SIRT1 Activation Using CRISPR/dCas9 Promotes Regeneration of Human Corneal Endothelial Cells through Inhibiting Senescence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Culture of Human Corneal Endothelial Cells (hCECs)
2.2. Cell Transfection
2.3. Cell Shape Evaluation
2.4. Cell Viability and Proliferation Assay
2.5. Mitochondrial Oxidative Stress Measurement
2.6. MitoTracker Red and Lysosome Staining
2.7. Senescence-Associated-β-Galactosidase (SA-β-gal) Assay
2.8. Western Blotting
2.9. Real Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.10. Cell Cycle Analysis
2.11. Mitochondrial Membrane Potential
2.12. Animal Study and In Vivo Transfection
2.13. Clinical Evaluation and Alizarin Red S Staining
2.14. Statistics
3. Results
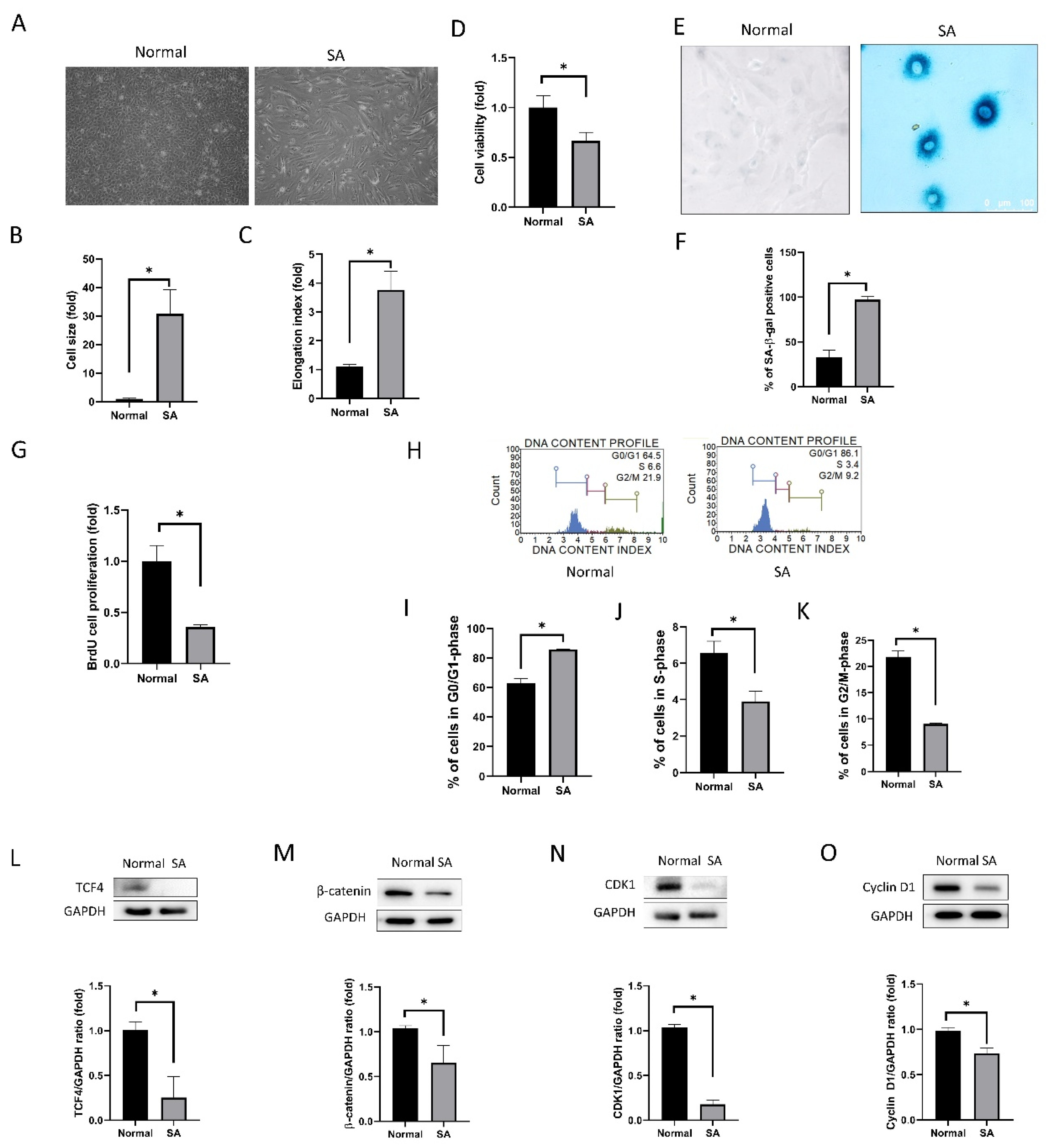
3.1. Senescence of Human Corneal Endothelial Cells
3.1.1. Cell Shape, Viability, Senescence, Proliferation, and Cell Cycle Analyses
3.1.2. Mitochondrial Oxidative Stress, MitoTracker Red Staining, and Lysosome Staining
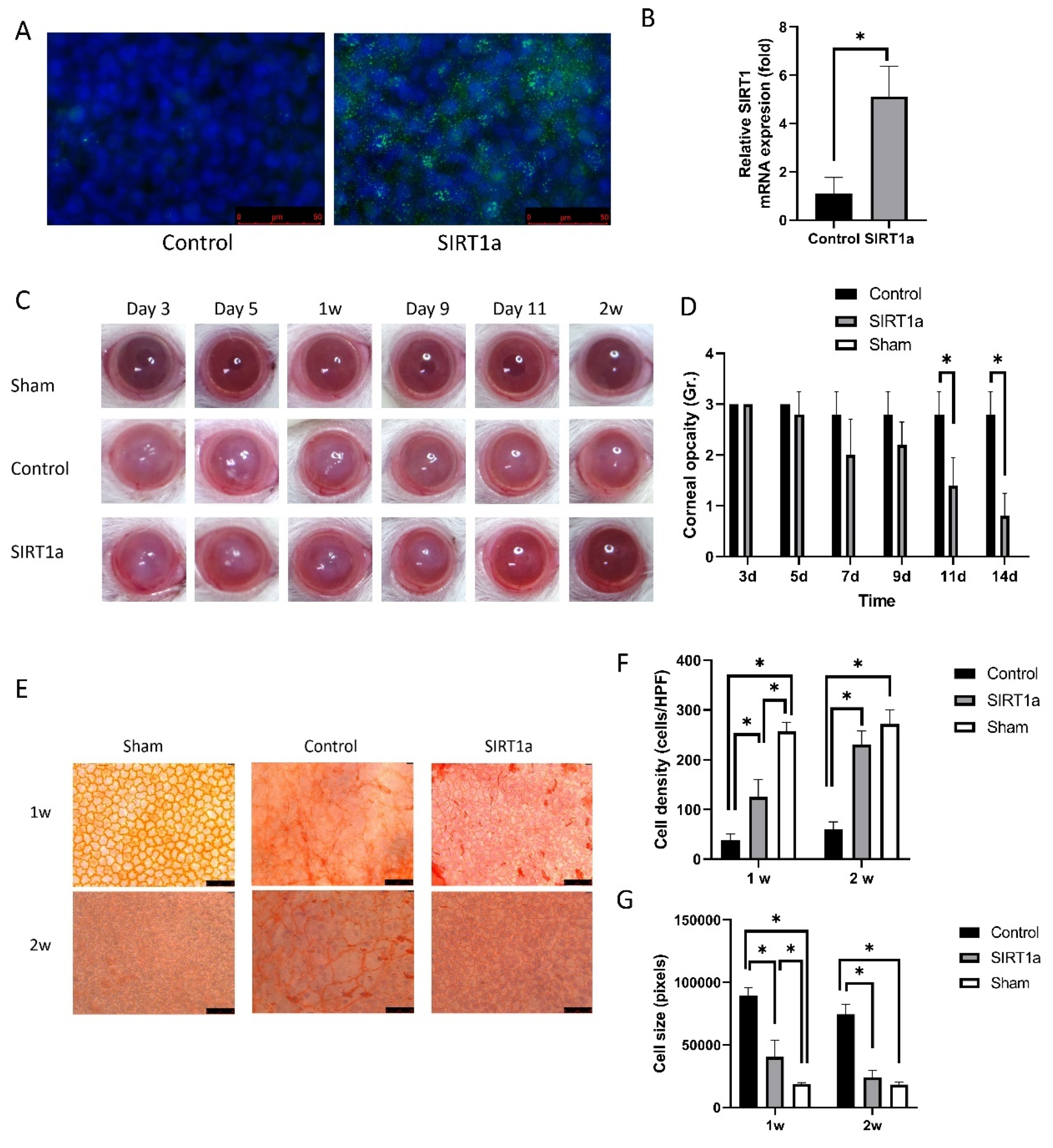
3.1.3. Animal Study of SIRT1 Activation Using CRISPR/dCas9 in Rat Corneal Endothelial Cells
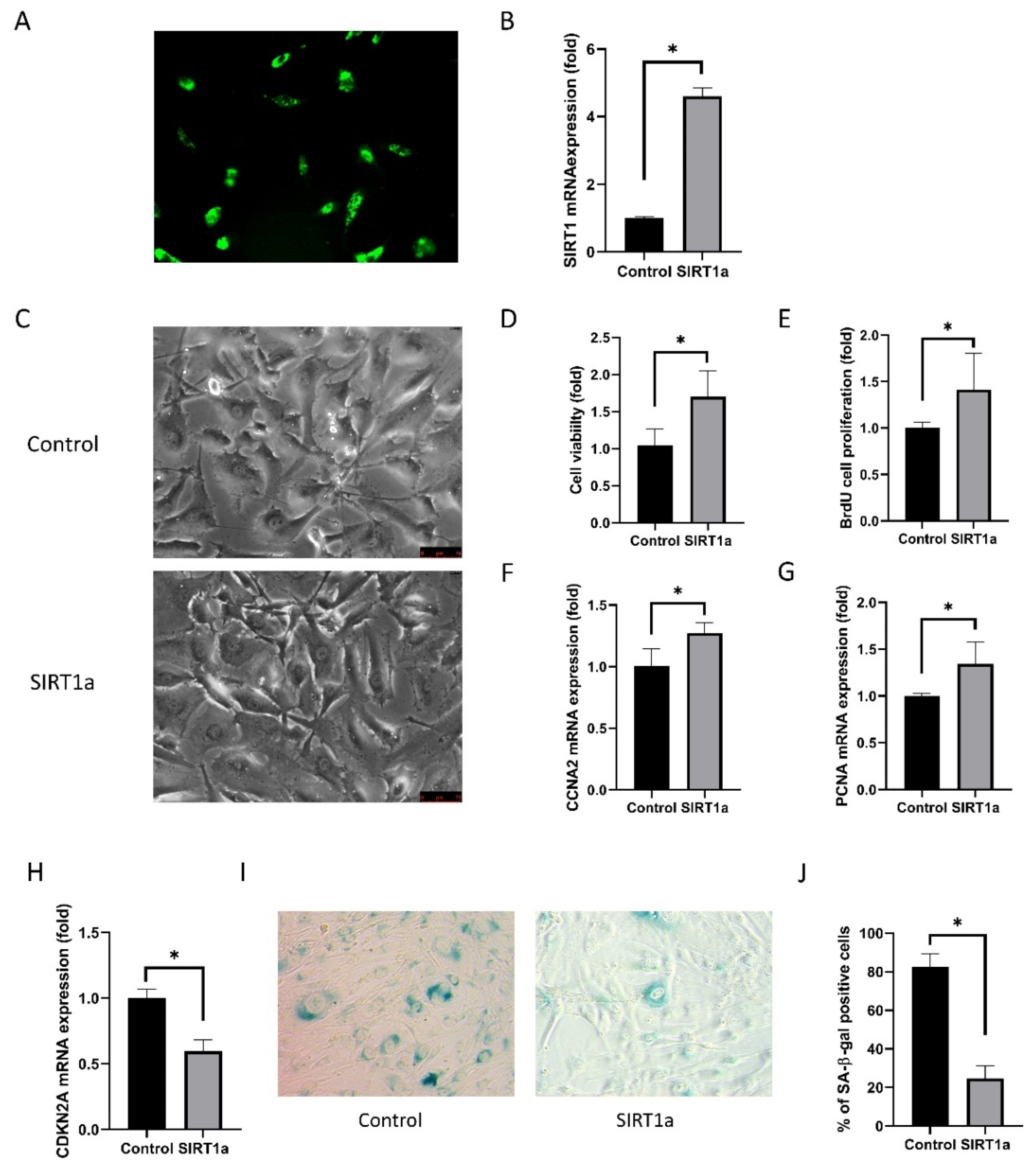
3.1.4. SIRT1 Activation Using CRISPR/dCas9 in Cultured Human Corneal Endothelial Cells
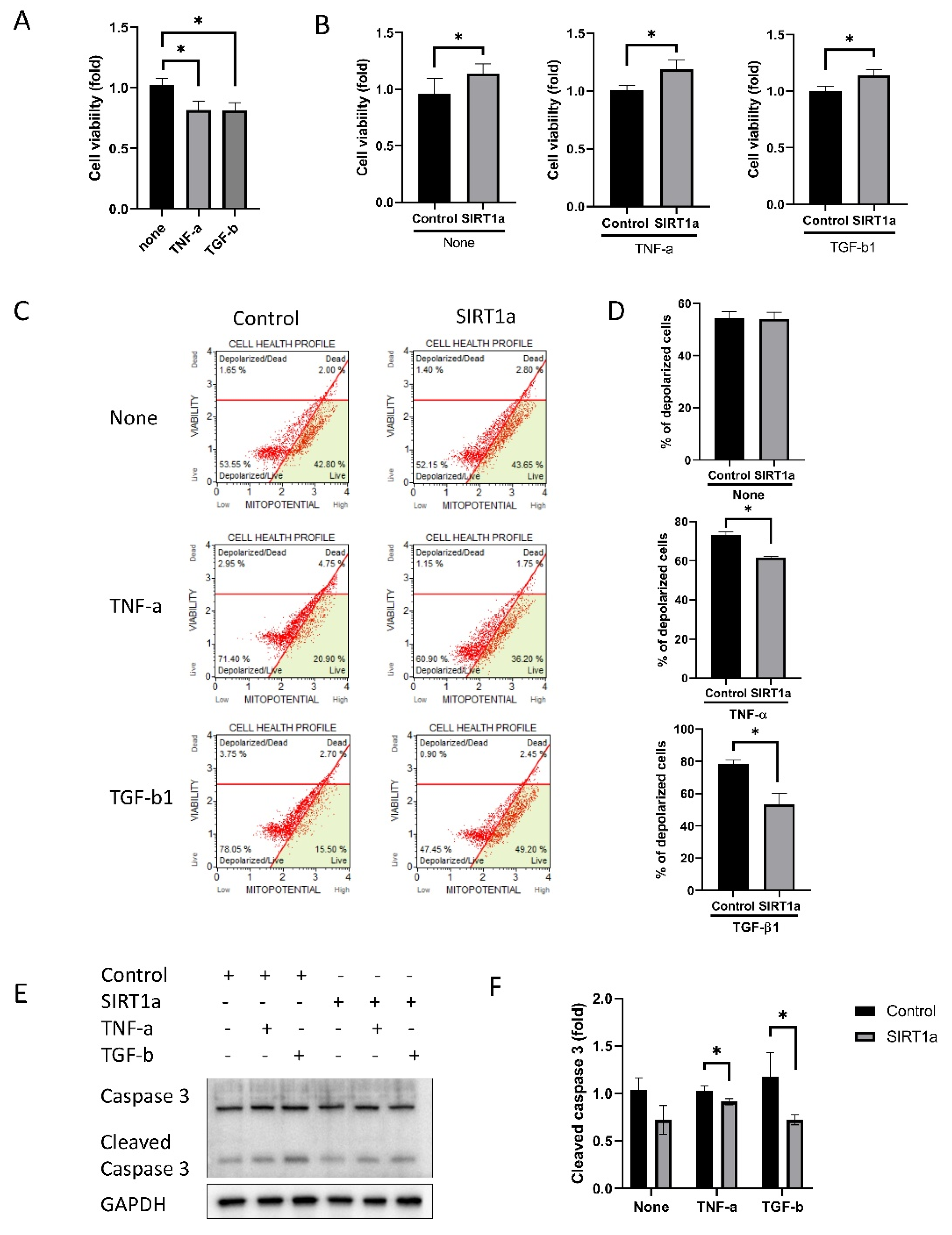
3.1.5. SIRT1 Inhibits Cytokine-Induced Cell Death
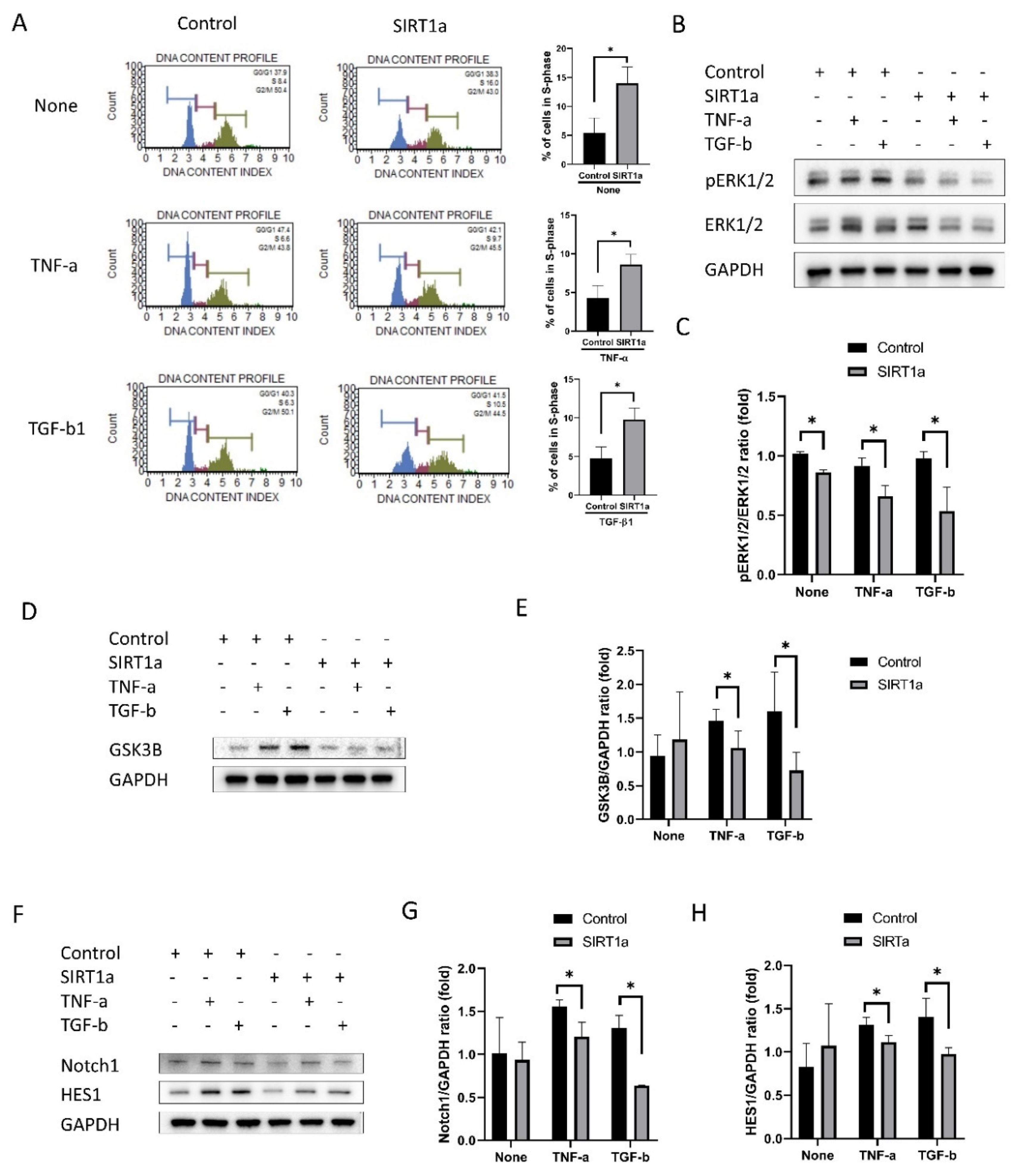
3.1.6. SIRT1 Inhibits Cytokine-Induced Cell Cycle Arrest
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Treffers, W.F. Human corneal endothelial wound repair. In vitro and in vivo. Ophthalmology 1982, 89, 605–613. [Google Scholar] [CrossRef]
- Nayak, S.K.; Binder, P.S. The growth of endothelium from human corneal rims in tissue culture. Invest. Ophthalmol. Vis. Sci 1984, 25, 1213–1216. [Google Scholar] [PubMed]
- Joyce, N.C. Proliferative capacity of the corneal endothelium. Prog. Retin. Eye Res. 2003, 22, 359–389. [Google Scholar] [CrossRef]
- Senoo, T.; Joyce, N.C. Cell cycle kinetics in corneal endothelium from old and young donors. Invest. Ophthalmol. Vis. Sci. 2000, 41, 660–667. [Google Scholar] [PubMed]
- Ueno, M.; Asada, K.; Toda, M.; Hiraga, A.; Montoya, M.; Sotozono, C.; Kinoshita, S.; Hamuro, J. MicroRNA Profiles Qualify Phenotypic Features of Cultured Human Corneal Endothelial Cells. Invest. Ophthalmol. Vis. Sci. 2016, 57, 5509–5517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mammone, T.; Gan, D.; Foyouzi-Youssefi, R. Apoptotic cell death increases with senescence in normal human dermal fibroblast cultures. Cell Biol. Int. 2006, 30, 903–909. [Google Scholar] [CrossRef] [PubMed]
- Tosato, M.; Zamboni, V.; Ferrini, A.; Cesari, M. The aging process and potential interventions to extend life expectancy. Clin. Interv. Aging 2007, 2, 401–412. [Google Scholar] [PubMed]
