The Critical Role of Small RNAs in Regulating Plant Innate Immunity
Abstract
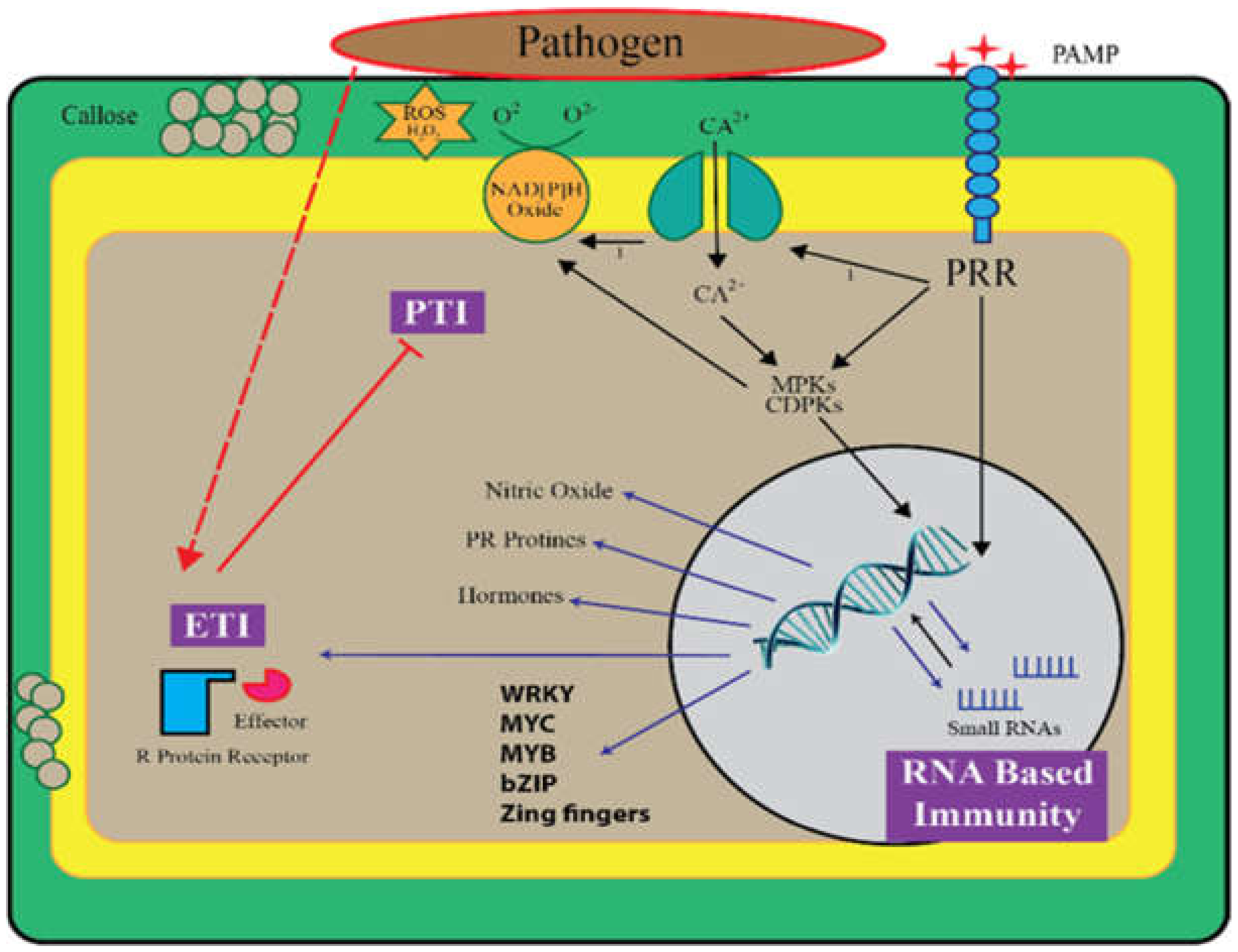
:1. Introduction
2. Small RNA Biogenesis and Mode of Action
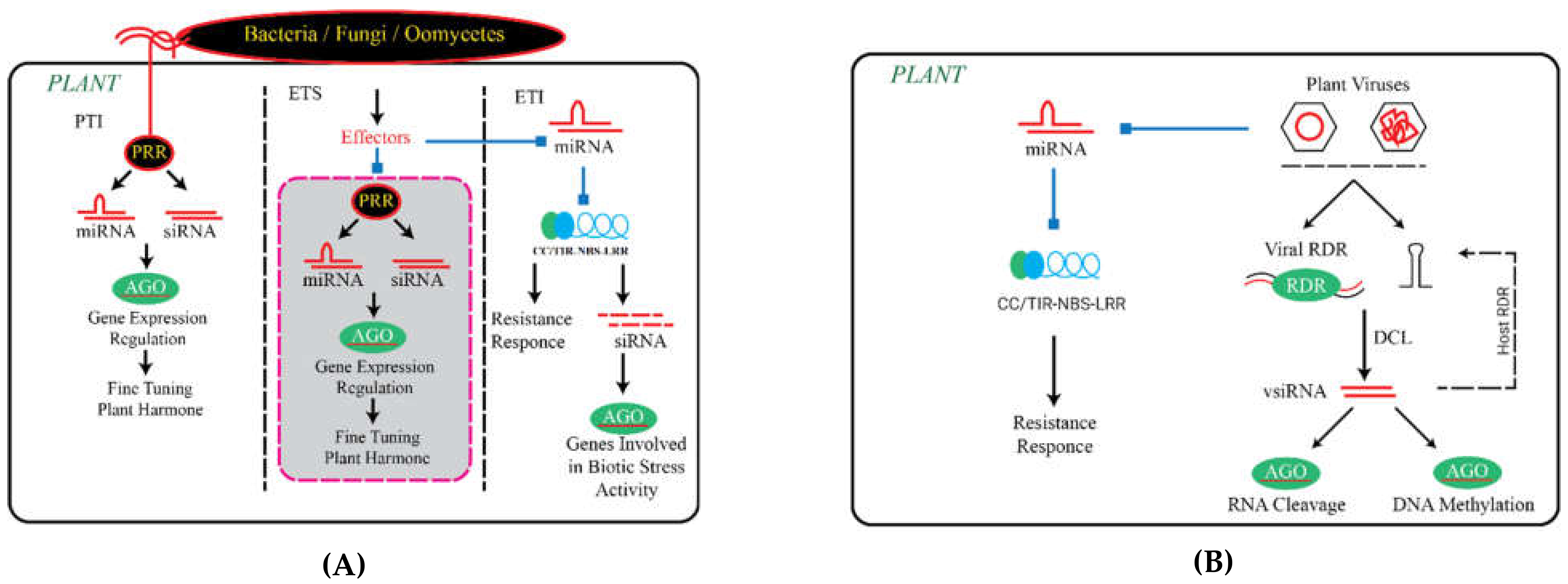
Function of Small RNAs in PTI
3. The Role of Small RNAs in ETI
4. miRNAs Involved in Environmental Stress Responses
5. miRNAs and Secondary Metabolites in the Defense Response of Plants
6. Role of miRNAs in Defense Priming
7. Perspectives and Biotechnological Applications of miRNAs
Host-Induced Gene Silencing
Funding
Conflicts of Interest
References
- Wang, J.; Chai, J. Structural insights into the plant immune receptors PRRs and NLRs. Plant Physiol. 2020, 182, 1566–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Zhang, X.; Zhou, J.; Zhu, S. Nitric oxide signaling in plants. In Postharvest Biology and Nanotechnology of Fruits, Vegetables and Flowers; Wiley: Davis, CA, USA, 2018; ISBN 9781119289470. [Google Scholar]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Mellersh, D.G.; Foulds, I.V.; Higgins, V.J.; Heath, M.C. H2O2plays different roles in determining penetration failure in three diverse plant-fungal interactions. Plant J. 2002, 29, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Morel, J.; Fromentin, J.; Blein, J.-P.; Simon-Plas, F.; Elmayan, T. Rac regulation of NtrbohD, the oxidase responsible for the oxidative burst in elicited tobacco cell. Plant J. 2003, 37, 282–293. [Google Scholar] [CrossRef]
- Rai, K.K.; Pandey, N.; Rai, S.P. Salicylic acid and nitric oxide signaling in plant heat stress. Physiol. Plant 2020, 168, 241–255. [Google Scholar] [CrossRef]
- Sinha, M.; Singh, R.P.; Kushwaha, G.S.; Iqbal, N.; Singh, A.; Kaushik, S.; Kaur, P.; Sharma, S.; Singh, T.P. Current overview of allergens of plant pathogenesis related protein families. Sci. World J. 2014, 2014, 543195. [Google Scholar] [CrossRef] [Green Version]
- Ellinger, D.; Voigt, C.A. Callose biosynthesis in arabidopsis with a focus on pathogen response: What we have learned within the last decade. Ann. Bot. 2014, 114, 1349–1358. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, L.; Birch, P.R.J. The zigzag model of plant-microbe interactions: Is it time to move on? Mol. Plant Pathol. 2014, 15, 865. [Google Scholar] [CrossRef]
- Balint-Kurti, P. The plant hypersensitive response: Concepts, control and consequences. Mol. Plant Pathol. 2019, 20, 1163–1178. [Google Scholar] [CrossRef] [Green Version]
- Seleem, D.; Pardi, V.; Murata, R.M. Review of flavonoids: A diverse group of natural compounds with anti-Candida albicans activity in vitro. Arch. Oral Biol. 2017, 76, 76–83. [Google Scholar] [CrossRef]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as important molecules of plant interactions with the environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef] [PubMed]
- Romeis, T.; Ludwig, A.A.; Martin, R.; Jones, J.D. Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 2001, 20, 5556–5567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Libo, S.; Meng, X.; Shan, L.; He, P. Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 2016, 19, 641–650. [Google Scholar] [CrossRef] [Green Version]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [Green Version]
- Pré, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.J.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [Green Version]
- Shan, T.; Rong, W.; Xu, H.; Du, L.; Liu, X.; Zhang, Z. The wheat R2R3-MYB transcription factor TaRIM1 participates in resistance response against the pathogen Rhizoctonia cerealis infection through regulating defense genes. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef]
- De Vleesschauwer, D.; Xu, J.; Höfte, M. Making sense of hormone-mediated defense networking: From rice to Arabidopsis. Front. Plant Sci. 2014, 5, 611. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Thaler, J.S.; Humphrey, P.T.; Whiteman, N.K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 2012, 17, 260–270. [Google Scholar] [CrossRef]
