Transcriptome Analysis of the Inhibitory Effect of Astaxanthin on Helicobacter pylori-Induced Gastric Carcinoma Cell Motility
Abstract
:1. Introduction
2. Results
2.1. Gene Expression Profile of H. pylori-Infected and/or ASTX-Treated AGS Cells
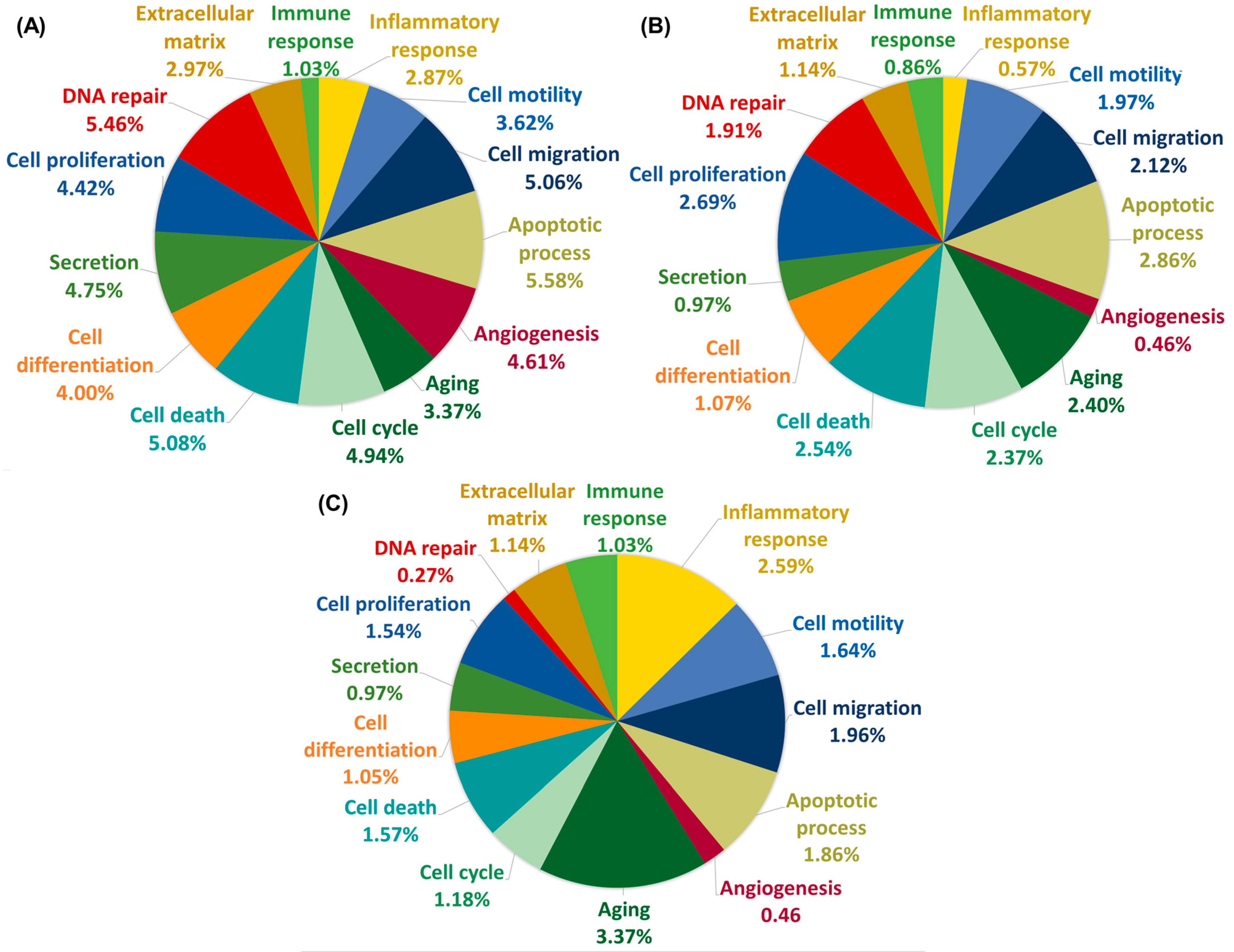
2.2. GO Analysis of DEGs
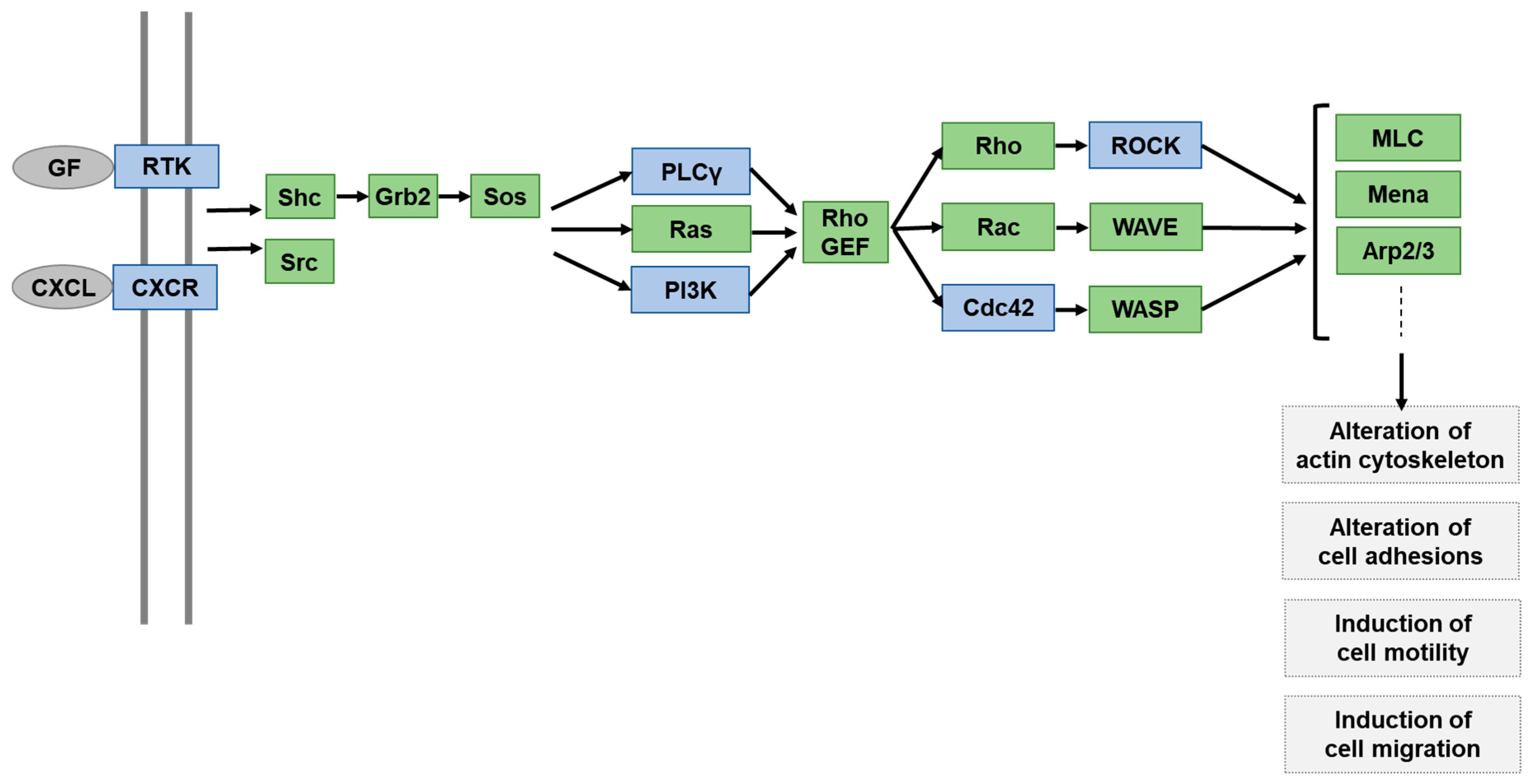
2.3. Functional Pathway Analysis of DEGs Altered by H. pylori Infection but Normalized by ASTX Pretreatment
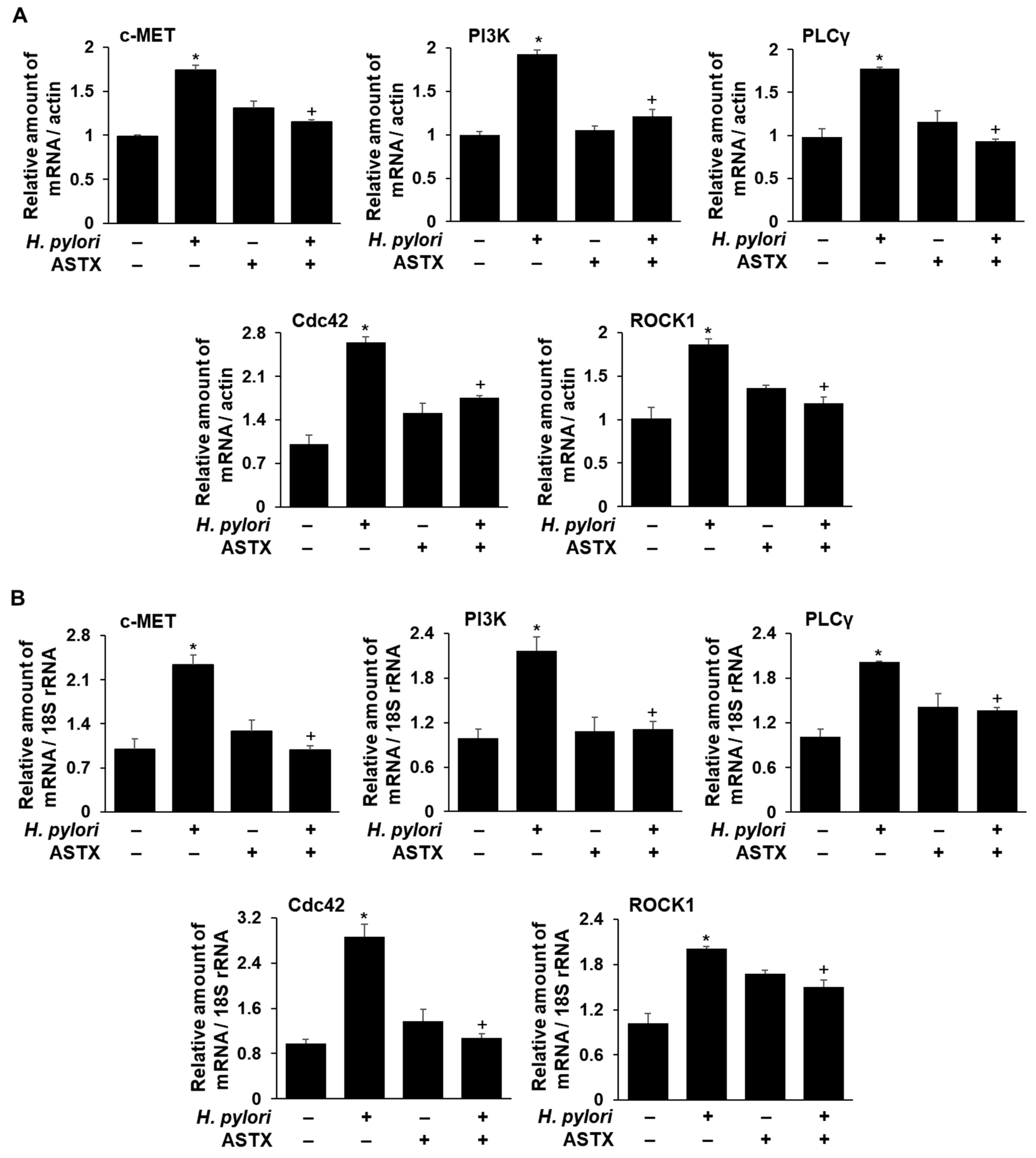
2.4. Determinations of c-MET, PI3KC2β, PLCγ1, Cdc42, and ROCK1 mRNA Levels in H. pylori-Infected AGS Cells, with and without ASTX Pretreatment
3. Discussion
4. Materials and Methods
4.1. Cell Line and Culture Conditions
4.2. Treatment of AGS Cells with ASTX
4.3. Bacterial Strain and H. pylori Infection
4.4. Preparation of Total RNA Extracts and Library Construction
4.5. RNA-Sequencing and Bioinformatics Analysis
4.6. Validation of DEGs by Real-Time Polymerase Chain Reaction (PCR)
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkin, D.M. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer 2006, 118, 3030–3044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riihimäki, M.; Hemminki, A.; Sundquist, K.; Sundquist, J.; Hemminki, K. Metastatic spread in patients with gastric cancer. Oncotarget 2016, 7, 52307–52310. [Google Scholar] [CrossRef] [Green Version]
- Gupta, G.P.; Massagué, J. Cancer metastasis: Building a framework. Cell 2006, 127, 679–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinberg, R.A. The Biology of Cancer, 2nd ed.; Norton & Company: New York, NY, USA, 2013; Chapter 14; ISBN 978-08-1534-528-2. [Google Scholar]
- Jouanneau, J.; Thiery, J.P. Tumor cell motility and invasion. In Encyclopedia of Cancer, 2nd ed.; Academic Press: Cambridge, MA, USA, 2002; pp. 467–473. ISBN 978-01-2227-555-5. [Google Scholar]
- Mitchison, T.J.; Cramer, L.P. Actin-based cell motility and cell locomotion. Cell 1996, 84, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Lauffenburger, D.A.; Horwitz, A.F. Cell migration: A physically integrated molecular process. Cell 1996, 84, 359–369. [Google Scholar] [CrossRef] [Green Version]
