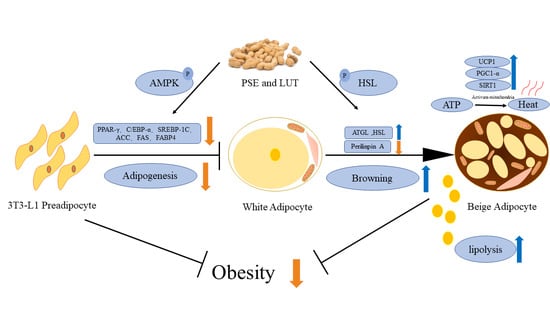
Peanut Shell Extract and Luteolin Regulate Lipid Metabolism and Induce Browning in 3T3-L1 Adipocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of the PSE
2.2. Cell Culture and Treatment
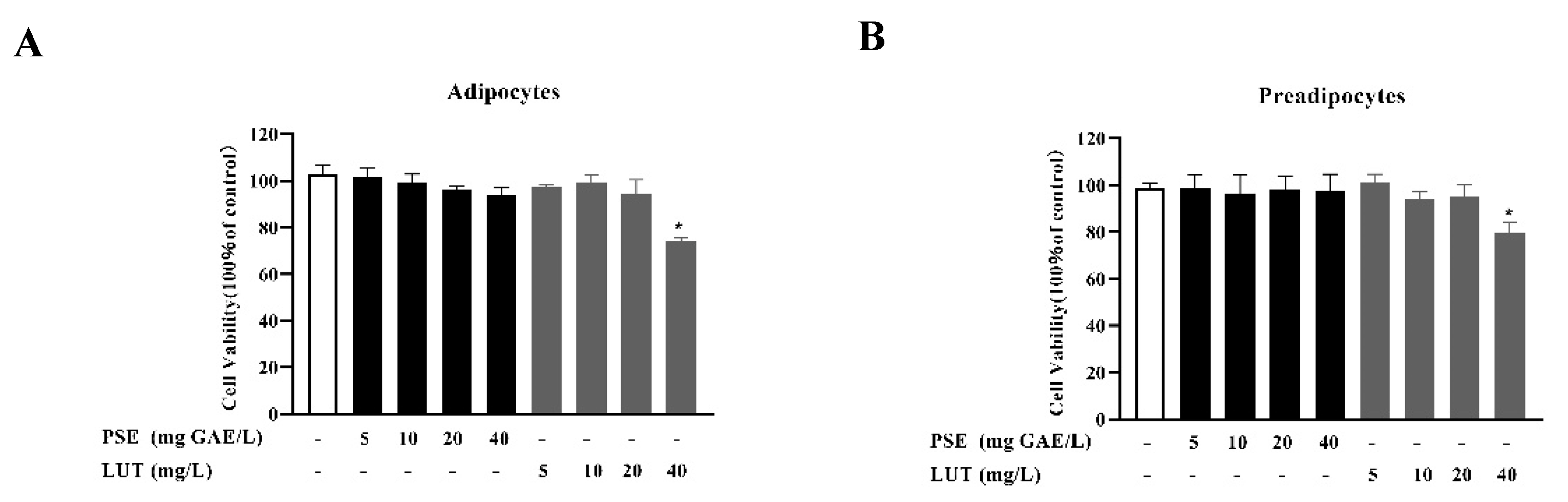
2.3. Cell Viability Assay
2.4. Oil Red O Staining
2.5. Lipolysis Assay
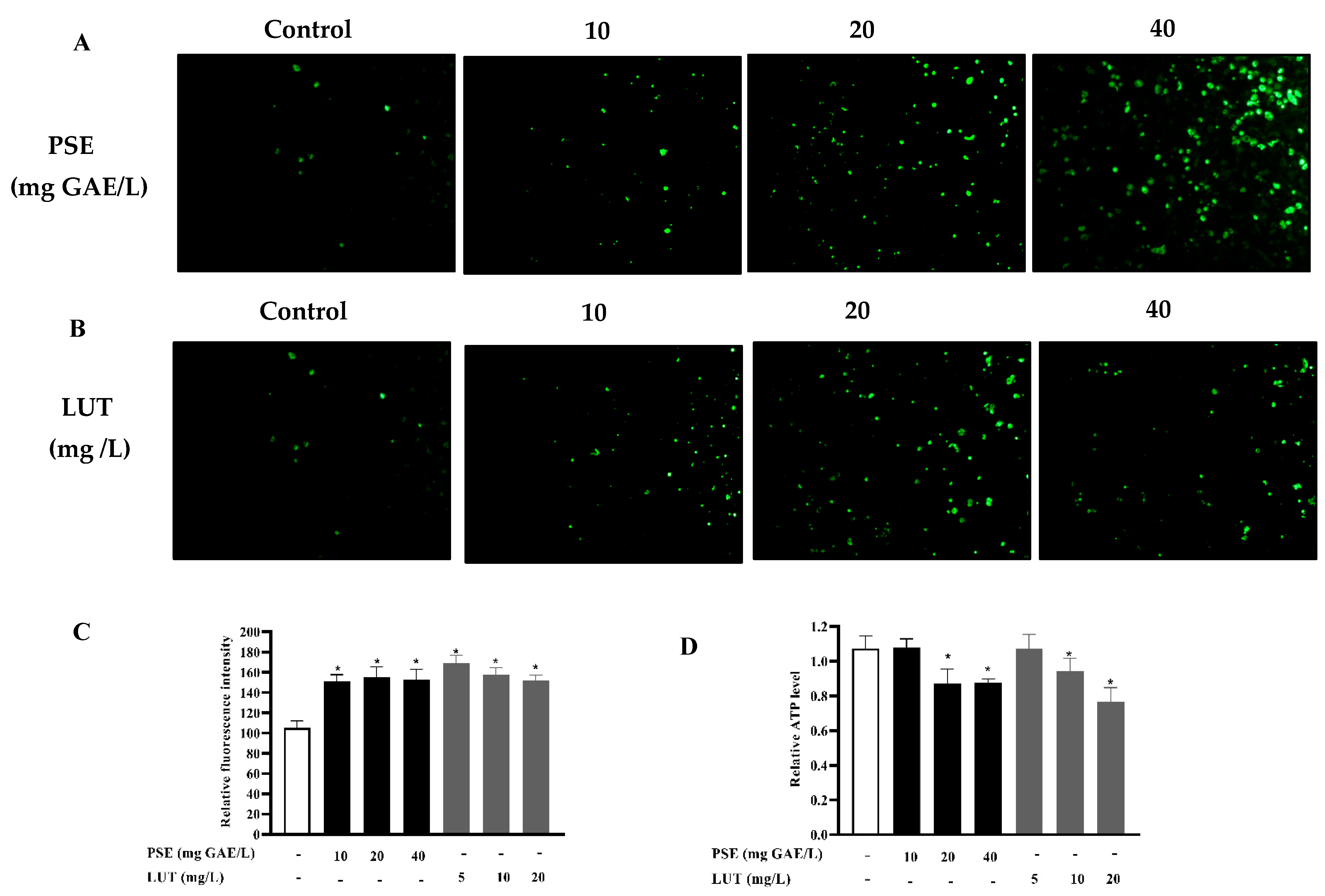
2.6. Intracellular ATP Determination
2.7. Mitochondrial Staining
2.8. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
2.9. Western Blot Analysis
2.10. Statistical Analysis
3. Results
3.1. The Effect of PSE and LUT on Cell Viability
3.2. The Effect of PSE and LUT on Intracellular Lipid Accumulation in Adipocytes
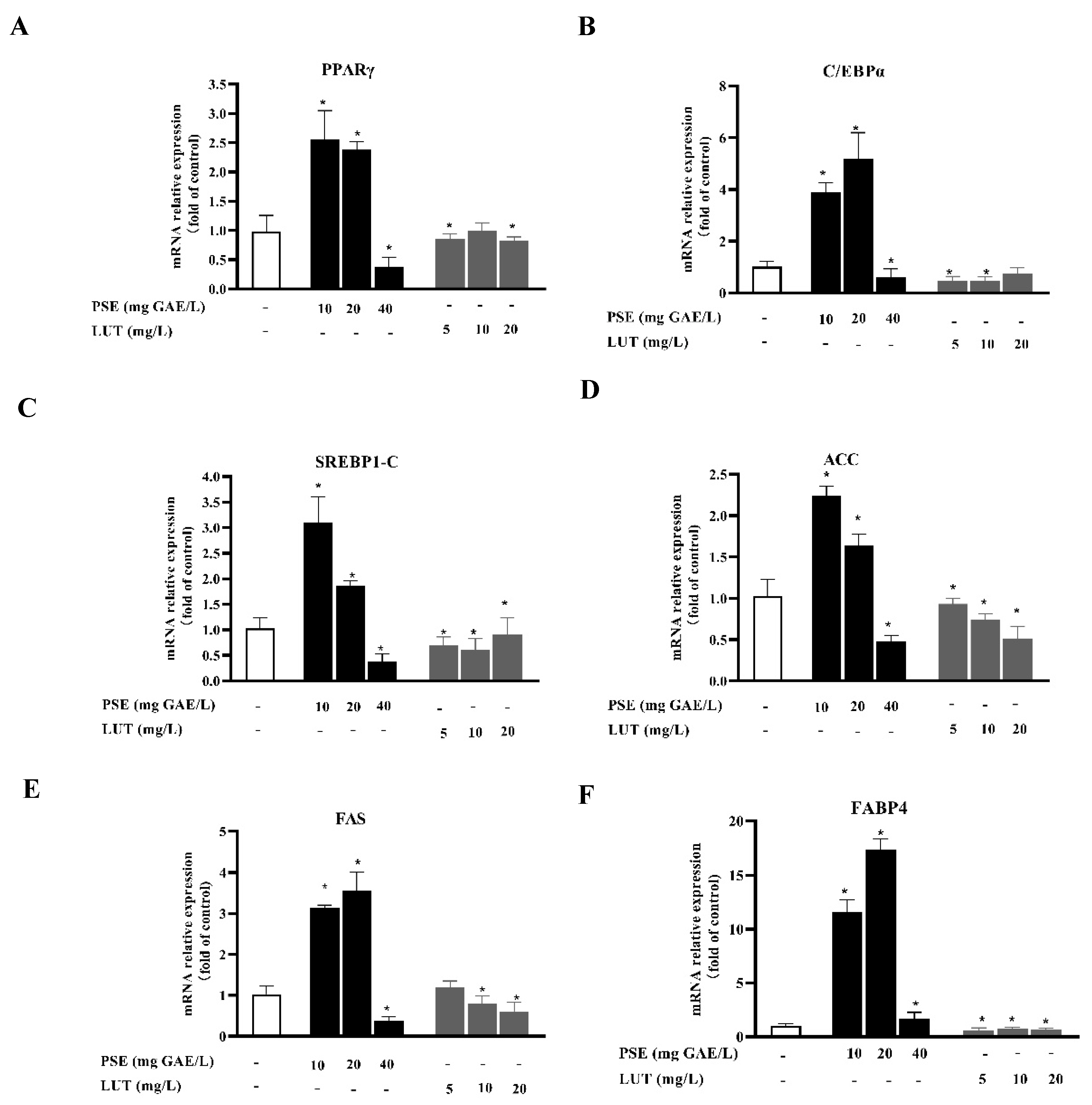
3.3. PSE and LUT Modulated the Expressions Levels of mRNA and Protein Involved in AdipoGenesis
3.4. The Effect of PSE and LUT on the Lipolysis in Differentiated 3T3-L1 Adipocytes
3.5. PSE and LUT Modulated the Expression Levels of mRNA and Protein Involved in Lipolysis
3.6. The Effect of PSE and LUT on Mitochondrial Biogenesis
3.7. PSE and LUT Modulated the Expression Levels of the mRNA and Protein Involved in Browning
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tseng, Y.; Cypess, A.M.; Kahn, C.R. Cellular bioenergetics as a target for obesity therapy. Nat. Rev. Drug Discov. 2010, 9, 465–482. [Google Scholar] [CrossRef] [PubMed]
- Zimmet, P.; Alberti, K.; Shaw, J. Global and societal implications of the diabetes epidemic. Nature 2001, 414, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, M.; Duncan, R.E.; Sul, H.S. The skinny on fat: Lipolysis and fatty acid utilization in adipocytes. Trends Endocrinol. Metab. 2009, 20, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.S.; Hong, J.H.; Cho, K.H.; Jung, H.K. Grape skin extract reduces adipogenesis-and lipogenesis-related gene expression in 3T3-L1 adipocytes through the peroxisome proliferator-activated receptor-γ signaling pathway. Nutr. Res. 2012, 32, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Ma, X.; Tian, W. Pomegranate husk extract, punicalagin and ellagic acid inhibit fatty acid synthase and adipogenesis of 3T3-L1 adipocyte. J. Funct. Foods. 2013, 5, 633–641. [Google Scholar] [CrossRef]