- De Vleesschauwer, D.; Van Buyten, E.; Satoh, K.; Balidion, J.; Mauleon, R.; Choi, I.-R.; Vera-Cruz, C.; Kikuchi, S.; Höfte, M. Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 2012, 158, 1833–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Vallet, A.; López, G.; Ramos, B.; Cerezo, M.M.D.; Riviere, M.-P.; Llorente, F.; Fernández, P.V.; Miedes, E.; Estevez, J.M.; Grant, M.; et al. Disruption of abscisic acid signaling constitutively activates arabidopsis resistance to the necrotrophic fungus plectosphaerella cucumerina. Plant Physiol. 2012, 160, 2109–2124. [Google Scholar] [CrossRef] [Green Version]
- Métraux, J.P. Systemic acquired resistance. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Elsevier: London, UK, 2013; ISBN 9780080961569. [Google Scholar]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A.H.M. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, H.; Li, X.; Dong, X. Generation of broad-spectrum disease resistance by everexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 1998, 95, 6531–6536. [Google Scholar] [CrossRef] [Green Version]
- Van Loon, L.C.; Bakker, P.A.H.M.; Pieterse, C.M.J. Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 1998, 36, 453–483. [Google Scholar] [CrossRef] [Green Version]
- Ton, J.; Mauch-Mani, B. β-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 2004, 38, 119–130. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, T.; Chang, Y.N.; Duan, C.G. Small RNA functions as a trafficking effector in plant immunity. Int. J. Mol. Sci. 2019, 20, 2816. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Li, W.; Zhang, J.; Wang, L.; Wu, J. Roles of small RNAs in virus-plant interactions. Viruses 2019, 11, 827. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Yang, M.; Zhang, X. The function of small RNAs in plant biotic stress response. J. Integr. Plant Biol. 2016, 58, 312–327. [Google Scholar] [CrossRef] [Green Version]
- Kuan, T.; Zhai, Y.; Ma, W. Small RNAs regulate plant responses to filamentous pathogens. Semin. Cell Dev. Biol. 2016, 56, 190–200. [Google Scholar] [CrossRef] [Green Version]
- Axtell, M.J. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Yang, M.; Lu, L.; Zhang, X. Diverse functions of small RNAs in different plant–pathogen communications. Front. Microbiol. 2016, 7, 1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodersen, P.; Sakvarelidze-Achard, L.; Bruun-Rasmussen, M.; Dunoyer, P.; Yamamoto, Y.Y.; Sieburth, L.; Voinnet, O. Widespread Translational Inhibition by Plant miRNAs and siRNAs. Science 2008, 320, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, L. Comparative analysis of MicroRNA promoters in arabidopsis and rice. Genom. Proteom. Bioinform. 2013, 11, 56–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurihara, Y.; Watanabe, Y. From the cover: Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 2004, 101, 12753–12758. [Google Scholar] [CrossRef] [Green Version]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef] [Green Version]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D.G. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Song, X.; Gu, L.; Li, X.; Cao, S.; Chu, C.; Cui, X.; Chen, X.; Cao, X. NOT2 proteins promote polymerase II–dependent transcription and interact with multiple MicroRNA biogenesis factors in arabidopsis. Plant Cell 2013, 25, 715–727. [Google Scholar] [CrossRef] [Green Version]
- Baldrich, P.; Campo, S.; Wu, M.T.; Liu, T.T.; Hsing, Y.I.C.; Segundo, B.S. MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biol. 2015, 12, 847–863. [Google Scholar] [CrossRef] [Green Version]