- Jacquemet, G.; Hamidi, H.; Ivaska, J. Filopodia in cell adhesion, 3D migration and cancer cell migration. Curr. Opin. Cell Biol. 2015, 36, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Parri, M.; Chiarugi, P. Rac and Rho GTPases in cancer cell motility control. Cell Commun. Signal. 2010, 8, 23. [Google Scholar] [CrossRef] [Green Version]
- Raftopoulou, M.; Hall, A. Cell migration: Rho GTPases lead the way. Dev. Biol. 2004, 265, 23–32. [Google Scholar] [CrossRef] [Green Version]
- Kedrin, D.; van Rheenen, J.; Hernandez, L.; Condeelis, J.; Segall, J.E. Cell motility and cytoskeletal regulation in invasion and metastasis. J. Mammary Gland Biol. 2007, 12, 143–152. [Google Scholar] [CrossRef]
- Normanno, N.; De Luca, A.; Bianco, C.; Strizzi, L.; Mancino, M.; Maiello, M.R.; Carotenuto, A.; De Feo, G.; Caponigro, F.; Salomon, D.S. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006, 366, 2–16. [Google Scholar] [CrossRef]
- Birchmeier, C.; Birchmeier, W.; Gherardi, E.; Woude, G.F.V. Met, metastasis, motility and more. Nat. Rev. Mol. Cell Biol. 2003, 4, 915–925. [Google Scholar] [CrossRef]
- Guillemin, K.; Salama, N.R.; Tompkins, L.S.; Falkow, S. Cag pathogenicity island-specific responses of gastric epithelial cells to Helicobacter pylori infection. Proc. Natl. Acad. Sci. USA 2002, 99, 15136–15141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peek, R.M.; Blaser, M.J. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2002, 2, 28–37. [Google Scholar] [CrossRef]
- Wessler, S.; Gimona, M.; Rieder, G. Regulation of the actin cytoskeleton in Helicobacter pylori-induced migration and invasive growth of gastric epithelial cells. Cell Commun. Signal. 2011, 9, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.Y.; Wang, C.J.; Tseng, C.C.; Chen, H.P.; Wu, M.S.; Lin, J.T.; Inoue, H.; Chen, G.H. Helicobacter pylori promote gastric cancer cells invasion through a NF-kB and COX-2-mediated pathway. World J. Gastroenterol. 2005, 11, 3197. [Google Scholar] [CrossRef] [PubMed]
- Segal, E.D.; Falkow, S.; Tompkins, L.S. Helicobacter pylori attachment to gastric cells induces cytoskeletal rearrangements and tyrosine phosphorylation of host cell proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 1259–1264. [Google Scholar] [CrossRef] [Green Version]