- Rupasinghe, H.V.; Sekhon-Loodu, S.; Mantso, T.; Panayiotidis, M.I. Phytochemicals in regulating fatty acid β-oxidation: Potential underlying mechanisms and their involvement in obesity and weight loss. Pharmacol. Ther. 2016, 165, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, Z.; Huang, J.; Luo, G.; Liang, Q.; Wang, D.; Ye, X.; Wu, C.; Wang, L.; Hu, J. Anti-obesity and hypolipidemic effects of Fuzhuan brick tea water extract in high-fat diet-induced obese rats. J. Sci. Food Agric. 2013, 93, 1310–1316. [Google Scholar] [CrossRef]
- Cao, Z.; Gu, D.; Lin, Q.; Xu, Z.; Huang, Q.; Rao, H.; Liu, E.; Jia, J.; Ge, C. Effect of pu-erh tea on body fat and lipid profiles in rats with diet-induced obesity. Phytother. Res. 2011, 25, 234–238. [Google Scholar] [CrossRef]
- Lee, S.G.; Parks, J.S.; Kang, H.W. Quercetin, a functional compound of onion peel, remodels white adipocytes to brown-like adipocytes. J. Nutr. Biochem. 2017, 42, 62–71. [Google Scholar] [CrossRef]
- Kim, H.; Jeon, Y.; Park, J.; Rim, H.; Jeong, M.; Lim, H.; Ko, S.; Jang, H.; Lee, B.; Lee, K. Corni fructus containing formulation attenuates weight gain in mice with diet-induced obesity and regulates adipogenesis through AMPK. Evid.-Based Complementary Altern. Med. 2013, 2013, 423741. [Google Scholar] [CrossRef] [Green Version]
- Ho, J.Y.; Choi, J.O.; Lim, W.H.; Kim, M.Y.; Lee, I.O.; Cho, H.O. Kefir inhibits 3T3-L1 adipocyte differentiation through down-regulation of adipogenic transcription factor expression. J. Sci. Food Agric. 2013, 93, 485–490. [Google Scholar] [CrossRef]
- Song, Z.; Xiaoli, A.; Yang, F. Regulation and Metabolic Significance of De Novo Lipogenesis in Adipose Tissues. Nutrients 2018, 10, 1383. [Google Scholar] [CrossRef]
- Frayn, K.N.; Karpe, F.; Fielding, B.A.; Macdonald, I.A.; Coppack, S.W. Integrative physiology of human adipose tissue. Int. J. Obes. 2003, 27, 875. [Google Scholar] [CrossRef]
- Mahadevaiah, M.V.; Santhekadur, P.K. Obesity, Cancer and Cachexia. Curr. Trends Biotechnol. Pharm. 2020, 15, 388–395. [Google Scholar] [CrossRef]
- Fenzl, A.; Kiefer, F.W. Brown adipose tissue and thermogenesis. Horm. Mol. Biol. Clin. Investig. 2014, 19, 25–37. [Google Scholar] [CrossRef]
- Cypess, A.M.; Kahn, C.R. Brown fat as a therapy for obesity and diabetes. Curr. Opin. Endocrinol. 2010, 17, 143. [Google Scholar] [CrossRef]
- Mcqueen, A.E.; Koliwad, S.K.; Wang, J.C. Fighting obesity by targeting factors regulating beige adipocytes. Curr. Opin. Clin. Nutr. 2018, 21, 1. [Google Scholar] [CrossRef] [PubMed]