- Prigione, A.; Fauler, B.; Lurz, R.; Lehrach, H.; Adjaye, J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem. Cells 2010, 28, 721–733. [Google Scholar] [CrossRef]
- Kryczka, T.; Szaflik, J.P.; Szaflik, J.; Midelfart, A. Influence of donor age, post-mortem time and cold storage on metabolic profile of human cornea. Acta Ophthalmol. 2013, 91, 83–87. [Google Scholar] [CrossRef]
- Pillarisetti, S. A review of Sirt1 and Sirt1 modulators in cardiovascular and metabolic diseases. Recent Pat. Cardiovasc. Drug Discov. 2008, 3, 156–164. [Google Scholar] [CrossRef]
- Nakagawa, T.; Guarente, L. Sirtuins at a glance. J. Cell Sci. 2011, 124, 833–838. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Qin, Q.; Chen, R.; Wei, C.; Mo, Q. SIRT1 promotes proliferation, migration, and invasion of breast cancer cell line MCF-7 by upregulating DNA polymerase delta1 (POLD1). Biochem. Biophys. Res. Commun. 2018, 502, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Rathbone, C.R.; Booth, F.W.; Lees, S.J. Sirt1 increases skeletal muscle precursor cell proliferation. Eur. J. Cell Biol. 2009, 88, 35–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, W.; Li, J.; Wang, Y.; Meng, J.; Zheng, G. miR-221 promotes lens epithelial cells apoptosis through interacting with SIRT1 and E2F3. Chem. Biol. Interact. 2019, 306, 39–46. [Google Scholar] [CrossRef]
- Jaliffa, C.; Ameqrane, I.; Dansault, A.; Leemput, J.; Vieira, V.; Lacassagne, E.; Provost, A.; Bigot, K.; Masson, C.; Menasche, M.; et al. Sirt1 involvement in rd10 mouse retinal degeneration. Invest. Ophthalmol. Vis. Sci. 2009, 50, 3562–3572. [Google Scholar] [CrossRef] [Green Version]
- Alcendor, R.R.; Gao, S.; Zhai, P.; Zablocki, D.; Holle, E.; Yu, X.; Tian, B.; Wagner, T.; Vatner, S.F.; Sadoshima, J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ. Res. 2007, 100, 1512–1521. [Google Scholar] [CrossRef]
- Tang, B.L. Sirt1 and the Mitochondria. Mol. Cells 2016, 39, 87–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aquilano, K.; Baldelli, S.; Pagliei, B.; Ciriolo, M.R. Extranuclear localization of SIRT1 and PGC-1alpha: An insight into possible roles in diseases associated with mitochondrial dysfunction. Curr. Mol. Med. 2013, 13, 140–154. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.T.; Kolesar, J.E.; Kaufman, B.A. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim. Biophys. Acta 2012, 1819, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, Y.; Burns, N.; Zhao, K.S.; Song, R. SIRT1 is required for mitochondrial biogenesis reprogramming in hypoxic human pulmonary arteriolar smooth muscle cells. Int. J. Mol. Med. 2017, 39, 1127–1136. [Google Scholar] [CrossRef] [Green Version]