- Shuai, P.; Liang, D.; Zhang, Z.; Yin, W.; Xia, X. Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis. BMC Genom. 2013, 14, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Xia, J.; Lii, Y.E.; Barrera-Figueroa, B.E.; Zhou, X.; Gao, S.; Lu, L.; Niu, D.; Chen, Z.; Leung, C.; et al. Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol. 2012, 13, R20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasschau, K.D.; Fahlgren, N.; Chapman, E.J.; Sullivan, C.M.; Cumbie, J.S.; Givan, S.A.; Carrington, J.C. Genome-wide profiling and analysis of arabidopsis siRNAs. PLoS Biol. 2007, 5, e57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fei, Q.; Xia, R.; Meyers, B.C. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 2013, 25, 2400–2415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vazquez, F.; Hohn, T. Biogenesis and biological activity of secondary siRNAs in plants. Science 2013, 2013, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Axtell, M.J. AGO4 is specifically required for heterochromatic siRNA accumulation at Pol V-dependent loci inArabidopsis thaliana. Plant J. 2017, 90, 37–47. [Google Scholar] [CrossRef] [Green Version]
- Curaba, J.; Singh, M.B.; Bhalla, P.L. miRNAs in the crosstalk between phytohormone signalling pathways. J. Exp. Bot. 2014, 65, 1425–1438. [Google Scholar] [CrossRef]
- Dempsey, D.A.; Klessig, D.F. How does the multifaceted plant hormone salicylic acid combat disease in plants and are similar mechanisms utilized in humans? BMC Biol. 2017, 15, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Spoel, S.H.; Johnson, J.S.; Dong, X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 2007, 104, 18842–18847. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhao, H.; Gao, S.; Wang, W.C.; Katiyar-Agarwal, S.; Huang, H.-D.; Raikhel, N.; Jin, H. Arabidopsis argonaute 2 regulates innate immunity via miRNA393∗-mediated silencing of a golgi-localized SNARE gene, MEMB12. Mol. Cell 2011, 42, 356–366. [Google Scholar] [CrossRef] [Green Version]
- Zhou, N.; Tootle, T.L.; Glazebrook, J. Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 1999, 11, 2419–2428. [Google Scholar] [PubMed] [Green Version]
- Nafisi, M.; Goregaoker, S.; Botanga, C.J.; Glawischnig, E.; Olsen, C.E.; Halkier, B.A.; Glazebrook, J. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of Indole-3-acetaldoxime in camalexin synthesis. Plant Cell 2007, 19, 2039–2052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calil, I.P.; Fontes, E.P.B. Plant immunity against viruses: Antiviral immune receptors in focus. Ann. Bot. 2017, 119, mcw200–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldrich, P.; Segundo, B.S. MicroRNAs in rice innate immunity. Rice 2016, 9, 6. [Google Scholar] [CrossRef] [Green Version]
- Weiberg, A.; Wang, M.; Lin, F.-M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef] [Green Version]
- Xia, K.; Wang, R.; Ou, X.; Fang, Z.; Tian, C.; Duan, J.; Wang, Y.; Zhang, M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 2012, 7, e30039. [Google Scholar] [CrossRef]
- Pandey, P.; Srivastava, P.K.; Pandey, S.P. Prediction of plant miRNA targets. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2019; pp. 99–107. [Google Scholar]
- Li, Y.; Zhang, Q.; Zhang, J.; Wu, L.; Qi, Y.; Zhou, J.-M. Identification of MicroRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010, 152, 2222–2231. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Lu, Y.-G.; Shi, Y.; Wu, L.; Xu, Y.-J.; Huang, F.; Guo, X.-Y.; Zhang, Y.; Fan, J.; Zhao, J.-Q.; et al. Multiple rice MicroRNAs are involved in immunity against the blast fungus magnaporthe oryzae. Plant Physiol. 2014, 164, 1077–1092. [Google Scholar] [CrossRef] [Green Version]
- Dunoyer, P.; Himber, C.; Voinnet, O. Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat. Genet. 2006, 38, 258–263. [Google Scholar] [CrossRef]
- Jagadeeswaran, G.; Saini, A.; Sunkar, R. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 2009, 229, 1009–1014. [Google Scholar] [CrossRef]