- Wessler, S.; Backert, S. Molecular mechanisms of epithelial-barrier disruption by Helicobacter pylori. Trends Microbiol. 2008, 16, 397–405. [Google Scholar] [CrossRef]
- Yin, Y.; Grabowska, A.M.; Clarke, P.A.; Whelband, E.; Robinson, K.; Argent, R.H.; Tobias, A.; Kumari, R.; Atherton, J.C.; Watson, S.A. Helicobacter pylori potentiates epithelial: Mesenchymal transition in gastric cancer: Links to soluble HB-EGF, gastrin and matrix metalloproteinase-7. Gut 2010, 59, 1037–1045. [Google Scholar] [CrossRef] [Green Version]
- Churin, Y.; Kardalinou, E.; Meyer, T.F.; Naumann, M. Pathogenicity island-dependent activation of Rho GTPases Rac1 and Cdc42 in Helicobacter pylori infection. Mol. Microbiol. 2001, 40, 815–823. [Google Scholar] [CrossRef]
- Higashi, H.; Nakaya, A.; Tsutsumi, R.; Yokoyama, K.; Fujii, Y.; Ishikawa, S.; Higuchi, M.; Takahashi, A.; Kurashima, Y.; Teishikata, Y.; et al. Helicobacter pylori CagA induces Ras-independent morphogenetic response through SHP-2 recruitment and activation. J. Biol. Chem. 2004, 279, 17205–17216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, M.; Mimuro, H.; Suzuki, T.; Park, M.; Yamamoto, T.; Sasakawa, C. Interaction of CagA with Crk plays an important role in Helicobacter pylori–induced loss of gastric epithelial cell adhesion. J. Exp. Med. 2005, 202, 1235–1247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagnoli, F.; Buti, L.; Tompkins, L.; Covacci, A.; Amieva, M.R. Helicobacter pylori CagA induces a transition from polarized to invasive phenotypes in MDCK cells. Proc. Natl. Acad. Sci. USA 2005, 102, 16339–16344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segal, E.D.; Cha, J.; Lo, J.; Falkow, S.; Tompkins, L.S. Altered states: Involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. USA 1999, 96, 14559–14564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keates, S.; Sougioultzis, S.; Keates, A.C.; Zhao, D.; Peek, R.M.; Shaw, L.M.; Kelly, C.P. cag+ Helicobacter pylori induce transactivation of the epidermal growth factor receptor in AGS gastric epithelial cells. J. Biol. Chem. 2001, 276, 48127–48134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mimuro, H.; Suzuki, T.; Tanaka, J.; Asahi, M.; Haas, R.; Sasakawa, C. Grb2 is a key mediator of Helicobacter pylori CagA protein activities. Mol. Cell 2002, 10, 745–755. [Google Scholar] [CrossRef]