- Miyai, T. Fuchs Endothelial Corneal Dystrophy and Mitochondria. Cornea 2018, 37 (Suppl. 1), S74–S77. [Google Scholar] [CrossRef] [PubMed]
- Aldrich, B.T.; Schlotzer-Schrehardt, U.; Skeie, J.M.; Burckart, K.A.; Schmidt, G.A.; Reed, C.R.; Zimmerman, M.B.; Kruse, F.E.; Greiner, M.A. Mitochondrial and Morphologic Alterations in Native Human Corneal Endothelial Cells Associated With Diabetes Mellitus. Invest. Ophthalmol. Vis. Sci. 2017, 58, 2130–2138. [Google Scholar] [CrossRef]
- Wang, J.; Fan, T.J.; Yang, X.X.; Chang, S.M. Transforming growth factor-beta2 induces morphological alteration of human corneal endothelial cells in vitro. Int. J. Ophthalmol. 2014, 7, 759–763. [Google Scholar] [CrossRef] [Green Version]
- Shivanna, M.; Rajashekhar, G.; Srinivas, S.P. Barrier dysfunction of the corneal endothelium in response to TNF-alpha: Role of p38 MAP kinase. Invest. Ophthalmol. Vis. Sci. 2010, 51, 1575–1582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajashekhar, G.; Shivanna, M.; Kompella, U.B.; Wang, Y.; Srinivas, S.P. Role of MMP-9 in the breakdown of barrier integrity of the corneal endothelium in response to TNF-alpha. Exp. Eye Res. 2014, 122, 77–85. [Google Scholar] [CrossRef]
- Yagi-Yaguchi, Y.; Yamaguchi, T.; Higa, K.; Suzuki, T.; Aketa, N.; Dogru, M.; Satake, Y.; Shimazaki, J. Association between corneal endothelial cell densities and elevated cytokine levels in the aqueous humor. Sci. Rep. 2017, 7, 13603. [Google Scholar] [CrossRef]
- Doll, D.N.; Rellick, S.L.; Barr, T.L.; Ren, X.; Simpkins, J.W. Rapid mitochondrial dysfunction mediates TNF-alpha-induced neurotoxicity. J. Neurochem. 2015, 132, 443–451. [Google Scholar] [CrossRef]
- Kastl, L.; Sauer, S.W.; Ruppert, T.; Beissbarth, T.; Becker, M.S.; Suss, D.; Krammer, P.H.; Gulow, K. TNF-alpha mediates mitochondrial uncoupling and enhances ROS-dependent cell migration via NF-kappaB activation in liver cells. FEBS Lett. 2014, 588, 175–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariappan, N.; Elks, C.M.; Fink, B.; Francis, J. TNF-induced mitochondrial damage: A link between mitochondrial complex I activity and left ventricular dysfunction. Free Radic Biol. Med. 2009, 46, 462–470. [Google Scholar] [CrossRef] [Green Version]
- Dimeloe, S.; Gubser, P.; Loeliger, J.; Frick, C.; Develioglu, L.; Fischer, M.; Marquardsen, F.; Bantug, G.R.; Thommen, D.; Lecoultre, Y.; et al. Tumor-derived TGF-beta inhibits mitochondrial respiration to suppress IFN-gamma production by human CD4+ T cells. Sci. Signal. 2019, 12. [Google Scholar] [CrossRef] [Green Version]
- Yoon, Y.S.; Lee, J.H.; Hwang, S.C.; Choi, K.S.; Yoon, G. TGF beta1 induces prolonged mitochondrial ROS generation through decreased complex IV activity with senescent arrest in Mv1Lu cells. Oncogene 2005, 24, 1895–1903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Niu, J.; Li, X.; Wang, X.; Guo, Z.; Zhang, F. TGF-beta1 induces senescence of bone marrow mesenchymal stem cells via increase of mitochondrial ROS production. BMC Dev. Biol. 2014, 14, 21. [Google Scholar] [CrossRef] [Green Version]
