The Sweetpotato Voltage-Gated K+ Channel β Subunit, KIbB1, Positively Regulates Low-K+ and High-Salinity Tolerance by Maintaining Ion Homeostasis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Sequence Analysis of KIbB1
2.3. Expression Analysis of KIbB1
2.4. Generation of KIbB1 Transgenic Plants
2.5. Low-K+ and High-Salinity Treatment Settings
2.6. Photosynthetic Characteristics Measurement
2.7. Expression Analysis of Stress-Responsive Gene Quantification
2.8. ROS Accumulation and Scavenging Assay
2.9. Na+ and K+ Content Measurement
2.10. Statistical Analysis
3. Results
3.1. Sequence Analysis of KIbB1
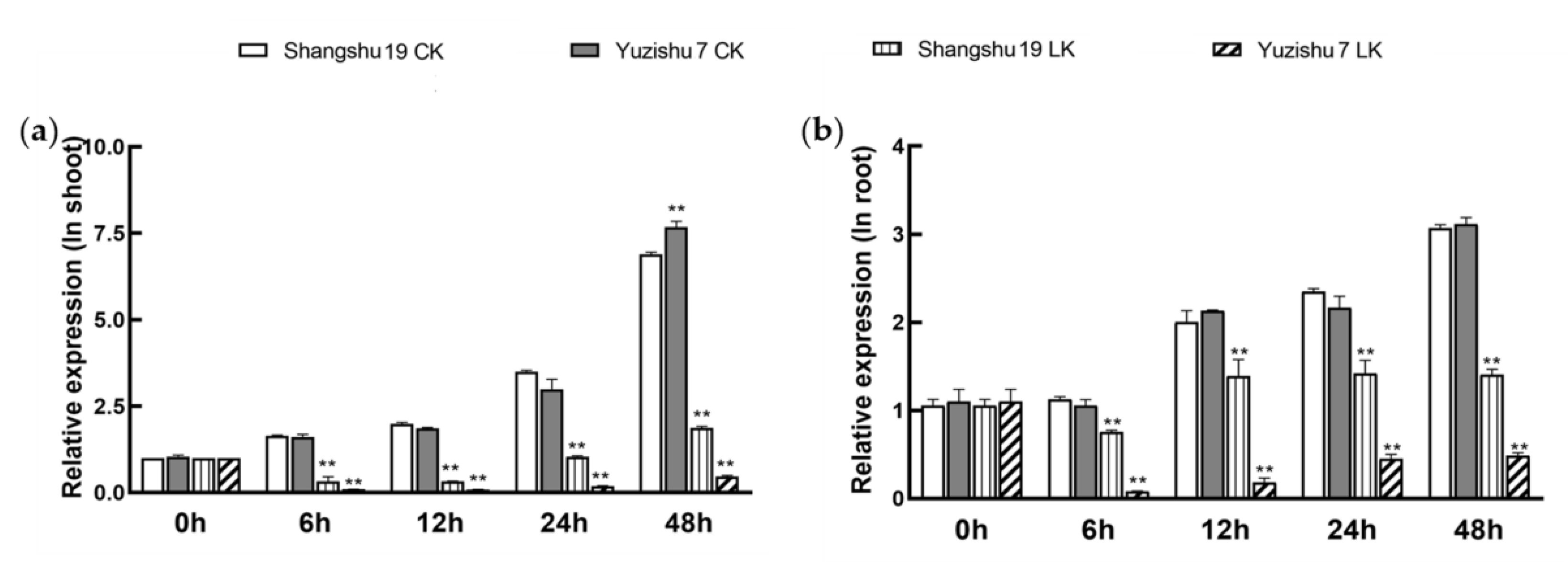
3.2. Expression Profiles of KIbB1
3.3. KIbB1 Positively Regulates Low-K+ Tolerance in Transgenic Plants
3.4. Overexpression of KIbB1 Enhances High-Salinity Resistance in Arabidopsis
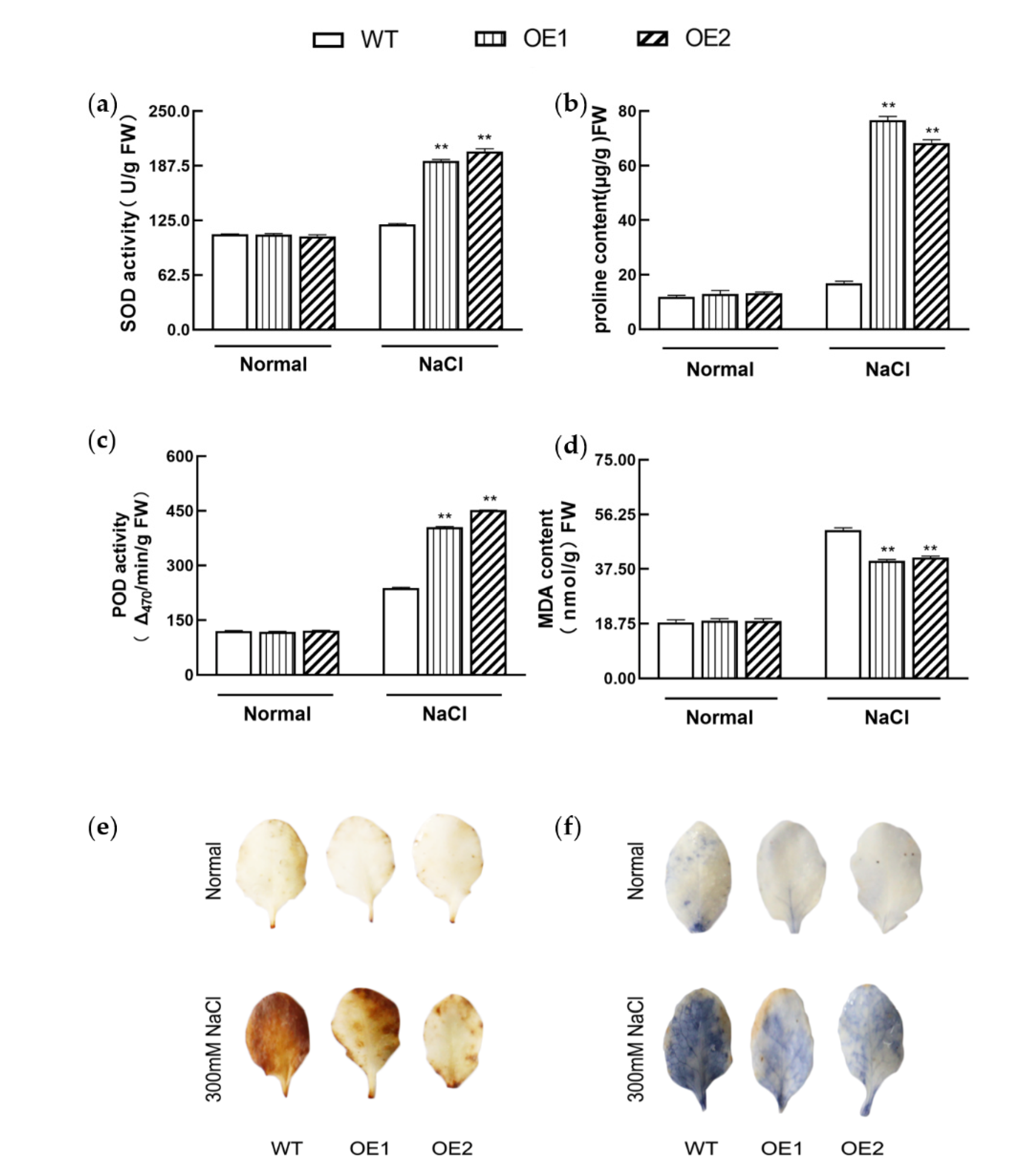
3.5. KIbB1 Promotes ROS Homeostasis under High-Salinity Conditions
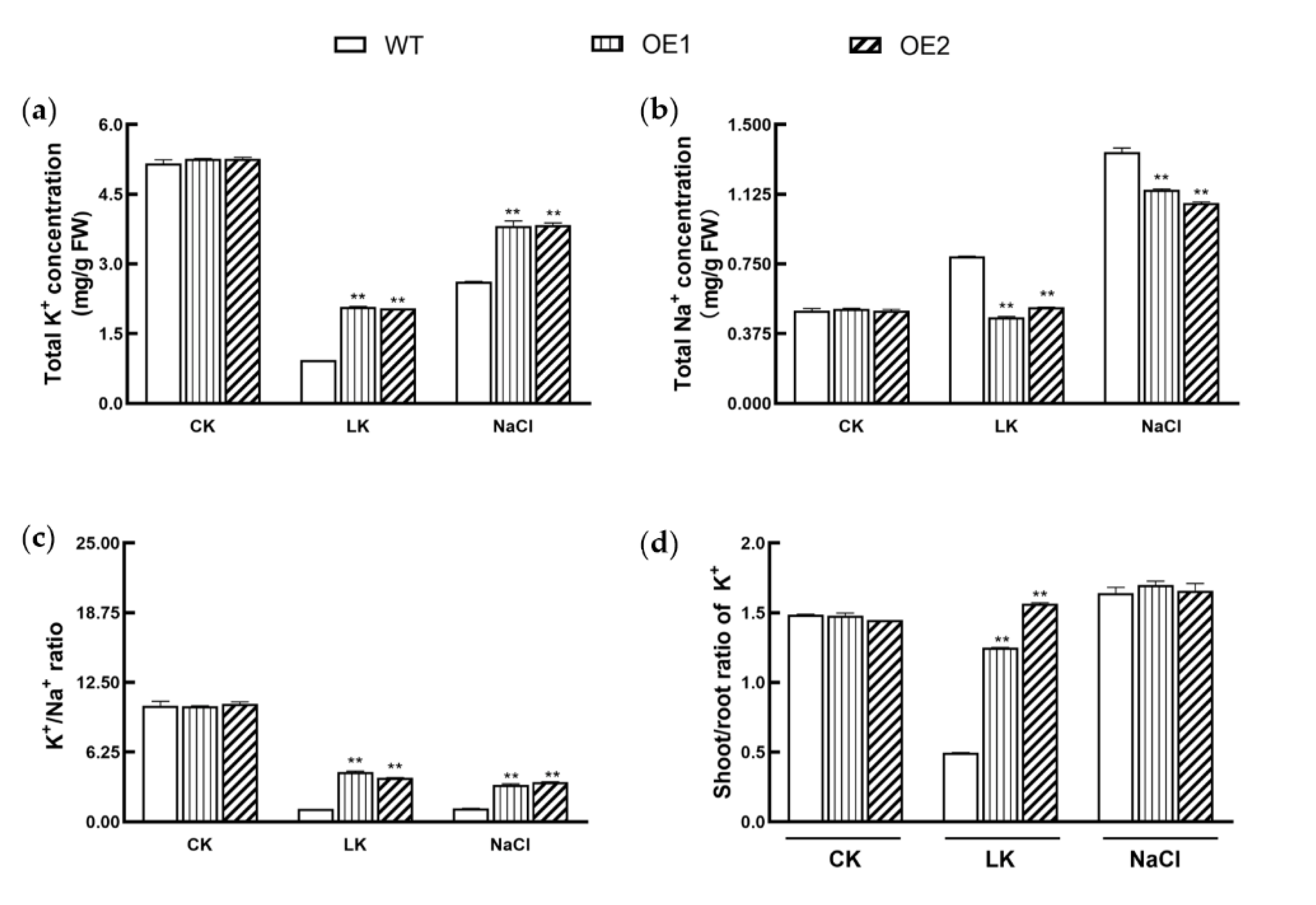
3.6. KIbB1 Enhances K+ Homeostasis under Low-K+ and High-Salinity Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gong, Z.Z.; Xiong, L.M.; Shi, H.Z.; Yang, S.H.; Herrera-Estrella, L.R.; Xu, G.H.; Chao, D.Y.; Li, J.R.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.Y.; Yang, T.Y.; Xing, C.H.; Dong, H.Z.; Qi, K.J.; Gao, J.Z.; Tao, S.T.; Wu, J.Y.; Wu, J.; Zhang, S.L.; et al. The β-amylase PbrBAM3 from pear (Pyrus betulaefolia) regulates soluble sugar accumulation and ROS homeostasis in response to cold stress. Plant Sci. 2019, 287, 110184. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chourasia, K.N.; More, S.J.; Kumar, A.; Kumar, D.; Singh, B.; Bhardwaj, V.; Kumar, A.; Das, S.K.; Singh, R.K.; Zinta, G.; et al. Salinity responses and tolerance mechanisms in underground vegetable crops: An integrative review. Planta 2022, 255, 68. [Google Scholar] [CrossRef] [PubMed]
- Chourasia, K.N.; Lal, M.K.; Tiwari, R.K.; Dev, D.; Kardile, H.B.; Patil, V.U.; Kumar, A.; Vanishree, G.; Kumar, D.; Bhardwaj, V.; et al. Salinity stress in potato: Understanding physiological, biochemical and molecular responses. Life 2021, 11, 545. [Google Scholar] [CrossRef] [PubMed]
