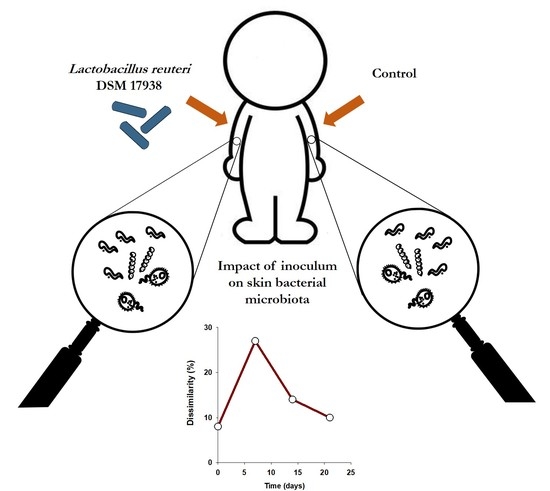
Human Skin Bacterial Community Response to Probiotic (Lactobacillus reuteri DSM 17938) Introduction
Abstract
1. Introduction
2. Materials and Methods
2.1. Inoculum Conception
2.2. Volunteer Recruitment and Inoculum Application
2.3. Sampling and DNA Extraction
2.4. Molecular Profiling of Bacterial Communities
2.5. Statistical Analysis
3. Results
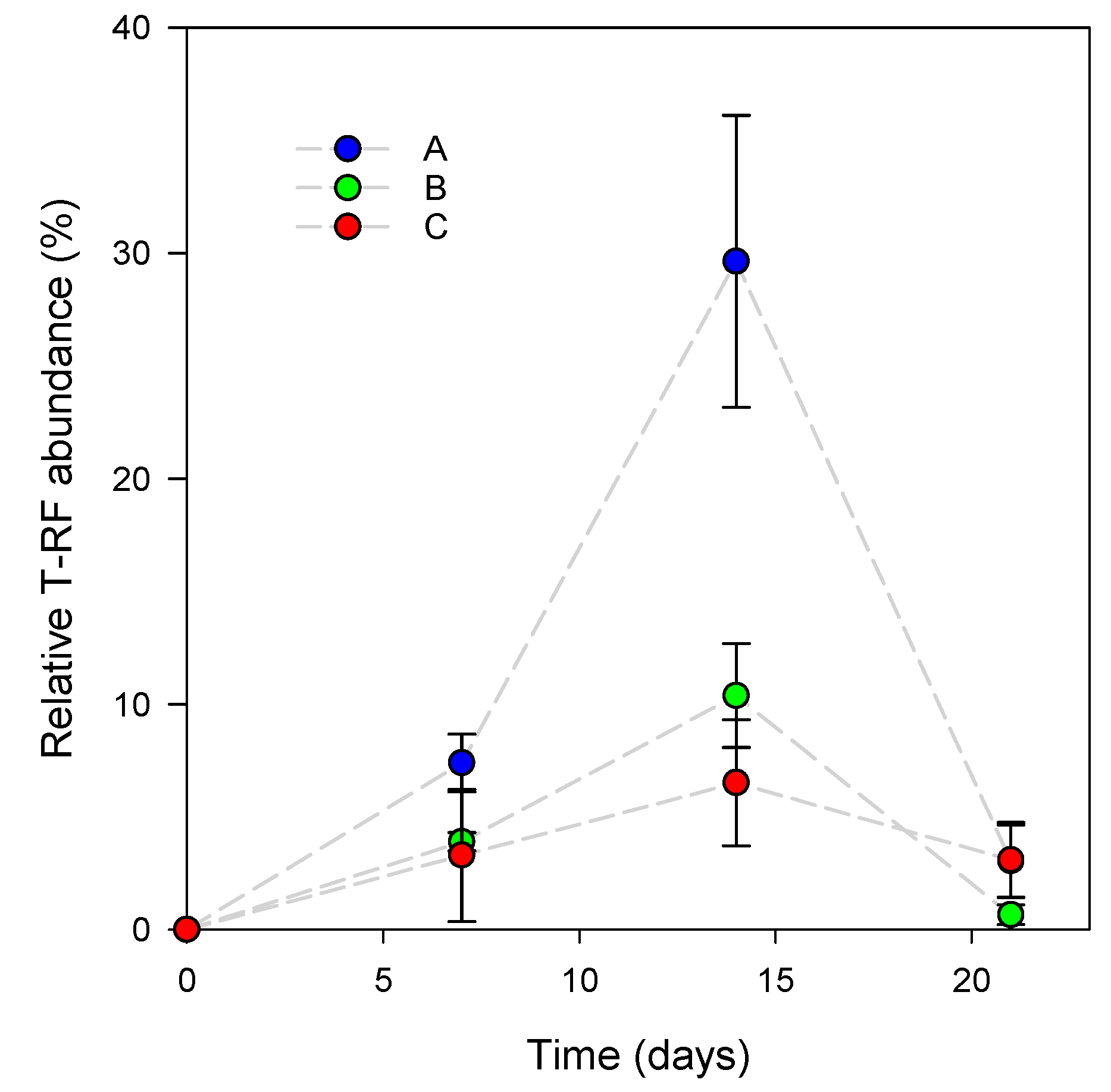
3.1. L. reuteri DSM 17938 Skin Colonization Dynamics
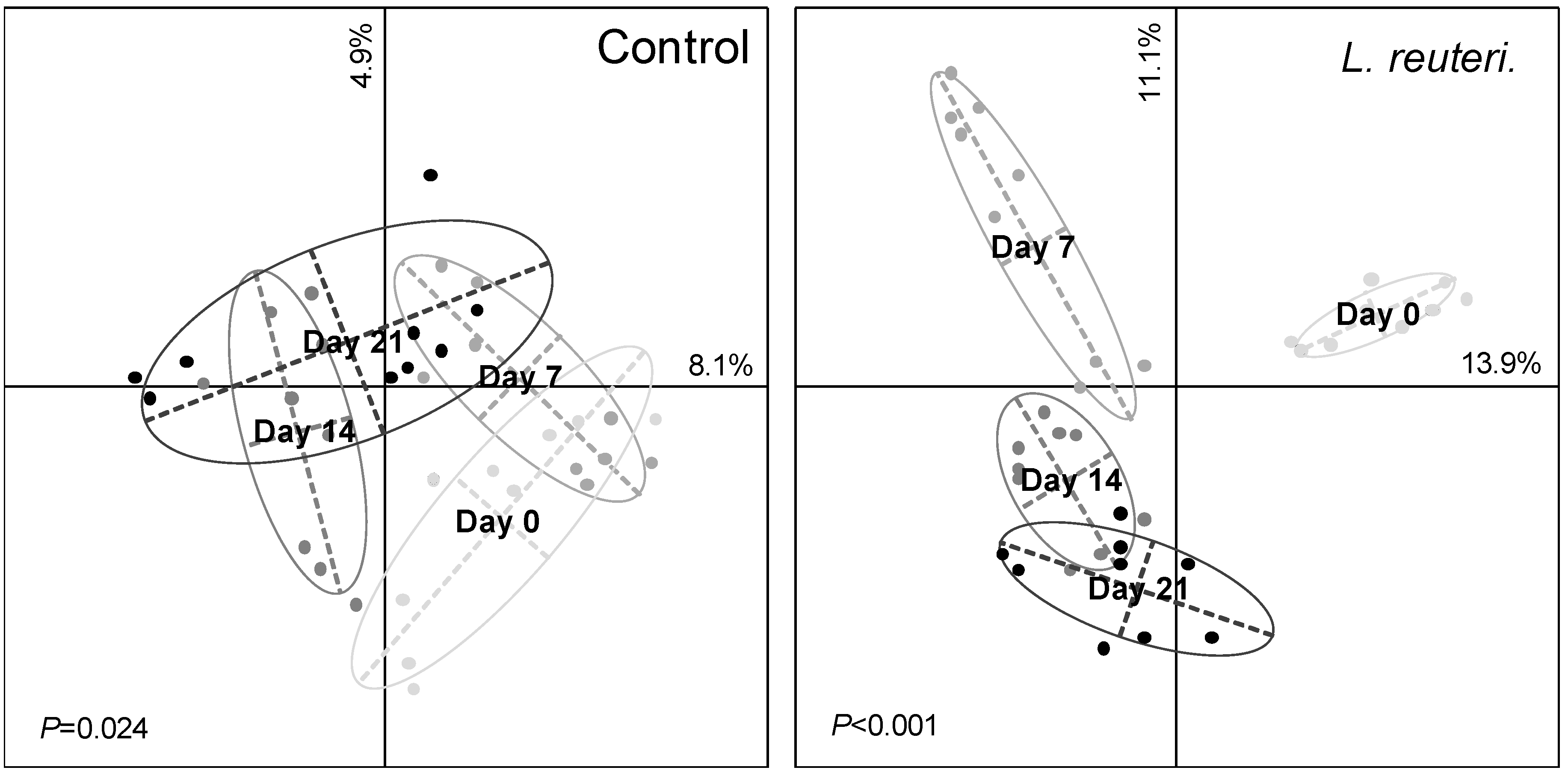
3.2. Effect of Probiotics on Skin Bacterial Community Structure
3.3. Resistance of Skin Microbiota to Probiotics Colonization
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Segre, J.A. Epidermal barrier formation and recovery in skin disorders. J. Clin. Investig. 2006, 116, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Walters, K.A.; Roberts, M.S. The structure and function of skin. In Design of Biomedical Devices and Systems, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2002; pp. 18–58. [Google Scholar]
- Leider, M. On the Weight of the Skin. J. Investig. Dermatol. 1949, 12, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Elston, D.M. Demodex mites: Facts and controversies. Clin. Dermatol. 2010, 28, 502–504. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; Green, E.D.; et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Hamady, M.; Lauber, C.L.; Knight, R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 17994. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Wolfsberg, T.G.; Turner, M.L.; Segre, J.A. A diversity profile of the human skin microbiota. Genome Res. 2008, 18, 1043–1050. [Google Scholar] [CrossRef]
- Dimitriu, P.A.; Iker, B.; Malik, K.; Leung, H.; Mohn, W.W.; Hillebrand, G.G. New Insights into the Intrinsic and Extrinsic Factors That Shape the Human Skin Microbiome. mBio 2019, 10, e00839-19. [Google Scholar] [CrossRef]
- Si, J.; Lee, S.; Park, J.M.; Sung, J.; Ko, G. Genetic associations and shared environmental effects on the skin microbiome of Korean twins. BMC Genom. 2015, 16, 992. [Google Scholar] [CrossRef]