- Shin, Y.J.; Cho, D.Y.; Chung, T.Y.; Han, S.B.; Hyon, J.Y.; Wee, W.R. Rapamycin reduces reactive oxygen species in cultured human corneal endothelial cells. Curr. Eye Res. 2011, 36, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Merlini, L.; Angelin, A.; Tiepolo, T.; Braghetta, P.; Sabatelli, P.; Zamparelli, A.; Ferlini, A.; Maraldi, N.M.; Bonaldo, P.; Bernardi, P. Cyclosporin A corrects mitochondrial dysfunction and muscle apoptosis in patients with collagen VI myopathies. Proc. Natl. Acad Sci. USA 2008, 105, 5225–5229. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Kim, J.J.; Hyon, J.Y.; Chung, E.S.; Chung, T.Y.; Yi, K.; Wee, W.R.; Shin, Y.J. The effects of different culture media on human corneal endothelial cells. Invest. Ophthalmol Vis. Sci. 2014, 55, 5099–5108. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.K.; Hwang, J.S.; Chung, T.Y.; Shin, Y.J. SOX2 Activation Using CRISPR/dCas9 Promotes Wound Healing in Corneal Endothelial Cells. Stem. Cells 2018, 36, 1851–1862. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Forest, F.; Gain, P.; Rageade, D.; Bernard, A.; Acquart, S.; Peoc’h, M.; Defoe, D.M.; Thuret, G. 3D map of the human corneal endothelial cell. Sci. Rep. 2016, 6, 29047. [Google Scholar] [CrossRef] [Green Version]
- Debacq-Chainiaux, F.; Erusalimsky, J.D.; Campisi, J.; Toussaint, O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 2009, 4, 1798–1806. [Google Scholar] [CrossRef]
- Leonard, A.P.; Cameron, R.B.; Speiser, J.L.; Wolf, B.J.; Peterson, Y.K.; Schnellmann, R.G.; Beeson, C.C.; Rohrer, B. Quantitative analysis of mitochondrial morphology and membrane potential in living cells using high-content imaging, machine learning, and morphological binning. Biochim. Biophys. Acta 2015, 1853, 348–360. [Google Scholar] [CrossRef] [Green Version]
- Demaria, M.; Desprez, P.Y.; Campisi, J.; Velarde, M.C. Cell Autonomous and Non-Autonomous Effects of Senescent Cells in the Skin. J. Invest. Derm. 2015, 135, 1722–1726. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.J.; Sato, T.; Matsubara, M.; Tanishima, T. Wound healing of the corneal endothelium in the bullous keratopathy after keratoplasty. Nippon Ganka Gakkai Zasshi 1983, 87, 701–707. [Google Scholar] [PubMed]
- Chen, H.; Li, Y.; Tollefsbol, T.O. Cell senescence culturing methods. Methods Mol. Biol. 2013, 1048, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laberge, R.M.; Awad, P.; Campisi, J.; Desprez, P.Y. Epithelial-mesenchymal transition induced by senescent fibroblasts. Cancer Microenviron. 2012, 5, 39–44. [Google Scholar] [CrossRef] [Green Version]