- Zhang, W.; Gao, S.; Zhou, X.; Chellappan, P.; Chen, Z.; Zhou, X.; Zhang, X.; Fromuth, N.; Coutino, G.; Coffey, M.; et al. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol. Biol. 2010, 75, 93–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.J.; Park, Y.J.; Kwak, K.J.; Kim, D.; Park, J.H.; Lim, J.Y.; Shin, C.; Yang, K.-Y.; Kang, H. MicroRNA844-guided downregulation of cytidinephosphate diacylglycerol synthase3 (CDS3) mRNA Affects the response of arabidopsis thaliana to bacteria and fungi. Mol. Plant Microbe Interact. 2015, 28, 892–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, O.P.; Permar, V.; Koundal, V.; Singh, U.D.; Praveen, S. MicroRNA regulated defense responses in Triticum aestivum L. during Puccinia graminis f.sp. tritici infection. Mol. Biol. Rep. 2011, 39, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Hammond-Kosack, K.E.; Kanyuka, K. Resistance genes (R genes) in plants. In eLS; Wiley: Hoboken, NJ, USA, 2007. [Google Scholar]
- Stergiopoulos, I.; Burg, H.A.V.D.; Okmen, B.; Beenen, H.G.; Van Liere, S.; Kema, G.H.J.; De Wit, P.J.G.M. Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc. Natl. Acad. Sci. USA 2010, 107, 7610–7615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.A.; Yeom, S.I. Plant NB-LRR proteins: Tightly regulated sensors in a complex manner. Brief. Funct. Genom. 2015, 14, 233–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, P.; Bhattacharya, A.; Char, B. Manipulating disease and pest resistance pathways in plants for enhanced crop improvement. Biosci. Biotechnol. Res. Commun. 2017, 10, 631–644. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Q.; Hao, L.; Wang, S.; Wang, S.; Zhang, W.; Xu, C.; Yu, Y.; Li, T. A novel miRNA negatively regulates resistance to Glomerella leaf spot by suppressing expression of an NBS gene in apple. Hortic. Res. 2019, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Yi, H.; Richards, E.J. A cluster of disease resistance genes in arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell 2007, 19, 2929–2939. [Google Scholar] [CrossRef] [Green Version]
- Thiebaut, F.; Grativol, C.; Carnavale-Bottino, M.; Rojas, C.A.; Tanurdzic, M.; Farinelli, L.; Martienssen, R.A.; Hemerly, A.S.; Ferreira, P.C.G. Computational identification and analysis of novel sugarcane microRNAs. BMC Genom. 2012, 13, 290. [Google Scholar] [CrossRef] [Green Version]
- Carrà, A.; Mica, E.; Gambino, G.; Pindo, M.; Moser, C.; Pè, M.E.; Schubert, A. Cloning and characterization of small non-coding RNAs from grape. Plant J. 2009, 59, 750–763. [Google Scholar] [CrossRef]
- Song, C.; Wang, C.; Zhang, C.; Kibet, K.N.; Yu, H.; Ma, Z.; Fang, J. Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genom. 2010, 11, 431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Lu, Y.; Bai, S.; Zhang, W.; Duan, X.; Meng, N.; Wang, Z.; Wang, A.; Zhou, Z.; Li, T.; et al. Cloning and characterization of miRNAs and their targets, including a novel miRNA-targeted NBS–LRR protein class gene in apple (golden delicious). Mol. Plant 2014, 7, 218–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivaprasad, P.V.; Chen, H.-M.; Patel, K.; Bond, D.M.; Santos, B.A.; Baulcombe, D.C. A MicroRNA superfamily regulates nucleotide binding site–leucine-rich repeats and other mRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef] [Green Version]
- Zhai, J.; Jeong, D.-H.; De Paoli, E.; Park, S.; Rosen, B.D.; Li, Y.; González, A.J.; Yan, Z.; Kitto, S.L.; Grusak, M.A.; et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011, 25, 2540–2553. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, S.; Park, G.; Atamian, H.S.; Han, C.S.; Stajich, J.E.; Kaloshian, I.; Borkovich, K.A. MicroRNAs suppress NB domain genes in tomato that confer resistance to fusarium oxysporum. PLOS Pathog. 2014, 10, e1004464. [Google Scholar] [CrossRef]
- Boccara, M.; Sarazin, A.; Thiébeauld, O.; Jay, F.; Voinnet, O.; Navarro, L.; Colot, V. The arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLOS Pathog. 2014, 10, e1003883. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Cheng, X.; Liu, D.; Xu, W.; Wise, R.; Shen, Q.H. The miR9863 family regulates distinct Mla alleles in barley to attenuate NLR receptor-triggered disease resistance and cell-death signaling. PLoS Genet. 2014, 10, e1004755. [Google Scholar] [CrossRef] [Green Version]