- Rosenwald, M.; Perdikari, A.; Rülicke, T.; Wolfrum, C. Bi-directional interconversion of brite and white adipocytes. Nat. Cell Biol. 2013, 15, 659–667. [Google Scholar] [CrossRef]
- Fedorenko, A.; Lishko, P.V.; Kirichok, Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 2012, 151, 400–413. [Google Scholar] [CrossRef]
- Kozak, L.P.; Anunciado-Koza, R. UCP1: Its involvement and utility in obesity. Int. J. Obes. 2008, 32, S32–S38. [Google Scholar] [CrossRef] [Green Version]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.H.; Doria, A. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009, 360, 1509. [Google Scholar] [CrossRef] [PubMed]
- Fisher, F.M.; Kleiner, S.; Douris, N.; Fox, E.C.; Mepani, R.J.; Verdeguer, F.; Wu, J.; Kharitonenkov, A.; Flier, J.S.; Maratos-Flier, E. FGF21 regulates PGC-1α and browning of white adipose tissues in adaptive thermogenesis. Genes Dev. 2012, 26, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Ohno, H.; Shinoda, K.; Spiegelman, B.M.; Kajimura, S. PPARγ agonists Induce a White-to-Brown Fat Conversion through Stabilization of PRDM16 Protein. Cell Metab. 2012, 15, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.W.; Lumeng, C.N. SirT1: A Guardian at the Gates of Adipose Tissue Inflammation. Diabetes 2011, 60, 3100–3102. [Google Scholar] [CrossRef]
- Choi, M.; Mukherjee, S.; Yun, J.W. Trigonelline induces browning in 3T3-L1 white adipocytes. Phytother. Res. 2021, 35, 1113–1124. [Google Scholar] [CrossRef]
- Manigandan, S.; Yun, J.W. Urolithin A induces brown-like phenotype in 3T3-L1 white adipocytes via β3-adrenergic receptor-p38 MAPK signaling pathway. Biotechnol. Bioprocess Eng. 2020, 25, 345–355. [Google Scholar] [CrossRef]
- Ge, S.; Wu, Y.; Peng, W.; Xia, C.; Mei, C.; Cai, L.; Shi, S.Q.; Sonne, C.; Lam, S.S.; Tsang, Y.F. High-pressure CO2 hydrothermal pretreatment of peanut shells for enzymatic hydrolysis conversion into glucose. Chem. Eng. J. 2020, 385, 123949. [Google Scholar] [CrossRef]
- Toomer, O.T. A comprehensive review of the value-added uses of peanut (Arachis hypogaea) skins and by-products. Crit. Rev. Food Sci. 2020, 60, 341–350. [Google Scholar] [CrossRef]
- Gao, F.; Ye, H.; Yu, Y.; Zhang, T.; Deng, X. Lack of toxicological effect through mutagenicity test of polyphenol extracts from peanut shells. Food Chem. 2011, 129, 920–924. [Google Scholar] [CrossRef]
- Zang, Y.; Igarashi, K.; Li, Y. Anti-diabetic effects of luteolin and luteolin-7-O-glucoside on KK-A y mice. Biosci. Biotechnol. Biochem. 2016, 80, 1580–1586. [Google Scholar] [CrossRef] [Green Version]