- Kasper, G.; Mao, L.; Geissler, S.; Draycheva, A.; Trippens, J.; Kuhnisch, J.; Tschirschmann, M.; Kaspar, K.; Perka, C.; Duda, G.N.; et al. Insights into mesenchymal stem cell aging: Involvement of antioxidant defense and actin cytoskeleton. Stem Cells 2009, 27, 1288–1297. [Google Scholar] [CrossRef]
- Levy, G.V.; Banuelos, C.P.; Nittolo, A.G.; Ortiz, G.E.; Mendiondo, N.; Moretti, G.; Tekiel, V.S.; Sanchez, D.O. Depletion of the SR-Related Protein TbRRM1 Leads to Cell Cycle Arrest and Apoptosis-Like Death in Trypanosoma brucei. PLoS ONE 2015, 10, e0136070. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.F.; Wang, N.N.; Xu, L.X.; Li, Z.H.; Li, X.L.; Xu, Y.Y.; Fang, F.; Li, M.; Qian, G.H.; Li, Y.H.; et al. Molecular mechanism of G1 arrest and cellular senescence induced by LEE011, a novel CDK4/CDK6 inhibitor, in leukemia cells. Cancer Cell Int. 2017, 17, 35. [Google Scholar] [CrossRef] [Green Version]
- Joyce, N.C. Proliferative capacity of corneal endothelial cells. Exp. Eye Res. 2012, 95, 16–23. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Wang, B.; Zang, W.; Wang, X.; Liu, Z.; Li, W.; Jia, J. Resveratrol inhibits the growth of gastric cancer by inducing G1 phase arrest and senescence in a Sirt1-dependent manner. PLoS ONE 2013, 8, e70627. [Google Scholar] [CrossRef]
- Sikora, E.; Mosieniak, G.; Sliwinska, M.A. Morphological and Functional Characteristic of Senescent Cancer Cells. Curr. Drug Targets 2016, 17, 377–387. [Google Scholar] [CrossRef]
- Cui, H.; Kong, Y.; Zhang, H. Oxidative stress, mitochondrial dysfunction, and aging. J. Signal. Transduct. 2012, 2012, 646354. [Google Scholar] [CrossRef] [Green Version]
- Bratic, I.; Trifunovic, A. Mitochondrial energy metabolism and ageing. Biochim. Biophys. Acta 2010, 1797, 961–967. [Google Scholar] [CrossRef] [Green Version]
- Maraldi, T.; Angeloni, C.; Giannoni, E.; Sell, C. Reactive Oxygen Species in Stem Cells. Oxid Med. Cell Longev. 2015, 2015, 159080. [Google Scholar] [CrossRef]
- Hamilton, M.L.; Guo, Z.; Fuller, C.D.; Van Remmen, H.; Ward, W.F.; Austad, S.N.; Troyer, D.A.; Thompson, I.; Richardson, A. A reliable assessment of 8-oxo-2-deoxyguanosine levels in nuclear and mitochondrial DNA using the sodium iodide method to isolate DNA. Nucleic. Acids Res. 2001, 29, 2117–2126. [Google Scholar] [CrossRef]
- Scott, I.; Youle, R.J. Mitochondrial fission and fusion. Essays Biochem. 2010, 47, 85–98. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Outschoorn, U.E.; Pavlides, S.; Sotgia, F.; Lisanti, M.P. Mitochondrial biogenesis drives tumor cell proliferation. Am. J. Pathol. 2011, 178, 1949–1952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prieto, J.; Leon, M.; Ponsoda, X.; Garcia-Garcia, F.; Bort, R.; Serna, E.; Barneo-Munoz, M.; Palau, F.; Dopazo, J.; Lopez-Garcia, C.; et al. Dysfunctional mitochondrial fission impairs cell reprogramming. Cell Cycle 2016, 15, 3240–3250. [Google Scholar] [CrossRef] [Green Version]