- Katiyar-Agarwal, S.; Morgan, R.; Dahlbeck, D.; Borsani, O.; Villegas, A.; Zhu, J.-K.; Staskawicz, B.J.; Jin, H. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 2006, 103, 18002–18007. [Google Scholar] [CrossRef] [Green Version]
- Katiyar-Agarwal, S.; Gao, S.; Vivian-Smith, A.; Jin, H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 2007, 21, 3123–3134. [Google Scholar] [CrossRef] [Green Version]
- Niu, D.; Lii, Y.E.; Chellappan, P.; Lei, L.; Peralta, K.; Jiang, C.; Guo, J.; Coaker, G.; Jin, H. miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nat. Commun. 2016, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, S.; Datta, S.K.; Datta, K. miRNA regulation of nutrient homeostasis in plants. Front. Plant Sci. 2015, 6, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Tao, Y.; Zhu, C. Emerging roles of microRNAs in the mediation of drought stress response in plants. J. Exp. Bot. 2013, 64, 3077–3086. [Google Scholar] [CrossRef] [PubMed]
- Megha, S.; Basu, U.; Kav, N.N. V Regulation of low temperature stress in plants by microRNAs. Plant Cell Environ. 2018, 41, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B. MicroRNA: A new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 2015, 66, 1749–1761. [Google Scholar] [CrossRef] [PubMed]
- Rajwanshi, R.; Devi, K.J.; Sharma, G.R.; Lal, B. Role of miRNAs in plant-microbe interaction. In In Vitro Plant Breeding towards Novel Agronomic Traits; Springer: Singapore, 2019; pp. 167–195. [Google Scholar]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant MicroRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Ai, Q.; Liang, G.; Zhang, H.; Yu, D. Control of sulfate concentration by miR395-targeted APS genes in Arabidopsis thaliana. Plant Divers. 2016, 38, 92–100. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Pan, X.P.; Wang, Q.L.; Cobb, G.P.; Anderson, T.A. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005, 15, 336–360. [Google Scholar] [CrossRef] [Green Version]
- Sunkar, R.; Zhu, J.K. Novel and stress regulated microRNAs and other small RNAs from Arabidopsis w inside box sign. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Sun, Y.-H.; Shi, R.; Clark, C.; Li, L.; Chiang, V.L. Novel and mechanical stress-responsive microRNAs in populus trichocarpa that are absent from Arabidopsis. Plant Cell 2005, 17, 2186–2203. [Google Scholar] [CrossRef] [Green Version]
- Achard, P.; Herr, A.; Baulcombe, D.C.; Harberd, N.P. Modulation of floral development by a gibberellin-regulated microRNA. Development 2004, 131, 3357–3365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waheed, S.; Zeng, L. The critical role of miRNAs in regulation of flowering time and flower development. Genes 2020, 11, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bej, S.; Basak, J. MicroRNAs: The potential biomarkers in plant stress response. Am. J. Plant Sci. 2014, 5, 748–759. [Google Scholar] [CrossRef] [Green Version]
- Omidvar, V.; Mohorianu, I.; Dalmay, T.; Zheng, Y.; Fei, Z.; Pucci, A.; Mazzucato, A.; Večeřová, V.; Sedlářova, M.; Fellner, M. Transcriptional regulation of male-sterility in 7B-1 male-sterile tomato mutant. PLoS ONE 2017, 12, e0170715. [Google Scholar] [CrossRef] [PubMed]
- Kulcheski, F.R.; De Oliveira, L.F.; Molina, L.G.; Almerão, M.P.; Rodrigues, F.A.; Marcolino, J.; Barbosa, J.F.; Stolf, R.; Nepomuceno, A.L.; Marcelino-Guimarães, F.C.; et al. Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genom. 2011, 12, 307–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuhegger, R.; Nafisi, M.; Mansourova, M.; Petersen, B.L.; Olsen, C.E.; Svatoš, A.; Halkier, B.A.; Glawischnig, E. CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 2006, 141, 1248–1254. [Google Scholar] [CrossRef] [Green Version]
- Robert-Seilaniantz, A.; MacLean, D.; Jikumaru, Y.; Hill, L.; Yamaguchi, S.; Kamiya, Y.; Jones, J.D. The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J. 2011, 67, 218–231. [Google Scholar] [CrossRef]
- Marcela, V.-H.; Gerardo, V.-M.; Agustín, A.-R.C.; Antonio, G.-M.M.; Oscar, R.; Diego, C.-P.; Cruz-Hernández, A. MicroRNAs associated with secondary metabolites production. In Plant Physiological Aspects of Phenolic Compounds; IntechOpen: London, UK, 2019. [Google Scholar]