- De Stefano, A.; Caporali, S.; Di Daniele, N.; Rovella, V.; Cardillo, C.; Schinzari, F.; Minieri, M.; Pieri, M.; Candi, E.; Bernardini, S. Anti-inflammatory and proliferative properties of luteolin-7-O-glucoside. Int. J. Mol. Sci. 2021, 22, 1321. [Google Scholar] [CrossRef]
- Gaihre, Y.R.; Iwamoto, A.; Oogai, S.; Hamajima, H.; Tsuge, K.; Nagata, Y.; Yanagita, T. Perilla pomace obtained from four different varieties have different levels and types of polyphenols and anti-allergic activity. Cytotechnology 2022, 74, 341–349. [Google Scholar] [CrossRef]
- Park, H.S.; Kim, S.H.; Kim, Y.S.; Ryu, S.Y.; Hwang, J.T.; Yang, H.J.; Kim, G.H.; Kwon, D.Y.; Kim, M.S. Luteolin inhibits adipogenic differentiation by regulating PPARγ activation. BioFactors 2009, 35, 373–379. [Google Scholar] [CrossRef]
- Zhang, L.; Han, Y.J.; Zhang, X.; Wang, X.; Bao, B.; Qu, W.; Liu, J. Luteolin reduces obesity-associated insulin resistance in mice by activating AMPKα1 signalling in adipose tissue macrophages. Diabetologia 2016, 59, 2219–2228. [Google Scholar] [CrossRef]
- Fang, C.; Kim, H.; Noratto, G.; Sun, Y.; Talcott, S.T.; Mertens-Talcott, S.U. Gallotannin derivatives from mango (Mangifera indica L.) suppress adipogenesis and increase thermogenesis in 3T3-L1 adipocytes in part through the AMPK pathway. J. Funct. Foods 2018, 46, 101–109. [Google Scholar]
- Iwamoto, K.; Kamo, S.; Takada, Y.; Ieda, A.; Yamashita, T.; Sato, T.; Zaima, N.; Moriyama, T. Soyasapogenols reduce cellular triglyceride levels in 3T3-L1 mouse adipocyte cells by accelerating triglyceride lipolysis. Biochem. Biophys. Rep. 2018, 16, 44–49. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, S.; Yang, X.; You, P.; Zhang, W. Myricetin suppresses differentiation of 3 T3-L1 preadipocytes and enhances lipolysis in adipocytes. Nutr. Res. 2015, 35, 317–327. [Google Scholar] [CrossRef]
- Rosen, E.D.; Spiegelman, B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006, 444, 847–853. [Google Scholar] [CrossRef]
- Trujillo, M.E.; Scherer, P.E. Adipose tissue-derived factors: Impact on health and disease. Endocr. Rev. 2006, 27, 762–778. [Google Scholar] [CrossRef]
- Hara, L.; Jinbong, P.; Hye-Lin, K.; Kang, J.W.; Mi-Young, J.; Dong-Hyun, Y.; Yunu, J.; Yong-Il, K.; Hyun-Ju, K.; Seok, A.K. Chrysophanic Acid Suppresses Adipogenesis and Induces Thermogenesis by Activating AMP-Activated Protein Kinase Alpha In vivo and In vitro. Front. Pharmacol. 2016, 7, 476. [Google Scholar]