- Ziegler, D.V.; Wiley, C.D.; Velarde, M.C. Mitochondrial effectors of cellular senescence: Beyond the free radical theory of aging. Aging Cell 2015, 14, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Kurz, T.; Terman, A.; Gustafsson, B.; Brunk, U.T. Lysosomes and oxidative stress in aging and apoptosis. Biochim. Biophys. Acta 2008, 1780, 1291–1303. [Google Scholar] [CrossRef]
- Carmona-Gutierrez, D.; Hughes, A.L.; Madeo, F.; Ruckenstuhl, C. The crucial impact of lysosomes in aging and longevity. Ageing Res. Rev. 2016, 32, 2–12. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Lee, S.S.; Arai, R.; Miki, K.; Fujii, M.; Ayusawa, D. ERK1/2 mediates unbalanced growth leading to senescence induced by excess thymidine in human cells. Biochem. Biophys. Res. Commun. 2012, 425, 897–901. [Google Scholar] [CrossRef]
- Settembre, C.; Zoncu, R.; Medina, D.L.; Vetrini, F.; Erdin, S.; Erdin, S.; Huynh, T.; Ferron, M.; Karsenty, G.; Vellard, M.C.; et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012, 31, 1095–1108. [Google Scholar] [CrossRef] [Green Version]
- Huber, L.A.; Teis, D. Lysosomal signaling in control of degradation pathways. Curr. Opin Cell Biol. 2016, 39, 8–14. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.J.; Lee, Y.M.; Kang, S.G.; Lim, H.W.; Shin, K.O.; Jeong, S.K.; Huh, Y.H.; Choi, S.; Kor, M.; Seo, H.S.; et al. Aquatide Activation of SIRT1 Reduces Cellular Senescence through a SIRT1-FOXO1-Autophagy Axis. Biomol. Ther. 2017, 25, 511–518. [Google Scholar] [CrossRef] [Green Version]
- Ferber, E.C.; Peck, B.; Delpuech, O.; Bell, G.P.; East, P.; Schulze, A. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death Differ. 2012, 19, 968–979. [Google Scholar] [CrossRef]
- Yang, W.; Restrepo-Perez, L.; Bengtson, M.; Heerema, S.J.; Birnie, A.; van der Torre, J.; Dekker, C. Detection of CRISPR-dCas9 on DNA with Solid-State Nanopores. Nano Lett. 2018, 18, 6469–6474. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, X.; Shi, D.; Chen, P.; Yu, Y.; Yang, L.; Xie, L. Overexpression of SIRT1 promotes high glucose-attenuated corneal epithelial wound healing via p53 regulation of the IGFBP3/IGF-1R/AKT pathway. Invest. Ophthalmol. Vis. Sci. 2013, 54, 3806–3814. [Google Scholar] [CrossRef] [Green Version]
- Loukil, A.; Cheung, C.T.; Bendris, N.; Lemmers, B.; Peter, M.; Blanchard, J.M. Cyclin A2: At the crossroads of cell cycle and cell invasion. World J. Biol. Chem. 2015, 6, 346–350. [Google Scholar] [CrossRef]