- Calzone, A.; Cotrozzi, L.; Pellegrini, E.; Lorenzini, G.; Nali, C.; Maathuis, F. Can the transcriptional regulation of NHX1, SOS1 and HKT1 genes handle the response of two pomegranate cultivars to moderate salt stress? Salt-tolerance of two pomegranate cultivars. Sci. Hortic. 2021, 188, 110309. [Google Scholar] [CrossRef]
- Maathuis, F.J.M.; Ahmad, I.; Patishtan, J. Regulation of Na+ fluxes in plants. Front. Plant Sci. 2014, 5, 467. [Google Scholar] [CrossRef] [Green Version]
- Leigh, R.A.; Wyn Jones, R.G. A hypothesis relating critical potassium concentrations for growth to the distribution and function of this ion in the plant cell. New Phytol. 1984, 97, 1–13. [Google Scholar] [CrossRef]
- Li, W.H.; Xu, G.H.; Alli, A.; Yu, L. Plant HAK/KUP/KT K+ transporters: Function and regulation. Semin. Cell Dev. Biol. 2018, 74, 133–141. [Google Scholar] [CrossRef]
- Ashley, M.K.; Grant, M.; Grabov, A. Plant responses to potassium deficiencies: A role for potassium transport proteins. J. Exp. Bot. 2006, 57, 425–436. [Google Scholar] [CrossRef] [Green Version]
- Flowers, T.J.; Yeo, A.R. Ion relations of plants under drought and salinity. Aust. J. Plant Physiol. 1986, 13, 75–91. [Google Scholar] [CrossRef]
- Maathuis, F.J. Physiological functions of mineral macronutrients. Curr. Opin. Plant Biol. 2009, 12, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hong, Y.C.; Zhu, G.T.; Li, Y.M.; Niu, Q.F.; Yao, J.J.; Hua, K.; Bai, J.J.; Zhu, Y.F.; Shi, H.Z.; et al. Loss of salt tolerance during tomato domestication conferred by variation in a Na+/K+ transporter. EMBO J. 2020, 39, e103256. [Google Scholar] [CrossRef]
- Han, M.; Wu, W.; Wu, W.H.; Wang, Y. Potassium transporter KUP7 is involved in K+ acquisition and translocation in Arabidopsis root under K+-limited conditions. Mol. Plant 2016, 9, 437–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebaudy, A.; Véry, A.A.; Sentenac, H. K+ channel activity in plants: Genes, regulations and functions. FEBS Lett. 2007, 581, 2357–2366. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, W.H. Potassium transport and signaling in higher plants. Annu. Rev. Plant Biol. 2013, 64, 451–476. [Google Scholar] [CrossRef] [Green Version]
- Dreyer, I.; Uozumi, N. Potassium channels in plant cells. FEBS J. 2011, 278, 4293–4303. [Google Scholar] [CrossRef]