- Belkaid, Y.; Tamoutounour, S. The influence of skin microorganisms on cutaneous immunity. Nat. Rev. Immunol. 2016, 16, 353–366. [Google Scholar] [CrossRef]
- Atlas, R.M.; Bartha, R. Microbial Ecology: Fundamentals and Applications; Menlo Park. Calif. Harlow Benjamin Cummings: Menlo Park, CA, USA, 1998. [Google Scholar]
- Gilbert, J.A.; Lynch, S.V. Community ecology as a framework for human microbiome research. Nat. Med. 2019, 25, 884–889. [Google Scholar] [CrossRef]
- Food and Agricultural Organization of United Nations and World Health Organization. Evaluation of Health and Nutritional Properties of Probiotics in Food, Including Powder Milk with Live Lactic Acid Bacteria; FAO/WHO Expert Consultation: Geneva, Switzerland, 2001. [Google Scholar]
- Wieërs, G.; Belkhir, L.; Enaud, R.; Leclercq, S.; Philippart de Foy, J.-M.; Dequenne, I.; de Timary, P.; Cani, P.D. How Probiotics Affect the Microbiota. Front. Cell. Infect. Microbiol. 2020, 9, 454. [Google Scholar] [CrossRef] [PubMed]
- McNulty, N.P.; Yatsunenko, T.; Hsiao, A.; Faith, J.J.; Muegge, B.D.; Goodman, A.L.; Henrissat, B.; Oozeer, R.; Cools-Portier, S.; Gobert, G.; et al. The Impact of a Consortium of Fermented Milk Strains on the Gut Microbiome of Gnotobiotic Mice and Monozygotic Twins. Sci. Transl. Med. 2011, 3, 106ra106. [Google Scholar] [CrossRef] [PubMed]
- Manichanh, C.; Reeder, J.; Gibert, P.; Varela, E.; Llopis, M.; Antolin, M.; Guigo, R.; Knight, R.; Guarner, F. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 2010, 20, 1411–1419. [Google Scholar] [CrossRef] [PubMed]
- Khoruts, A.; Dicksved, J.; Jansson, J.K.; Sadowsky, M.J. Changes in the Composition of the Human Fecal Microbiome after Bacteriotherapy for Recurrent Clostridium difficile-associated Diarrhea. J. Clin. Gastroenterol. 2010, 44, 354–360. [Google Scholar] [CrossRef]
- Paetzold, B.; Willis, J.R.; Pereira de Lima, J.; Knodlseder, N.; Bruggemann, H.; Quist, S.R.; Gabaldon, T.; Guell, M. Skin microbiome modulation induced by probiotic solutions. Microbiome 2019, 7, 95. [Google Scholar] [CrossRef]