- Ito, T.; Teo, Y.V.; Evans, S.A.; Neretti, N.; Sedivy, J.M. Regulation of Cellular Senescence by Polycomb Chromatin Modifiers through Distinct DNA Damage- and Histone Methylation-Dependent Pathways. Cell Rep. 2018, 22, 3480–3492. [Google Scholar] [CrossRef] [Green Version]
- Correia-Melo, C.; Jurk, D.; Passos, J.F. Robust multiparametric assessment of cellular senescence. Methods Mol. Biol. 2013, 965, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Park, S.R.; Park, H.J.; Choi, B.H.; Min, B.H. Potential predictive markers for proliferative capacity of cultured human articular chondrocytes: PCNA and p21. Artif. Organs 2005, 29, 393–398. [Google Scholar] [CrossRef]
- Itahana, K.; Campisi, J.; Dimri, G.P. Methods to detect biomarkers of cellular senescence: The senescence-associated beta-galactosidase assay. Methods Mol. Biol. 2007, 371, 21–31. [Google Scholar] [CrossRef]
- Kim, J.; Xu, M.; Xo, R.; Mates, A.; Wilson, G.L.; Pearsall, A.W.t.; Grishko, V. Mitochondrial DNA damage is involved in apoptosis caused by pro-inflammatory cytokines in human OA chondrocytes. Osteoarthr. Cartil. 2010, 18, 424–432. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Gan, Y.; Xu, Y.; Song, L.; Wang, L.; Ouyang, B.; Zhang, C.; Zhou, Q. The inflammatory cytokine TNF-alpha promotes the premature senescence of rat nucleus pulposus cells via the PI3K/Akt signaling pathway. Sci. Rep. 2017, 7, 42938. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Gu, Z.T.; Li, L.; Maegele, M.; Zhou, B.Y.; Li, F.; Zhao, M.; Zhao, K.S. SIRT1 plays a neuroprotective role in traumatic brain injury in rats via inhibiting the p38 MAPK pathway. Acta Pharm. Sin. 2017, 38, 168–181. [Google Scholar] [CrossRef] [Green Version]
- Martin, S.A.; Souder, D.C.; Miller, K.N.; Clark, J.P.; Sagar, A.K.; Eliceiri, K.W.; Puglielli, L.; Beasley, T.M.; Anderson, R.M. GSK3beta Regulates Brain Energy Metabolism. Cell Rep. 2018, 23, 1922–1931.e4. [Google Scholar] [CrossRef]
- Li, X.H.; Chen, C.; Tu, Y.; Sun, H.T.; Zhao, M.L.; Cheng, S.X.; Qu, Y.; Zhang, S. Sirt1 promotes axonogenesis by deacetylation of Akt and inactivation of GSK3. Mol. Neurobiol. 2013, 48, 490–499. [Google Scholar] [CrossRef]
- Xie, M.; Liu, M.; He, C.S. SIRT1 regulates endothelial Notch signaling in lung cancer. PLoS ONE 2012, 7, e45331. [Google Scholar] [CrossRef] [Green Version]
- Noseda, M.; Chang, L.; McLean, G.; Grim, J.E.; Clurman, B.E.; Smith, L.L.; Karsan, A. Notch activation induces endothelial cell cycle arrest and participates in contact inhibition: Role of p21Cip1 repression. Mol. Cell Biol. 2004, 24, 8813–8822. [Google Scholar] [CrossRef] [Green Version]
- Hisahara, S.; Chiba, S.; Matsumoto, H.; Tanno, M.; Yagi, H.; Shimohama, S.; Sato, M.; Horio, Y. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc. Natl. Acad Sci. USA 2008, 105, 15599–15604. [Google Scholar] [CrossRef] [Green Version]