- Jegla, T.; Busey, G.; Assmann, S.M. Evolution and structural characteristics of plant voltage-gated K+ channels. Plant Cell 2018, 30, 2898–2909. [Google Scholar] [CrossRef] [Green Version]
- Musavizadeh, Z.; Najafi-Zarrini, H.; Kazemitabar, S.K.; Hashemi, S.H.; Faraji, S.; Barcaccia, G.; Heidari, P. Genome-Wide Analysis of Potassium Channel Genes in Rice: Expression of the OsAKT and OsKAT Genes under Salt Stress. Genes 2021, 12, 784. [Google Scholar] [CrossRef]
- Pongs, O.; Schwarz, J.R. Ancillary subunits associated with voltage-dependent K+ channels. Physiol. Rev. 2010, 90, 755–796. [Google Scholar] [CrossRef] [Green Version]
- Kamb, A.; Iverson, L.E.; Tanouye, M.A. Molecular characterization of Shaker, a Drosophila gene that encodes a potassium channel. Cell 1987, 50, 405–413. [Google Scholar] [CrossRef]
- Sharma, T.; Dreyer, I.; Riedelsberger, J. The role of K+ channels in uptake and redistribution of potassium in the model plant Arabidopsis thaliana. Front. Plant Sci. 2013, 4, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dreyer, I.; Sussmilch, F.C.; Fukushima, K.; Riadi, G.; Becker, D.; Schultz, J.; Rainer, H. How to Grow a Tree: Plant Voltage-Dependent Cation Channels in the Spotlight of Evolution. Trends Plant Sci. 2021, 26, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, I.; Vergara-Jaque, A.; Riedelsberger, J.; González, W. Exploring the fundamental role of potassium channels in novel model plants. J. Exp. Bot. 2019, 70, 5985–5989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reintanz, B.; Szyroki, A.; Ivashikina, N.; Ache, P.; Godde, M.; Becker, D.; Palme, K.; Hedrich, R. AtKC1, a silent Arabidopsis potassium channel α-subunit modulates root hair K+ influx. Proc. Natl. Acad. Sci. USA 2002, 99, 4079–4084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, J.A.; Huprikar, S.S.; Kochian, L.V.; Lucas, W.J.; Gaber, R.F. Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1992, 89, 3736–3740. [Google Scholar] [CrossRef] [Green Version]
- Schachtman, D.P.; Schroeder, J.I.; Lucas, W.J.; Anderson, J.A.; Gaber, R.F. Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science 1992, 258, 1654–1658. [Google Scholar] [CrossRef]