- Padhan, J.K.; Kumar, P.; Sood, H.; Chauhan, R.S. Prospecting NGS-transcriptomes to assess regulation of miRNA-mediated secondary metabolites biosynthesis in Swertia chirayita, a medicinal herb of the North-Western Himalayas. Med. Plants 2016, 8, 219–228. [Google Scholar] [CrossRef]
- Qiao, Y.; Zhang, J.; Zhang, J.; Wang, Z.; Ran, A.; Guo, H.; Wang, D.; Zhang, J. Integrated RNA-seq and sRNA-seq analysis reveals miRNA effects on secondary metabolism in Solanum tuberosum L. Mol. Genet. Genom. 2017, 292, 37–52. [Google Scholar] [CrossRef]
- Jian, H.; Yang, B.; Zhang, A.; Ma, J.-Q.; Ding, Y.; Chen, Z.; Li, J.-N.; Xu, X.; Liu, L. Genome-wide identification of micrornas in response to cadmium stress in oilseed rape (Brassica napus L.) using high-throughput sequencing. Int. J. Mol. Sci. 2018, 19, 1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aires, A.; Mota, V.R.; Saavedra, M.J.; Monteiro, A.A.; Simões, M.; Rosa, E.A.S.; Bennett, R.N. Initial in vitro evaluations of the antibacterial activities of glucosinolate enzymatic hydrolysis products against plant pathogenic bacteria. J. Appl. Microbiol. 2009, 106, 2096–2105. [Google Scholar] [CrossRef] [PubMed]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biała, W.; Jasiński, M. The phenylpropanoid case–it is transport that matters. Front. Plant Sci. 2018, 9, 1610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Yuan, Y.; Wang, Y.; Jiang, C.; Chen, T.; Zhu, F.; Zhao, Y.; Zhou, J.; Huang, L. Regulation of fatty acid and flavonoid biosynthesis by miRNAs in Lonicera japonica. RSC Adv. 2017, 7, 35426–35437. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.; Sharma, A. Turmeric (Curcuma longa): miRNAs and their regulating targets are involved in development and secondary metabolite pathways. Comptes Rendus Biol. 2017, 340, 481–491. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Avramenko, T.V. New opportunities for the regulation of secondary metabolism in plants: Focus on microRNAs. Biotechnol. Lett. 2015, 37, 1719–1727. [Google Scholar] [CrossRef]
- Mouradov, A.; Spangenberg, G. Flavonoids: A metabolic network mediating plants adaptation to their real estate. Front. Plant Sci. 2014, 5, 620. [Google Scholar] [CrossRef]
- Camargo-Ramírez, R.; Val-Torregrosa, B.; San Segundo, B. MiR858-mediated regulation of flavonoid-specific MYB transcription factor genes controls resistance to pathogen infection in Arabidopsis. Plant Cell Physiol. 2018, 59, 190–204. [Google Scholar] [CrossRef]
- Srivastava, S.; Singh, R.; Srivastava, G.; Sharma, A. Comparative study of withanolide biosynthesis-related miRNAs in root and leaf tissues of withania somnifera. Appl. Biochem. Biotechnol. 2018, 185, 1145–1159. [Google Scholar] [CrossRef]
- Zhang, M.; Dong, Y.; Nie, L.; Lu, M.; Fu, C.; Yu, L.J. High-throughput sequencing reveals miRNA effects on the primary and secondary production properties in long-term subcultured Taxus cells. Front. Plant Sci. 2015, 6, 604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Medina, A.; Flors, V.; Heil, M.; Mauch-Mani, B.; Pieterse, C.M.J.; Pozo, M.J.; Ton, J.; Van Dam, N.M.; Conrath, U. Recognizing plant defense priming. Trends Plant Sci. 2016, 21, 818–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luna, E.; Bruce, T.J.; Roberts, M.R.; Flors, V.; Ton, J. Next-generation systemic acquired resistance. Plant Physiol. 2011, 158, 844–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, C.; Yu, J.; Bai, J.; Zhu, Z.; Wang, X. Induced defense responses in rice plants against small brown planthopper infestation. Crop. J. 2014, 2, 55–62. [Google Scholar] [CrossRef] [Green Version]