- Delley, M.; Bruttin, A.; Richard, M.; Affolter, M.; Rezzonico, E.; Brück, W.M. In Vitro activity of commercial probiotic Lactobacillus strains against uropathogenic Escherichia coli. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef]
- Górska, A.; Przystupski, D.; Niemczura, M.J.; Kulbacka, J. Probiotic Bacteria: A Promising Tool in Cancer Prevention and Therapy. Curr. Microbiol. 2019, 76, 939–949. [Google Scholar] [CrossRef]
- Rosander, A.; Connolly, E.; Roos, S. Removal of Antibiotic Resistance Gene-Carrying Plasmids from Lactobacillus reuteri ATCC 55730 and Characterization of the Resulting Daughter Strain, L. reuteri DSM 17938. Appl. Environ. Microbiol. 2008, 74, 6032. [Google Scholar] [CrossRef]
- Axelsson, L.T.; Chung, T.C.; Dobrogosz, W.J.; Lindgren, S.E. Production of a Broad Spectrum Antimicrobial Substance by Lactobacillus reuteri. Microb. Ecol. Health Dis. 1989, 2, 131–136. [Google Scholar] [CrossRef]
- Khmaladze, I.; Butler, E.; Fabre, S.; Gillbro, J.M. Lactobacillus reuteri DSM 17938-A comparative study on the effect of probiotics and lysates on human skin. Exp. Dermatol. 2019, 28, 822–828. [Google Scholar] [CrossRef]
- Lauber, C.L.; Zhou, N.; Gordon, J.I.; Knight, R.; Fierer, N. Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol. Lett. 2010, 307, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Magurran, A.E. Ecological Diversity and Its Measurement; Princeton University Press: Princeton, NJ, USA, 1988; ISBN 0-691-08491-2. [Google Scholar]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Bauer, M.A.; Kainz, K.; Carmona-Gutierrez, D.; Madeo, F. Microbial wars: Competition in ecological niches and within the microbiome. Microb. Cell Graz Austria 2018, 5, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Valeur, N.; Engel, P.; Carbajal, N.; Connolly, E.; Ladefoged, K. Colonization and Immunomodulation by Lactobacillus reuteri ATCC 55730 in the Human Gastrointestinal Tract. Appl. Environ. Microbiol. 2004, 70, 1176. [Google Scholar] [CrossRef]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science 2009, 326, 1694. [Google Scholar] [CrossRef]
- Lee, H.J.; Jeong, S.E.; Lee, S.; Kim, S.; Han, H.; Jeon, C.O. Effects of cosmetics on the skin microbiome of facial cheeks with different hydration levels. MicrobiologyOpen 2018, 7, e00557. [Google Scholar] [CrossRef]