- Sandoval, R.; Lazcano, P.; Ferrari, F.; Pinto-Pardo, N.; Gonzalez-Billault, C.; Utreras, E. TNF-alpha Increases Production of Reactive Oxygen Species through Cdk5 Activation in Nociceptive Neurons. Front. Physiol. 2018, 9, 65. [Google Scholar] [CrossRef] [Green Version]
- Krstic, J.; Trivanovic, D.; Mojsilovic, S.; Santibanez, J.F. Transforming Growth Factor-Beta and Oxidative Stress Interplay: Implications in Tumorigenesis and Cancer Progression. Oxid Med. Cell Longev. 2015, 2015, 654594. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, T.; Mitoma, H.; Harashima, S.; Tsukamoto, H.; Shimoda, T. Transmembrane TNF-alpha: Structure, function and interaction with anti-TNF agents. Rheumatology 2010, 49, 1215–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wajant, H.; Pfizenmaier, K.; Scheurich, P. Tumor necrosis factor signaling. Cell Death Differ. 2003, 10, 45–65. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.K.; Sheppard, D.; Chapman, H.A. TGF-beta1 Signaling and Tissue Fibrosis. Cold Spring Harb. Perspect Biol. 2018, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boice, A.; Bouchier-Hayes, L. Targeting apoptotic caspases in cancer. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118688. [Google Scholar] [CrossRef]
- Jin, X.; Wei, Y.; Xu, F.; Zhao, M.; Dai, K.; Shen, R.; Yang, S.; Zhang, N. SIRT1 promotes formation of breast cancer through modulating Akt activity. J. Cancer 2018, 9, 2012–2023. [Google Scholar] [CrossRef] [Green Version]

| Gene | Forward | Reverse |
|---|---|---|
| SIRT1 (NM_012238) | TCGCAACTATACCCAGAACATAGACA | CTGTTGCAAAGGAACCATGACA |
| COL8A2 (NM_005202.) | GGCAAAGGCCAGTACCTG | CCCCTCGTATTCCTGGCT |
| CDKN2A (NM_000077) | CATAGATGCCGCGGAAGGT | CTAAGTTTCCCGAGGTTTCTCAGA |
| PCNA (NM_002592) | GCGTGAACCTCACCAGTATGT | TCTTCGGCCCTTAGTGTAATGAT |
| CDC25B (NM_021873) | GGCTGAGGAACCTAAAGCCC | CTTTCCGTCTACTGTCTGTAGGA |
| ACTB (NM_001101) | AGAGCTACGCTGCCTGAC | AGCACTGTTGGCGTACAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joo, H.J.; Ma, D.J.; Hwang, J.S.; Shin, Y.J. SIRT1 Activation Using CRISPR/dCas9 Promotes Regeneration of Human Corneal Endothelial Cells through Inhibiting Senescence. Antioxidants 2020, 9, 1085. https://doi.org/10.3390/antiox9111085
Joo HJ, Ma DJ, Hwang JS, Shin YJ. SIRT1 Activation Using CRISPR/dCas9 Promotes Regeneration of Human Corneal Endothelial Cells through Inhibiting Senescence. Antioxidants. 2020; 9(11):1085. https://doi.org/10.3390/antiox9111085
Chicago/Turabian StyleJoo, Hye Jun, Dae Joong Ma, Jin Sun Hwang, and Young Joo Shin. 2020. "SIRT1 Activation Using CRISPR/dCas9 Promotes Regeneration of Human Corneal Endothelial Cells through Inhibiting Senescence" Antioxidants 9, no. 11: 1085. https://doi.org/10.3390/antiox9111085