- Tsutsumi, R.; Takahashi, A.; Azuma, T.; Higashi, H.; Hatakeyama, M. Focal adhesion kinase is a substrate and downstream effector of SHP-2 complexed with Helicobacter pylori CagA. Mol. Cell. Biol. 2006, 26, 261–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palovuori, R.; Perttu, A.; Yan, Y.; Karttunen, R.; Eskelinen, S.; Karttunen, T.J. Helicobacter pylori induces formation of stress fibers and membrane ruffles in AGS cells by rac activation. Biochem. Biophys. Res. Commun. 2001, 269, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Wroblewski, L.E.; Noble, P.J.; Pagliocca, A.; Pritchard, D.M.; Hart, C.A.; Campbell, F.; Dodson, A.R.; Dockray, G.J.; Varro, A. Stimulation of MMP-7 (matrilysin) by Helicobacter pylori in human gastric epithelial cells: Role in epithelial cell migration. J. Cell Sci. 2003, 116, 3017–3026. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.P.; Peng, J.; Yin, K.; Wang, J.H. Potential health-promoting effects of astaxanthin: A high-value carotenoid mostly from microalgae. Mol. Nutr. Food Res. 2001, 55, 150–165. [Google Scholar] [CrossRef]
- Higuera-Ciapara, I.; Felix-Valenzuela, L.; Goycoolea, F.M. Astaxanthin: A review of its chemistry and applications. Crit. Rev. Food Sci. Nutr. 2006, 46, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Ambati, R.R.; Phang, S.M.; Ravi, S.; Aswathanarayana, R.G. Astaxanthin: Sources, extraction, stability, biological activities and its commercial applications—A review. Mar. Drugs 2014, 12, 128–152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, H. Multiple mechanisms of anti-cancer effects exerted by astaxanthin. Mar. Drugs 2015, 13, 4310–4330. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Park, J.J.; Lee, B.J.; Joo, M.K.; Chun, H.J.; Lee, S.W.; Bak, Y.T. Astaxanthin inhibits proliferation of human gastric cancer cell lines by interrupting cell cycle progression. Gut Liver. 2016, 10, 369. [Google Scholar] [CrossRef] [Green Version]
- Kozuki, Y.; Miura, Y.; Yagasaki, K. Inhibitory effects of carotenoids on the invasion of rat ascites hepatoma cells in culture. Cancer Lett. 2000, 151, 111–115. [Google Scholar] [CrossRef]