- Griffiths, B.S.; Hallett, P.D.; Kuan, H.L.; Gregory, A.S.; Watts, C.W.; Whitmore, A.P. Functional resilience of soil microbial communities depends on both soil structure and microbial community composition. Biol. Fertil. Soils 2008, 44, 745–754. [Google Scholar] [CrossRef]
- Berga, M.; Székely, A.J.; Langenheder, S. Effects of Disturbance Intensity and Frequency on Bacterial Community Composition and Function. PLoS ONE 2012, 7, e36959. [Google Scholar] [CrossRef]
- Trabelsi, D.; Mhamdi, R. Microbial Inoculants and Their Impact on Soil Microbial Communities: A Review. BioMed. Res. Int. 2013, 2013, 863240. [Google Scholar] [CrossRef]
- Gibson, M.K.; Pesesky, M.W.; Dantas, G. The yin and yang of bacterial resilience in the human gut microbiota. J. Mol. Biol. 2014, 426, 3866–3876. [Google Scholar] [CrossRef]
- Sommer, F.; Anderson, J.M.; Bharti, R.; Raes, J.; Rosenstiel, P. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 2017, 15, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Chien, A.L.; Tsai, J.; Leung, S.; Mongodin, E.F.; Nelson, A.M.; Kang, S.; Garza, L.A. Association of Systemic Antibiotic Treatment of Acne with Skin Microbiota Characteristics. JAMA Dermatol. 2019, 155, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, T.A.; Naeem, S.; Howe, K.M.; Knops, J.M.H.; Tilman, D.; Reich, P. Biodiversity as a barrier to ecological invasion. Nature 2002, 417, 636–638. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini, A.; de Souza, R.; Passaglia, L.M.P. Ecological role of bacterial inoculants and their potential impact on soil microbial diversity. Plant Soil 2016, 400, 193–207. [Google Scholar] [CrossRef]
- Lee, S.M.; Donaldson, G.P.; Mikulski, Z.; Boyajian, S.; Ley, K.; Mazmanian, S.K. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 2013, 501, 426–429. [Google Scholar] [CrossRef]
- Seth, E.C.; Taga, M.E. Nutrient cross-feeding in the microbial world. Front. Microbiol. 2014, 5, 350. [Google Scholar] [CrossRef]