- Hirsch, R.E.; Lewis, B.D.; Spalding, E.P.; Sussman, M.R. A role for the AKT1 potassium channel in plant nutrition. Science 1998, 280, 918–921. [Google Scholar] [CrossRef]
- Xu, J.; Li, H.D.; Chen, L.Q.; Wang, Y.; Liu, L.L.; He, L.; Wu, W.H. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 2006, 125, 1347–1360. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, I.; Stolzle, S.; Ivashikina, N.; Hedrich, R. Rice K+ uptake channel OsAKT1 is sensitive to salt stress. Planta 2005, 221, 212–221. [Google Scholar] [CrossRef]
- Boscari, A.; Clement, M.; Volkov, V.; Golldack, D.; Hybiak, J.; Miller, A.J.; Amtmann, A.; Fricke, W. Potassium channels in barley: Cloning, functional characterization and expression analyses in relation to leaf growth and development. Plant Cell Environ. 2009, 32, 1761–1777. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.S.; Zhao, J.L.; Fang, Q.W.; Chang, X.C.; Sun, M.Y.; Li, W.B.; Li, Y.G. GmAKT1 is involved in K+ uptake and Na+ /K+ homeostasis in Arabidopsis and soybean plants. Plant Sci. 2021, 304, 110736. [Google Scholar] [CrossRef] [PubMed]
- Ardie, S.W.; Liu, S.K.; Takano, T. Expression of the AKT1-type K+ channel gene from Puccinellia tenuiflora, PutAKT1, enhances salt tolerance in Arabidopsis. Plant Cell Rep. 2010, 29, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.M. Functional bases for interpreting amino acid sequences of voltage-dependent K+ channels. Annu. Rev. Biophys. Biomol. Struct. 1993, 22, 173–198. [Google Scholar] [CrossRef] [PubMed]
- Scott, V.E.S.; Rettig, J.; Parcej, D.N.; Keen, J.N.; Findaly, J.B.C.; Pongs, O. Primary structure of a subunit of a-dendrotoxin-sensitive K+ channels from bovine brain. Proc. Natl. Acad. Sci. USA 1994, 91, 163–1641. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Vasconcelos, A.C.; Berkowitz, G.A. Physical Association of KABl with Plant K+ Channel a Subunits. Plant Cell 1996, 8, 1545–1553. [Google Scholar] [CrossRef] [Green Version]
- Fang, Z.W.; Kamasani, U.; Berkowitz, G.A. Molecular cloning and expression characterization of a rice K+ channel β subunit. Plant Mol. Biol. 1998, 37, 597–606. [Google Scholar] [CrossRef]
