The First Plastid Genome of the Holoparasitic Genus Prosopanche (Hydnoraceae)
Abstract
1. Introduction
2. Results
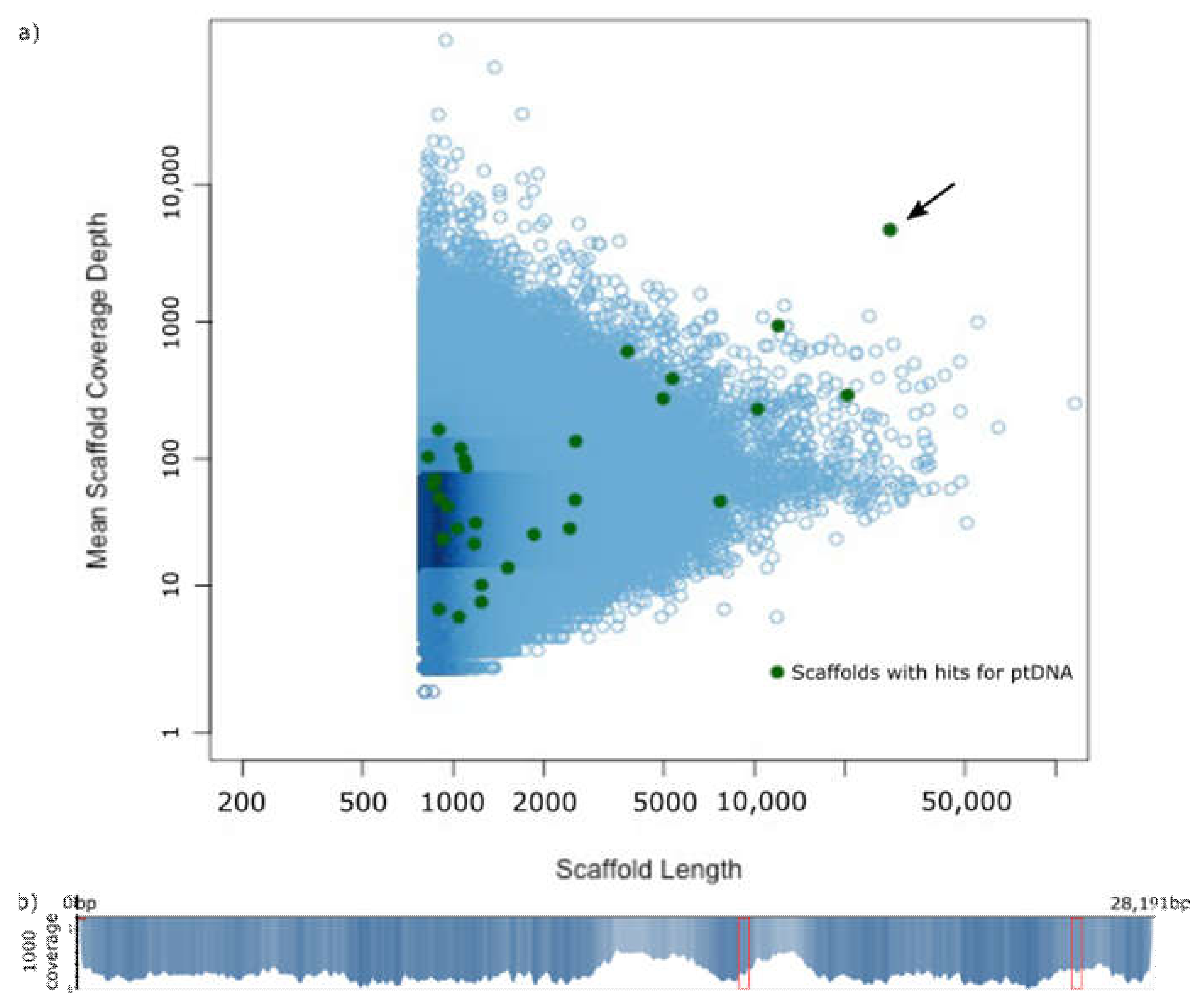