- Lakshminarayanan, B.; Guinane, C.M.; O’Connor, P.M.; Coakley, M.; Hill, C.; Stanton, C.; O’Toole, P.W.; Ross, R.P. Isolation and characterization of bacteriocin-producing bacteria from the intestinal microbiota of elderly Irish subjects. J. Appl. Microbiol. 2013, 114, 886–898. [Google Scholar] [CrossRef]
- O’Sullivan, J.N.; Rea, M.C.; O’Connor, P.M.; Hill, C.; Ross, R.P. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 2018, 95, fiy241. [Google Scholar] [CrossRef]
- Allison, S.D.; Martiny, J.B.H. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512. [Google Scholar] [CrossRef]

| Degree of Freedom | T-RF Richness | T-RF Evenness | |
|---|---|---|---|
| Lactobacillus (L) | 1 | 66.036 * | 171.927 *** |
| Time (T) | 3 | 9.567 * | 21.345 *** |
| Subject (S) | 2 | 4.136 | 14.287 ** |
| L × T | 3 | 8.745 * | 20.323 *** |
| L × S | 2 | 4.964 | 2.873 |
| T × S | 6 | 4.184 * | 8.142 * |
| L × T × S | 6 | 2.089 | 8.081 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frerejacques, M.; Rousselle, C.; Gauthier, L.; Cottet-Emard, S.; Derobert, L.; Roynette, A.; Lerch, T.Z.; Changey, F. Human Skin Bacterial Community Response to Probiotic (Lactobacillus reuteri DSM 17938) Introduction. Microorganisms 2020, 8, 1223. https://doi.org/10.3390/microorganisms8081223
Frerejacques M, Rousselle C, Gauthier L, Cottet-Emard S, Derobert L, Roynette A, Lerch TZ, Changey F. Human Skin Bacterial Community Response to Probiotic (Lactobacillus reuteri DSM 17938) Introduction. Microorganisms. 2020; 8(8):1223. https://doi.org/10.3390/microorganisms8081223
Chicago/Turabian StyleFrerejacques, Marie, Camille Rousselle, Loüen Gauthier, Salomé Cottet-Emard, Léa Derobert, Anne Roynette, Thomas Z. Lerch, and Frédérique Changey. 2020. "Human Skin Bacterial Community Response to Probiotic (Lactobacillus reuteri DSM 17938) Introduction" Microorganisms 8, no. 8: 1223. https://doi.org/10.3390/microorganisms8081223
APA StyleFrerejacques, M., Rousselle, C., Gauthier, L., Cottet-Emard, S., Derobert, L., Roynette, A., Lerch, T. Z., & Changey, F. (2020). Human Skin Bacterial Community Response to Probiotic (Lactobacillus reuteri DSM 17938) Introduction. Microorganisms, 8(8), 1223. https://doi.org/10.3390/microorganisms8081223