- Su, X.Z.; Chen, R.; Wang, C.B.; Ouyang, X.L.; Jiang, Y.; Zhu, M.Y. Astaxanthin combine with human serum albumin to abrogate cell proliferation, migration, and drug-resistant in human ovarian carcinoma SKOV3 cells. Anti-Cancer Agents Med. Chem. 2019, 19, 792–801. [Google Scholar] [CrossRef]
- McCall, B.; McPartland, C.K.; Moore, R.; Frank-Kamenetskii, A.; Booth, B.W. Effects of astaxanthin on the proliferation and migration of breast cancer cells in vitro. Antioxidants 2018, 7, 135. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.Y.; Kim, Y.M.; Hong, S. Astaxanthin suppresses the metastasis of colon cancer by inhibiting the MYC-mediated downregulation of microRNA-29a-3p and microRNA-200a. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Chen, Y.T.; Kao, C.J.; Huang, H.Y.; Huang, S.Y.; Chen, C.Y.; Lin, Y.S.; Wen, Z.H.; Wang, H.M.D. Astaxanthin reduces MMP expressions, suppresses cancer cell migrations, and triggers apoptotic caspases of in vitro and in vivo models in melanoma. J. Funct. Foods 2017, 31, 20–31. [Google Scholar] [CrossRef]
- Stadelman, K.M. Astaxanthin Decreases Invasion and MMP Activity in Triple Negative Breast Cancer Cells. Doctoral Dissertation, Wake Forest University, Winston-Salem, NC, USA, 2012. [Google Scholar]
- Kowshik, J.; Nivetha, R.; Ranjani, S.; Venkatesan, P.; Selvamuthukumar, S.; Veeravarmal, V.; Nagini, S. Astaxanthin inhibits hallmarks of cancer by targeting the PI3K/NF-κΒ/STAT3 signalling axis in oral squamous cell carcinoma models. IUBMB Life 2019, 71, 1595–1610. [Google Scholar] [CrossRef]
- Al-Ghoul, L.; Wessler, S.; Hundertmark, T.; Krüger, S.; Fischer, W.; Wunder, C.; Haas, R.; Roessner, A.; Naumann, M. Analysis of the type IV secretion system-dependent cell motility of Helicobacter pylori-infected epithelial cells. Biochem. Biophys. Res. Commun. 2004, 322, 860–866. [Google Scholar] [CrossRef] [PubMed]
- Moese, S.; Selbach, M.; Kwok, T.; Brinkmann, V.; König, W.; Meyer, T.F.; Backert, S. Helicobacter pylori induces AGS cell motility and elongation via independent signaling pathways. Infect. Immun. 2004, 72, 3646–3649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.E.; Khoi, P.N.; Xia, Y.; Park, J.S.; Joo, Y.E.; Kim, K.K.; Choi, S.Y.; Jung, Y.D. Helicobacter pylori and interleukin-8 in gastric cancer. World J. Gastroenterol. 2013, 19, 8192. [Google Scholar] [CrossRef] [PubMed]