- Francisco, M.; Resurreccion, A. Functional Components in Peanuts. Crit. Rev. Food. Sci. Nutr. 2008, 48, 715–746. [Google Scholar] [CrossRef]
- Seelinger, G.; Merfort, I.; Schempp, C. Anti-Oxidant, Anti-Inflammatory and Anti-Allergic Activities of Luteolin. Planta Med. 2008, 74, 1667–1677. [Google Scholar] [CrossRef]
- Xu, N.; Zhang, L.; Dong, J.; Zhang, X.; Chen, Y.G.; Bao, B.; Liu, J. Low-dose diet supplement of a natural flavonoid, luteolin, ameliorates diet-induced obesity and insulin resistance in mice. Mol. Nutr. Food Res. 2014, 58, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Bhaskar, N.; Kaur, H.; Pandey, R.; Manhas, S. LipidDroplets: A Dynamic Organelle in Health and Disease. Indian J. Pharm. Res. 2015, 5, 173–184. [Google Scholar]
- Daigle, D.J.; Conkerton, E.J.; Sanders, T.H.; Mixon, A.C. Peanut hull flavonoids: Their relationship with peanut maturity. J. Agric. Food Chem. 1988, 36, 1179–1181. [Google Scholar] [CrossRef]
- Hwang, J.M.; Lee, M.; Lee, J.; Lee, J.H. Agastache rugosa Extract and Its Bioactive Compound Tilianin Suppress Adipogenesis and Lipogenesis on 3T3-L1 Cells. Appl. Sci. 2021, 11, 7679. [Google Scholar] [CrossRef]
- Ma, Q.; Cui, Y.; Xu, S.; Zhao, Y.; Yuan, H.; Piao, G. Synergistic Inhibitory Effects of Acacetin and 11 Other Flavonoids Isolated from Artemisia sacrorum on Lipid Accumulation in 3T3-L1 Cells. J. Agric. Food Chem. 2018, 66, 12931–12940. [Google Scholar] [CrossRef]
- Bo, H.; Hai, D.Y.; Kim, D.Y.; Hai, Y.Q.; Chung, S.H. Cinnamaldehyde prevents adipocyte differentiation and adipogenesis via regulation of peroxisome proliferator-activated receptor-γ (PPARγ) and AMP-activated protein kinase (AMPK) pathways. J. Agric. Food Chem. 2011, 59, 3666. [Google Scholar]
- Ono, M.; Fujimori, K. Antiadipogenic effect of dietary apigenin through activation of AMPK in 3T3-L1 cells. J. Agric. Food Chem. 2011, 59, 13346–13352. [Google Scholar] [CrossRef]
- Ahn, J.; Lee, H.; Kim, S.; Park, J.; Ha, T. The anti-obesity effect of quercetin is mediated by the AMPK and MAPK signaling pathways. Biochem. Biophys. Res. Commun. 2008, 373, 545–549. [Google Scholar] [CrossRef]
- Ardévol, A.; Bladé, C.; Salvadó, M.J.; Arola, L. Changes in lipolysis and hormone-sensitive lipase expression caused by procyanidins in 3T3-L1 adipocytes. Int. J. Obes. Relat. Metab. Disord. 2000, 24, 319–324. [Google Scholar] [CrossRef]