- Ardie, S.W.; Nishiuchi, S.; Liu, S.; Takano, T. Ectopic Expression of the K+ Channel β Subunits from Puccinellia tenuiflora (KputB1) and Rice (KOB1) Alters K+ Homeostasis of Yeast and Arabidopsis. Mol. Biotechnol. 2011, 48, 76–86. [Google Scholar] [CrossRef]
- Rettig, J.; Heinenmann, S.H.; Wunder, F.; Lorra, C.; Parcej, D.N.; Dolly, J.O.; Pongs, O. Inactivation properties of voltage-gated K+ channels altered by presence of beta-subunit. Nature 1994, 369, 289–294. [Google Scholar] [CrossRef]
- Sewing, S.; Roeper, J.; Pongs, O. Kvb1 subunit binding specific for Shaker-related potassium channel a subunits. Neuron 1996, 16, 455–463. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Li, M. Auxiliary subunits of Shaker-type potassium channels. Trends Cardiovasc. Med. 1998, 8, 229–234. [Google Scholar] [CrossRef]
- Park, S.U.; Lee, C.J.; Kim, S.E.; Lim, Y.H.; Lee, H.U.; Nam, S.S.; Kim, H.S.; Kwak, S.S. Selection of flooding stress tolerant sweetpotato cultivars based on biochemical and phenotypic characterization. Plant Physiol. Biochem. 2020, 155, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Zhang, A.J.; Sun, J.; Chen, X.G.; Liu, M.; Zhao, P.; Jiang, W.; Tang, Z.G. Identification of Shaker K+ channel family members in sweetpotato and functional exploration of IbAKT1. Gene 2021, 768, 145311. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhou, Y.Y.; Zhai, H.; He, S.Z.; Zhao, N.; Liu, Q.C. A novel sweetpotato WRKY transcription factor, IbWRKY2, positively regulates drought and salt tolerance in transgenic Arabidopsis. Biomolecules 2020, 10, 506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Du, B.; Nie, N.; Sun, S.F.; Hu, Y.F.; Bai, Y.D.; He, S.Z.; Zhao, N.; Liu, Q.C.; Zhai, H. A novel sweetpotato RING-H2 type E3 ubiquitin ligase gene IbATL38 enhances salt tolerance in transgenic Arabidopsis. Plant Sci. 2021, 304, 110802. [Google Scholar] [CrossRef]
- Zhao, L.N.; Liu, F.X.; Xu, W.Y.; Di, C.; Zhou, S.X.; Xue, Y.B.; Yu, J.J.; Su, Z. Increased expression of OsSPX1 enhances cold/subfreezing tolerance in tobacco and Arabidopsis thaliana. Plant Biotechnol. J. 2009, 7, 550–561. [Google Scholar] [CrossRef]