- Takeshima, E.; Tomimori, K.; Kawakami, H.; Ishikawa, C.; Sawada, S.; Tomita, M.; Senba, M.; Kinjo, F.; Mimuro, H.; Sasakawa, C.; et al. NF-κB activation by Helicobacter pylori requires Akt-mediated phosphorylation of p65. BMC Microbiol. 2009, 9, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eftang, L.L.; Esbensen, Y.; Tannæs, T.M.; Bukholm, I.R.; Bukholm, G. Interleukin-8 is the single most up-regulated gene in whole genome profiling of H. pylori exposed gastric epithelial cells. BMC Microbiol. 2012, 12, 9. [Google Scholar] [CrossRef] [Green Version]
- Churin, Y.; Al-Ghoul, L.; Kepp, O.; Meyer, T.F.; Birchmeier, W.; Naumann, M. Helicobacter pylori CagA protein targets the c-MET receptor and enhances the motogenic response. J. Cell Biol. 2003, 161, 249–255. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.J.; Wu, M.S.; Lin, J.T.; Chen, C.C. Helicobacter pylori-induced invasion and angiogenesis of gastric cells is mediated by cyclooxygenase-2 induction through TLR2/TLR9 and promoter regulation. J. Immunol. 2005, 175, 8242–8252. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Yang, Z.; Hu, Y.; Cao, X.; Chen, J.; Zhu, Y.; Lu, N. Expression of c-Met and hepatocyte growth factor in various gastric pathologies and its association with Helicobacter pylori infection. Oncol. Lett. 2017, 14, 6151–6155. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, M.J.; Costa, A.C.; Costa, A.M.; Henriques, L.; Suriano, G.; Atherton, J.C.; Machado, J.C.; Carneiro, F.; Seruca, R.; Mareel, M.; et al. Helicobacter pylori induces gastric epithelial cell invasion in a c-Met and type IV secretion system-dependent manner. J. Biol. Chem. 2006, 281, 34888–34896. [Google Scholar] [CrossRef] [Green Version]
- Xiang, C.; Chen, J.; Fu, P. HGF/Met signaling in cancer invasion: The impact on cytoskeleton remodeling. Cancers 2017, 9, 44. [Google Scholar] [CrossRef] [Green Version]