- Nielsen, T.S.; Jessen, N.; Jørgensen, J.O.L.; Møller, N.; Lund, S. Dissecting adipose tissue lipolysis: Molecular regulation and implications for metabolic disease. J. Mol. Endocrinol. 2014, 52, R199–R222. [Google Scholar] [CrossRef]
- Sztalryd, C.; Brasaemle, D.L. The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1221–1232. [Google Scholar] [CrossRef]
- Huang, W.C.; Chen, C.Y.; Wu, S.J. Almond Skin Polyphenol Extract Inhibits Inflammation and Promotes Lipolysis in Differentiated 3T3-L1 Adipocytes. J. Med. Food 2017, 20, 103. [Google Scholar] [CrossRef]
- Kim, H.J.; Lee, M.; Jang, J.; Lee, S. Allium hookeri root extract inhibits adipogenesis by promoting lipolysis in high fat diet-induced obese mice. Nutrients 2019, 11, 2262. [Google Scholar] [CrossRef] [PubMed]
- Anthonsen, M.W.; Ronnstrand, L.; Wernstedt, C.; Degerman, E.; Holm, C. Identification of Novel Phosphorylation Sites in Hormone-sensitive Lipase That Are Phosphorylated in Response to Isoproterenol and Govern Activation Properties in Vitro. J. Biol. Chem. 1998, 273, 215. [Google Scholar] [CrossRef]
- Ho, J.N.; Park, S.J.; Choue, R.; Lee, J. Standardized Ethanol Extract of Curcuma longa L. Fermented by Aspergillus oryzae Promotes Lipolysis via Activation of cAMP-Dependent PKA in 3T3-L1 Adipocytes. J. Food Biochem. 2013, 37, 595–603. [Google Scholar] [CrossRef]
- Hwan, K.Y.; Kon, K.K.; Jung, K.D.; Myeon, C. Antiobesity effects of the water-soluble fraction of the ethanol extract of Smilax china L. leaf in 3T3-L1 adipocytes. Nutr. Res. Pract. 2015, 9, 606–612. [Google Scholar]
- Rydén, M.; Arvidsson, E.; Blomqvist, L.; Perbeck, L.; Dicker, A.; Arner, P. Targets for TNF-α-induced lipolysis in human adipocytes. Biochem. Bioph. Res. Commun. 2004, 318, 168–175. [Google Scholar] [CrossRef]
- Arner, P.; Langin, D. Lipolysis in lipid turnover, cancer cachexia, and obesity-induced insulin resistance. Trends Endocrinol. Metab. 2014, 25, 255–262. [Google Scholar] [CrossRef]
- Harms, M.; Seale, P. Brown and beige fat: Development, function and therapeutic potential. Nat. Med. 2013, 19, 1252–1263. [Google Scholar] [CrossRef] [PubMed]