- Soto-Suárez, M.; Baldrich, P.; Weigel, D.; Rubio-Somoza, I.; Segundo, B.S. The Arabidopsis miR396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens. Sci. Rep. 2017, 7, 44898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holt, D.B.; Gupta, V.; Meyer, D.; Abel, N.B.; Andersen, S.U.; Stougaard, J.; Markmann, K. MicroRNA 172 (miR172) signals epidermal infection and is expressed in cells primed for bacterial invasion in Lotus japonicus roots and nodules. New Phytol. 2015, 208, 241–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chojak-Koźniewska, J.; Kuźniak, E.; Zimny, J. The effects of combined abiotic and pathogen stress in plants: Insights from salinity and pseudomonas syringae pv lachrymans interaction in cucumber. Front. Plant Sci. 2018, 9, 1691. [Google Scholar] [CrossRef]
- Wingard, S.A. Hosts and Symptoms of Ring Spot, A Virus Disease of Plants; Journal of Agriculture Research; Authority of the Secretary of Agriculture with the Cooperation of Lapid-Grant Colleges and Universities: Washington, DC, USA, 1928. [Google Scholar]
- Sanford, J.; Johnston, S. The concept of parasite-derived resistance—Deriving resistance genes from the parasite’s own genome. J. Theor. Biol. 1985, 113, 395–405. [Google Scholar] [CrossRef]
- Nunes, C.C.; Dean, R.A. Host-induced gene silencing: A tool for understanding fungal host interaction and for developing novel disease control strategies. Mol. Plant Pathol. 2011, 13, 519–529. [Google Scholar] [CrossRef]
- Koch, A.; Kogel, K.H. New wind in the sails: Improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol. J. 2014, 12, 821–831. [Google Scholar] [CrossRef]
- Gonsalves, D. Control of papaya ringspot virus in papaya: A case study. Annu. Rev. Phytopathol. 1998, 36, 415–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basso, M.F.; Ferreira, P.C.G.; Kobayashi, A.K.; Harmon, F.G.; Nepomuceno, A.L.; Molinari, H.B.C.; Grossi-de-Sa, M.F. Micro RNA s and new biotechnological tools for its modulation and improving stress tolerance in plants. Plant Biotechnol. J. 2019, 17, 1482–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Zhang, N.; Shen, W.; Li, J.-F. Engineered artificial MicroRNA precursors facilitate cloning and gene silencing in arabidopsis and rice. Int. J. Mol. Sci. 2019, 20, 5620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Song, X.; Wang, G.; Yang, K.; Wang, Y.; Niu, L.; Chen, X.; Fang, R. Plant-generated artificial small RNAs mediated aphid resistance. PLoS ONE 2014, 9, e97410. [Google Scholar] [CrossRef] [Green Version]
- Guo, Q.; Liu, Q.; Smith, N.A.; Liang, G.; Wang, M.-B. RNA silencing in plants: Mechanisms, technologies and applications in horticultural crops. Curr. Genom. 2016, 17, 476–489. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Fu, B.Y.; Gao, Y.M.; Wang, W.S.; Xu, J.L.; Zhang, F.; Zhao, X.Q.; Zheng, T.Q.; Zhou, Y.L.; Zhang, G.; et al. The 3000 rice genomes project. Gigascience 2014, 3, 2047–217X. [Google Scholar]
- Ferdous, J.; Whitford, R.; Nguyen, M.; Brien, C.; Langridge, P.; Tricker, P.J. Drought-inducible expression of Hv-miR827 enhances drought tolerance in transgenic barley. Funct. Integr. Genom. 2017, 17, 279–292. [Google Scholar] [CrossRef]
- Liu, S.R.; Zhou, J.J.; Hu, C.G.; Wei, C.L.; Zhang, J.Z. MicroRNA-mediated gene silencing in plant defense and viral counter-defense. Front. Microbiol. 2017, 8, 1801. [Google Scholar] [CrossRef]
- Kamthan, A.; Chaudhuri, A.; Kamthan, M.; Datta, A. Small RNAs in plants: Recent development and application for crop improvement. Front. Plant Sci. 2015, 6, 208. [Google Scholar] [CrossRef] [Green Version]
- Manghwar, H.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/cas system: Recent advances and future prospects for genome editing. Trends Plant Sci. 2019, 24, 1102–1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schenke, D.; Cai, D. Applications of CRISPR/cas to improve crop disease resistance: Beyond inactivation of susceptibility factors. iScience 2020, 23, 101478. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, C.; Liu, W.; Gao, W.; Liu, C.; Song, G.; Li, W.-X.; Mao, L.; Chen, B.; Xu, Y. An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci. Rep. 2016, 6, 23890. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Buchner, J. Hsp90: Chaperoning signal transduction. J. Cell. Physiol. 2001, 188, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Will, T.; Vilcinskas, A. Aphid-proof plants: Biotechnology-based approaches for aphid control. In Yellow Biotechnology II; Springer: Berlin/Heidelberg, Germany, 2013; pp. 179–203. [Google Scholar]