- Ma, P.C.; Maulik, G.; Christensen, J.; Salgia, R. c-MET: Structure, functions and potential for therapeutic inhibition. Cancer Metastasis Rev. 2003, 22, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Huang, B.; Li, Z.; Li, H.; Sun, L.; Zhang, Q.; Qui, X.; Wang, E. MicroRNA-449a is downregulated in non-small cell lung cancer and inhibits migration and invasion by targeting c-MET. PLoS ONE 2013, 8, e64759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, H.L.; Janmey, P.A. Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 2003, 65, 761–789. [Google Scholar] [CrossRef] [PubMed]
- Rei, K.; Nobes, C.D.; Thomas, G.; Hall, A.; Cantrell, D.A. Phosphatidylinositol 3-kinase signals activate a selective subset of Rac/Rho-dependent effector pathways. Curr. Biol. 1996, 6, 1445–1455. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, G.A.; Falasca, M.; Zhang, Z.; Ong, S.H.; Schlessinger, J. A novel positive feedback loop mediated by the docking protein Gab1 and phosphatidylinositol 3-kinase in epidermal growth factor receptor signaling. Mol. Cell. Biol. 2000, 20, 1448–1459. [Google Scholar] [CrossRef] [Green Version]
- Nagy, T.A.; Frey, M.R.; Yan, F.; Israel, D.A.; Polk, D.B.; Peek, R.M. Helicobacter pylori regulates cellular migration and apoptosis by activation of phosphatidylinositol 3-kinase signaling. J. Infect. Dis. 2009, 199, 641–651. [Google Scholar] [CrossRef] [Green Version]
- Wells, A.; Grandis, J.R. Phospholipase C-γ1 in tumor progression. Clin. Exp. Metastasis 2003, 20, 285. [Google Scholar] [CrossRef]
- Khoshyomn, S.; Penar, P.L.; Rossi, J.; Wellsge, A.; Abramson, D.L.; Bhushan, A. Inhibition of phospholipase C-γ1 activation blocks glioma cell motility and invasion of fetal rat brain aggregates. Neurosurgery 1999, 44, 568–577. [Google Scholar] [CrossRef]
- Mouneimne, G.; Soon, L.; DesMarais, V.; Sidani, M.; Song, X.; Yip, S.C.; Ghosh, M.; Eddy, R.; Backer, J.M.; Condeelis, J. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J. Cell Biol. 2004, 166, 697–708. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Lim, J.W.; Kim, H. Astaxanthin Inhibits Mitochondrial Dysfunction and Interleukin-8 Expression in Helicobacter pylori-Infected Gastric Epithelial Cells. Nutrients 2018, 10, 1320. [Google Scholar] [CrossRef] [Green Version]

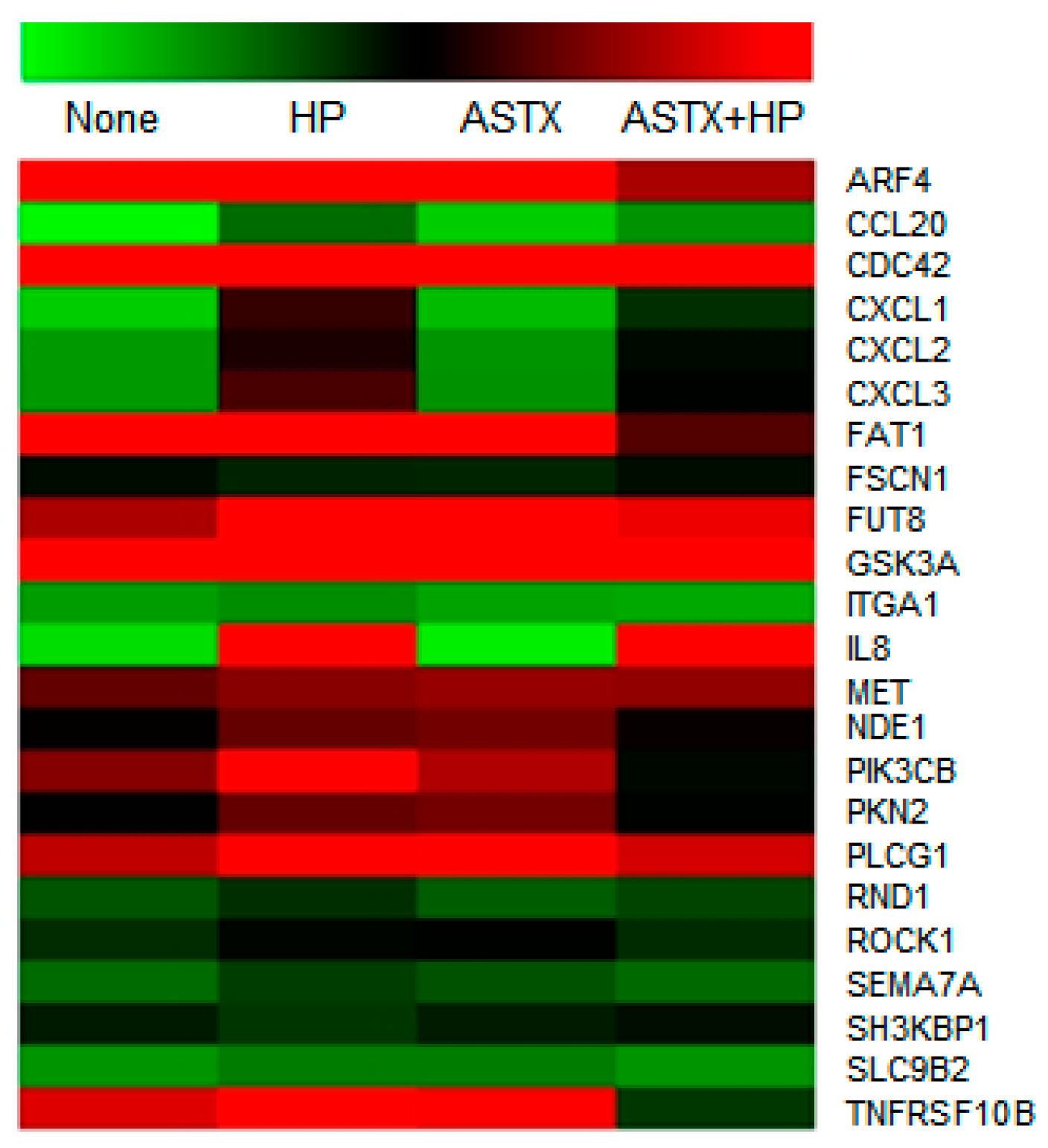
| Gene ID | Normalized (log2) Read Count | Encoded Protein | |||
|---|---|---|---|---|---|
| None | HP | ASTX | ASTX + HP | ||
| ARF4 | 7.424 | 8.152 | 8.192 | 6.187 | ADP-ribosylation factor 4 |