- Fu, X.Z.; Khan, E.U.; Hu, S.S.; Fan, Q.J.; Liu, J.H. Overexpression of the betaine aldehyde dehydrogenase gene from Atriplex hortensis enhances salt tolerance in the transgenic trifoliate orange (Poncirus trifoliata L. Raf.). Environ. Exp. Bot. 2011, 74, 106–113. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Breusegem, F.V. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Clarkson, D.T.; Hanson, J.B. The mineral nutrition of higher plants. Annu. Rev. Plant Physiol. 1980, 31, 239–298. [Google Scholar] [CrossRef]
- Tang, H.; Vasconcelos, A.C.; Berkowitz, G.A. Evidence that plant K+ channel proteins have two different types of subunits. Plant Physiol. 1995, 109, 327–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafsi, C.; Debez, A.; Abdelly, C. Potassium deficiency in plants: Effects and signaling cascades. Acta Physiol. Plant 2014, 36, 1055–1070. [Google Scholar] [CrossRef]
- Thaler, P.; Pages, L. Modeling the influence of assimilates availability on root growth and architecture. Plant Soil 1998, 201, 307–320. [Google Scholar] [CrossRef]
- Shin, R.; Schachtman, D.P. Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl. Acad. Sci. USA 2004, 101, 8827–8832. [Google Scholar] [CrossRef] [Green Version]
- Xiao-Li, T.; Gang-Wei, W.; Rui, Z.; Pei-Zhu, Y.; Liu-Sheng, D.; Zhao-Hu, L. Conditions and indicators for screening cotton (Gossypium hirsutum L.) varieties tolerant to low potassium. Acta Agron. Sin. 2008, 34, 1435–1443. [Google Scholar] [CrossRef]
- Han, B.H.; Zhao, H.L.; Xiao, L.T. Mechanism of tolerance to potassium deficiency between Liaomian 18 and NuCOTN99B at seedling stage. Acta Agron. Sin. 2009, 35, 475–482. [Google Scholar] [CrossRef]
- Gierth, M.; Mäser, P.; Schroeder, J.I. The potassium transporter HAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 2005, 137, 1105–1114. [Google Scholar] [CrossRef] [Green Version]
- Flowers, T.J. Improving crop salt tolerance. J. Exp. Bot. 2004, 55, 307–319. [Google Scholar] [CrossRef]
- Kronzucker, H.J.; Britto, D.T. Sodium transport in plants: A critical review. New Phytol. 2011, 189, 54–81. [Google Scholar] [CrossRef]
- Ganie, S.A.; Molla, K.A.; Henry, R.J.; Bhat, K.V.; Mondal, T.K. Advances in understanding salt tolerance in rice. Theor. Appl. Genet. 2019, 132, 851–870. [Google Scholar] [CrossRef]
