0.010 vs. 0.087 (p = 0.007) | x | |||||
| NC_000010.11:g.41055940C>T | Intergenic | 0 vs. 0.083 (p = 0.019) | 0.025 vs. 0.102 (p = 0.004) | 0.015 vs. 0.096 (p = 0.003) | x | |||
| NC_000011.10:g.53074815C>T | Intergenic | 0 vs. 0.098 (p = 0.012) | x | |||||
| NC_000012.12:g.95148880_95148887del | Intron (FGD6) | 0.015 vs. 0.077 (p = 0.013) | x | |||||
| NC_000013.11:g.71735277T>C | Intron (DACH1) | 0.025 vs. 0.085 (p = 0.018) | x | |||||
| NC_000015.10:g.92047192C>T | Intron (SLCO3A1) | 0 vs. 0.113 (p = 0.004) | 0.062 vs. 0.136 (p = 0.029) | 0.046 vs. 0.135 (p = 0.009) | x | |||
| NC_000022.11:g.18388073G>A | Upstream intergenic | 0 vs. 0.098 (p = 0.012) | x | |||||
| NC_000022.11:g.18732630G>A | Intergenic | 0.025 vs. 0.080 (p = 0.029) | 0.015 vs. 0.077 (p = 0.013) | x | ||||
| NC_000003.12:g.49939469C>T | Intron (RBM6) | 0.161 vs. 0.049 (p = 0.006) | 0.023 vs. 0.091 (p = 0.013) | x | ||||
| NC_000009.12:g.17119108_17119116del | Intergenic | 0.128 vs. 0.058 (p = 0.046) | 0.179 vs. 0.056 (p = 0.005) | 0.015 vs. 0.120 (p = 3 × 10−4) | x |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Foden, C.J.; Durant, K.; Mellet, J.; Joubert, F.; van Rensburg, J.; Masemola, K.; Velaphi, S.C.; Nakwa, F.L.; Horn, A.R.; Pillay, S.; et al. Genetic Variants Associated with Suspected Neonatal Hypoxic Ischaemic Encephalopathy: A Study in a South African Context. Int. J. Mol. Sci. 2025, 26, 2075. https://doi.org/10.3390/ijms26052075
Foden CJ, Durant K, Mellet J, Joubert F, van Rensburg J, Masemola K, Velaphi SC, Nakwa FL, Horn AR, Pillay S, et al. Genetic Variants Associated with Suspected Neonatal Hypoxic Ischaemic Encephalopathy: A Study in a South African Context. International Journal of Molecular Sciences. 2025; 26(5):2075. https://doi.org/10.3390/ijms26052075
Chicago/Turabian StyleFoden, Caroline J., Kevin Durant, Juanita Mellet, Fourie Joubert, Jeanne van Rensburg, Khomotso Masemola, Sithembiso C. Velaphi, Firdose L. Nakwa, Alan R. Horn, Shakti Pillay, and et al. 2025. "Genetic Variants Associated with Suspected Neonatal Hypoxic Ischaemic Encephalopathy: A Study in a South African Context" International Journal of Molecular Sciences 26, no. 5: 2075. https://doi.org/10.3390/ijms26052075
APA StyleFoden, C. J., Durant, K., Mellet, J., Joubert, F., van Rensburg, J., Masemola, K., Velaphi, S. C., Nakwa, F. L., Horn, A. R., Pillay, S., Kali, G., Coetzee, M., Ballot, D. E., Kalua, T., Babbo, C., & Pepper, M. S., on behalf of the NESHIE Working Group. (2025). Genetic Variants Associated with Suspected Neonatal Hypoxic Ischaemic Encephalopathy: A Study in a South African Context. International Journal of Molecular Sciences, 26(5), 2075. https://doi.org/10.3390/ijms26052075