| CCL20 | 0.116 | 2.928 | 0.000 | 2.179 | Chemokine C-C motif ligand 20 |
| CDC42 | 9.142 | 9.921 | 9.389 | 6.982 | Cell division control protein 42 homolog |
| CXCL1 | 1.005 | 5.432 | 1.348 | 4.080 | Chemokine C-X-C motif ligand 1 |
| CXCL2 | 2.008 | 5.274 | 2.124 | 4.697 | Chemokine C-X-C motif ligand 2 |
| CXCL3 | 2.023 | 5.557 | 2.193 | 4.950 | Chemokine C-X-C motif ligand 3 |
| FAT1 | 7.348 | 8.019 | 8.158 | 5.615 | FAT atypical cadherin 1 |
| FUT8 | 6.201 | 7.140 | 7.199 | 6.646 | Fucosyltransferase 8 alpha 1,6 fucosyltransferase |
| GSK3A | 7.385 | 8.294 | 8.267 | 8.362 | Glycogen synthase kinase 3 alpha |
| ITGA1 | 1.946 | 2.266 | 1.831 | 1.728 | Integrin alpha 1 |
| IL8 | 0.704 | 7.118 | 0.341 | 6.954 | Interleukin 8 |
| MET | 5.712 | 6.961 | 6.059 | 6.022 | Met proto-oncogene |
| NDE1 | 5.108 | 5.734 | 5.819 | 5.165 | Nude neurodevelopment protein 1 |
| PIK3CB | 5.937 | 7.222 | 6.233 | 4.757 | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta |
| PKN2 | 4.999 | 5.727 | 5.828 | 4.947 | Protein kinase N2 |
| PLCG1 | 6.315 | 6.806 | 6.962 | 6.451 | Phospholipase C gamma-1 |
| RND1 | 3.343 | 4.082 | 3.228 | 3.647 | Rho family GTPase 1 |
| ROCK1 | 4.122 | 4.820 | 4.957 | 4.123 | Rho-associated coiled-coil containing protein kinase 1 |
| SEMA7A | 2.916 | 3.803 | 3.406 | 2.948 | Semaphoring 7A GPI membrane anchor |
| SLC9B2 | 2.137 | 2.614 | 2.601 | 2.130 | Solute carrier family 9 subfamily B2 |
| TNFRSF10B | 6.533 | 7.260 | 6.763 | 3.900 | Tumor necrosis factor receptor superfamily member 10b |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.H.; Kim, H. Transcriptome Analysis of the Inhibitory Effect of Astaxanthin on Helicobacter pylori-Induced Gastric Carcinoma Cell Motility. Mar. Drugs 2020, 18, 365. https://doi.org/10.3390/md18070365
Kim SH, Kim H. Transcriptome Analysis of the Inhibitory Effect of Astaxanthin on Helicobacter pylori-Induced Gastric Carcinoma Cell Motility. Marine Drugs. 2020; 18(7):365. https://doi.org/10.3390/md18070365
Chicago/Turabian StyleKim, Suhn Hyung, and Hyeyoung Kim. 2020. "Transcriptome Analysis of the Inhibitory Effect of Astaxanthin on Helicobacter pylori-Induced Gastric Carcinoma Cell Motility" Marine Drugs 18, no. 7: 365. https://doi.org/10.3390/md18070365