- Muchate, N.S.; Nikalje, G.C.; Rajurkar, N.S.; Suprasanna, P.; Nikam, T.D. Plant salt stress adaptive responses, tolerance mechanism and bioengineering for salt tolerance. Bot. Rev. 2016, 82, 371–406. [Google Scholar] [CrossRef]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanella, M.; Borghi, G.L.; Pirone, C.; Thalmann, M.; Pazmino, D.; Costa, A.; Santelia, D.; Trost, P.; Sparla, F. β-Amylase 1 (BAM1) degrades transitory starch to sustain proline biosynthesis during drought stress. J. Exp. Bot. 2016, 67, 1819–1826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Chen, X.; Gu, H.; Wu, B.; Zhang, H.; Yuan, X.; Cui, X. GmHKT1;4, a novel soybean gene regulating Na+/K+ ratio in roots enhances salt tolerance in transgenic plants. Plant Growth Regul. 2014, 73, 299–308. [Google Scholar] [CrossRef]
- Wu, H.; Shabala, L.; Zhou, M.; Shabala, S. Chloroplast-generated ROS dominate NaCl-induced K+ efflux in wheat leaf mesophyll. Plant Signal. Behav. 2015, 10, e1013793. [Google Scholar] [CrossRef] [Green Version]
- Mangal, V.; Lal, M.K.; Tiwari, R.K.; Altal, M.A.; Sood, S.; Kumar, D.; Bharadwaj, V.; Singh, B.; Singh, R.K.; Aftab, T. Molecular insights into the role of reactive oxygen, nitrogen and sulphur species in conferring salinity stress tolerance in plants. J. Plant Growth Regul. 2022. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, H.; Yang, X.; Li, Q.; Guo, J.; Ma, T.; Liu, S.; Lin, S.; Zhou, Y.; Zhao, C.; Wang, J.; et al. The Sweetpotato Voltage-Gated K+ Channel β Subunit, KIbB1, Positively Regulates Low-K+ and High-Salinity Tolerance by Maintaining Ion Homeostasis. Genes 2022, 13, 1100. https://doi.org/10.3390/genes13061100
Zhu H, Yang X, Li Q, Guo J, Ma T, Liu S, Lin S, Zhou Y, Zhao C, Wang J, et al. The Sweetpotato Voltage-Gated K+ Channel β Subunit, KIbB1, Positively Regulates Low-K+ and High-Salinity Tolerance by Maintaining Ion Homeostasis. Genes. 2022; 13(6):1100. https://doi.org/10.3390/genes13061100
Chicago/Turabian StyleZhu, Hong, Xue Yang, Qiyan Li, Jiayu Guo, Tao Ma, Shuyan Liu, Shunyu Lin, Yuanyuan Zhou, Chunmei Zhao, Jingshan Wang, and et al. 2022. "The Sweetpotato Voltage-Gated K+ Channel β Subunit, KIbB1, Positively Regulates Low-K+ and High-Salinity Tolerance by Maintaining Ion Homeostasis" Genes 13, no. 6: 1100. https://doi.org/10.3390/genes13061100