- Ploumi, C.; Daskalaki, I.; Tavernarakis, N. Mitochondrial biogenesis and clearance: A balancing act. FEBS J. 2017, 284, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Lee, M.; Chang, E.; Shin, Y.; Oh, S.; Kim, I.; Kim, Y. Rutin increases muscle mitochondrial biogenesis with AMPK activation in high-fat diet-induced obese rats. Nutrients 2015, 7, 8152–8169. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Salmeán, G.; Ortiz-Vilchis, P.; Vacaseydel, C.M.; Garduño-Siciliano, L.; Chamorro-Cevallos, G.; Meaney, E.; Villafaña, S.; Villarreal, F.; Ceballos, G.; Ramírez-Sánchez, I. Effects of (−)-epicatechin on a diet-induced rat model of cardiometabolic risk factors. Eur. J. Pharmacol. 2014, 728, 24–30. [Google Scholar] [CrossRef]
- Ricquier, D. Uncoupling Protein 1 of Brown Adipocytes, the Only Uncoupler: A Historical Perspective. Front. Endocrinol. 2011, 2, 85. [Google Scholar] [CrossRef] [Green Version]
- Rebollo-Hernanz, M.; Zhang, Q.; Aguilera, Y.; Martín-Cabrejas, M.A.; Gonzalez De Mejia, E. Phenolic compounds from coffee by-products modulate adipogenesis-related inflammation, mitochondrial dysfunction, and insulin resistance in adipocytes, via insulin/PI3K/AKT signaling pathways. Food Chem. Toxicol. 2019, 132, 110672. [Google Scholar] [CrossRef]
- Zhang, H.Q.; Chen, S.Y.; Wang, A.S.; Yao, A.J.; Fu, J.F.; Zhao, J.S.; Chen, F.; Zou, Z.Q.; Zhang, X.H.; Shan, Y.J.; et al. Sulforaphane induces adipocyte browning and promotes glucose and lipid utilization. Mol. Nutr. Food. Res. 2016, 60, 2185–2197. [Google Scholar] [CrossRef]
- Bertholet, A.M.; Kazak, L.; Chouchani, E.T.; Bogaczyńska, M.G.; Paranjpe, I.; Wainwright, G.L.; Bétourné, A.; Kajimura, S.; Spiegelman, B.M.; Kirichok, Y. Mitochondrial patch clamp of beige adipocytes reveals UCP1-positive and UCP1-negative cells both exhibiting futile creatine cycling. Cell Metab. 2017, 25, 811–822. [Google Scholar] [CrossRef]
- Shabalina, I.G.; Petrovic, N.; de Jong, J.M.; Kalinovich, A.V.; Cannon, B.; Nedergaard, J. UCP1 in brite/beige adipose tissue mitochondria is functionally thermogenic. Cell Rep. 2013, 5, 1196–1203. [Google Scholar] [CrossRef]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C. Mechanisms Controlling Mitochondrial Biogenesis and Respiration through the Thermogenic Coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef]
- Wilson-Fritch, L.; Nicoloro, S.M.; Chouinard, M.T.; Lazar, M.A.; Corvera, S. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J. Clin. Investig. 2004, 114, 1281–1289. [Google Scholar] [CrossRef]
- Rodgers, J.; Lerin, C.; Haas, W.; Gygi, S.; Spiegelman, B.; Puigserver, P. Nutrient control of glucose homeostasis through a complex of PGC-1 alpha and SIRT1. Nature 2005, 434, 113–118. [Google Scholar] [CrossRef]