- Macedo, L.; Antonino, J.D.; Coelho, R.; Fonseca, F.C.D.A.; Firmino, A.; Silva, M.; Fragoso, R.; Albuquerque, E.; Engler, J.D.A.; Terra, W.; et al. Knocking down chitin synthase 2 by RNAi is lethal to the cotton boll weevil. Biotechnol. Res. Innov. 2017, 1, 72–86. [Google Scholar] [CrossRef]
- Lilley, C.J.; Bakhetia, M.; Charlton, W.L.; Urwin, P.E. Recent progress in the development of RNA interference for plant parasitic nematodes. Mol. Plant Pathol. 2007, 8, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.; Gheysen, G.; Subramaniam, K. RNA interference in Pratylenchus coffeae: Knock down of Pc-pat-10 and Pc-unc-87 impedes migration. Mol. Biochem. Parasitol. 2012, 186, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.A.C.; Jones, M.G.; Fosu-Nyarko, J. Gene silencing in root lesion nematodes (Pratylenchus spp.) significantly reduces reproduction in a plant host. Exp. Parasitol. 2013, 133, 166–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, B.C.; Veluthambi, K.; Subramaniam, K. Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Mol. Biochem. Parasitol. 2006, 148, 219–222. [Google Scholar] [CrossRef]
- Ghag, S.B. Host induced gene silencing, an emerging science to engineer crop resistance against harmful plant pathogens. Physiol. Mol. Plant Pathol. 2017, 100, 242–254. [Google Scholar] [CrossRef]
- Sivamani, E.; Brey, C.W.; Dyer, W.E.; Talbert, L.E.; Qu, R. Resistance to wheat streak mosaic virus in transgenic wheat expressing the viral replicase (NIb) gene. Mol. Breed. 2000, 6, 469–477. [Google Scholar] [CrossRef]
- Nowara, D.; Gay, A.; Lacomme, C.; Shaw, J.; Ridout, C.; Douchkov, D.; Hensel, G.; Kumlehn, J.; Schweizer, P. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen blumeria graminis. Plant Cell 2010, 22, 3130–3141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saurabh, S.; Vidyarthi, A.S.; Prasad, D. RNA interference: Concept to reality in crop improvement. Planta 2014, 239, 543–564. [Google Scholar] [CrossRef] [Green Version]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, S.-D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef] [Green Version]
- Khatri, M.; Rajam, M. V Targeting polyamines of Aspergillus nidulans by siRNA specific to fungal ornithine decarboxylase gene. Sabouraudia 2007, 45, 211–220. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Weiberg, A.; Lin, F.-M.; Thomma, B.P.H.J.; Huang, H.-D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 1–10. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Liu, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2011, 22, 107–126. [Google Scholar] [CrossRef]
- Zhang, J.; Khan, S.A.; Heckel, D.G.; Bock, R. Next-generation insect-resistant plants: RNAi-mediated crop protection. Trends Biotechnol. 2017, 35, 871–882. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waheed, S.; Anwar, M.; Saleem, M.A.; Wu, J.; Tayyab, M.; Hu, Z. The Critical Role of Small RNAs in Regulating Plant Innate Immunity. Biomolecules 2021, 11, 184. https://doi.org/10.3390/biom11020184
Waheed S, Anwar M, Saleem MA, Wu J, Tayyab M, Hu Z. The Critical Role of Small RNAs in Regulating Plant Innate Immunity. Biomolecules. 2021; 11(2):184. https://doi.org/10.3390/biom11020184
Chicago/Turabian StyleWaheed, Saquib, Muhammad Anwar, Muhammad Asif Saleem, Jinsong Wu, Muhammad Tayyab, and Zhangli Hu. 2021. "The Critical Role of Small RNAs in Regulating Plant Innate Immunity" Biomolecules 11, no. 2: 184. https://doi.org/10.3390/biom11020184
APA StyleWaheed, S., Anwar, M., Saleem, M. A., Wu, J., Tayyab, M., & Hu, Z. (2021). The Critical Role of Small RNAs in Regulating Plant Innate Immunity. Biomolecules, 11(2), 184. https://doi.org/10.3390/biom11020184