- Altshuler-Keylin, S.; Kajimura, S. Mitochondrial homeostasis in adipose tissue remodeling. Sci. Signal. 2017, 10, i9248. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, L.; Dong, L.; Hu, X.; Feng, F.; Chen, F. 6-Gingerol, a Functional Polyphenol of Ginger, Promotes Browning through an AMPK-Dependent Pathway in 3T3-L1 Adipocytes. J. Agric. Food Chem. 2019, 67, 14056–14065. [Google Scholar] [CrossRef]
- Csiszar, A.; Labinskyy, N.; Pinto, J.T.; Ballabh, P.; Zhang, H.; Losonczy, G.; Pearson, K.; De Cabo, R.; Pacher, P.; Zhang, C. Resveratrol induces mitochondrial biogenesis in endothelial cells. Am. J. Physiol.-Heart Circ. Physiol. 2009, 297, H13–H20. [Google Scholar] [CrossRef] [Green Version]







| Target Gene | Forward | Reverse |
|---|---|---|
| PPARγ | GACGCGGAAGAAGAGACCTG | GTGTGACTTCTCCTCAGCCC |
| C/ebp-α | AATGGCAGTGTGCACGTCTA | CCCCAGCCGTTAGTGAAGAG |
| SREBP1 | CAGACTCACTGCTGCTGACA | GATGGTCCCTCCACTCACCA |
| ACC | GCCTCAGGAGGATTTGCTGT | AGGATCTACCCAGGCCACAT |
| FAS | GAGGGTGTGCCATTCTGTCA | GCTATTCTCTACCGCTGGGG |
| FABP4 | GGATTTGGTCACCATCCGGT | TTCCATCCCACTTCTGCACC |
| HSL | AAAAGCTGAACCTGGGGGAG | TGCCATCTGCTGGGAAAACA |
| ATGL | AACGCCACTCACATCTACGG | TGGCAGGCATGGGACATAAA |
| Perilipin A | CTCAGCTCTCCTGTTAGGCG | GTAGAGCTCACCAAGGGCAG |
| UCP1 | ACGTCCCCTGCCATTTACTG | GGTACGCTTGGGTACTGTCC |
| Ppargc1α | ACTCTCAGTAAGGGGCTGGT | ACATGTCCCAAGCCATCCAG |
| Sirt1 | CGGCTACGAGGTCCATATAC | ACAATCTGCCACAGCGTCAT |
| β-actin | GAGCGCAAGTACTCTGTGTG | AACGCAGCTCAGTAACAGTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Wang, L.; Zhang, J. Peanut Shell Extract and Luteolin Regulate Lipid Metabolism and Induce Browning in 3T3-L1 Adipocytes. Foods 2022, 11, 2696. https://doi.org/10.3390/foods11172696
Liu W, Wang L, Zhang J. Peanut Shell Extract and Luteolin Regulate Lipid Metabolism and Induce Browning in 3T3-L1 Adipocytes. Foods. 2022; 11(17):2696. https://doi.org/10.3390/foods11172696
Chicago/Turabian StyleLiu, Wenrui, Lihua Wang, and Jie Zhang. 2022. "Peanut Shell Extract and Luteolin Regulate Lipid Metabolism and Induce Browning in 3T3-L1 Adipocytes" Foods 11, no. 17: 2696. https://doi.org/10.3390/foods11172696
APA StyleLiu, W., Wang, L., & Zhang, J. (2022). Peanut Shell Extract and Luteolin Regulate Lipid Metabolism and Induce Browning in 3T3-L1 Adipocytes. Foods, 11(17), 2696. https://doi.org/10.3390/foods11172696