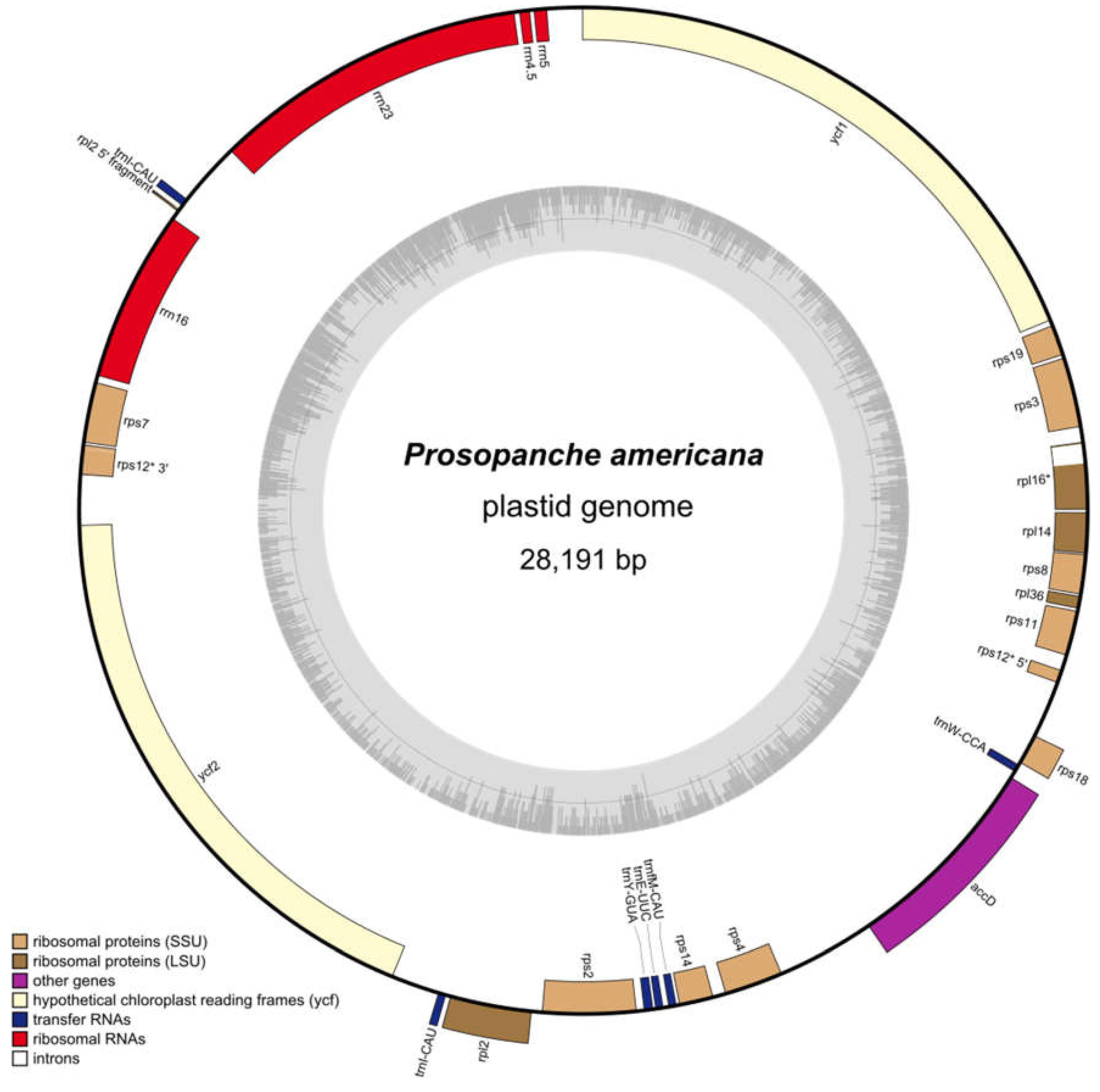
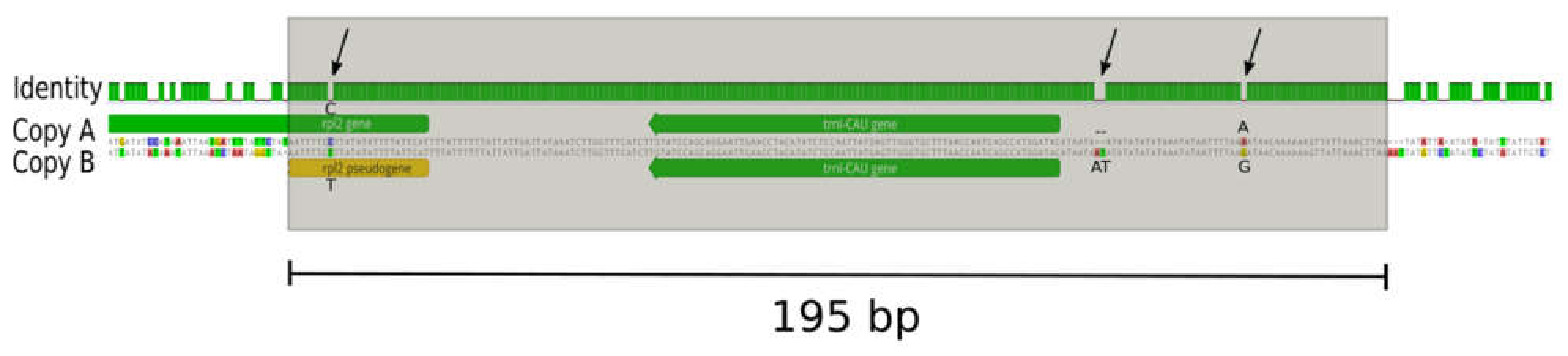
2.1. The Plastome of Prosopanche Americana
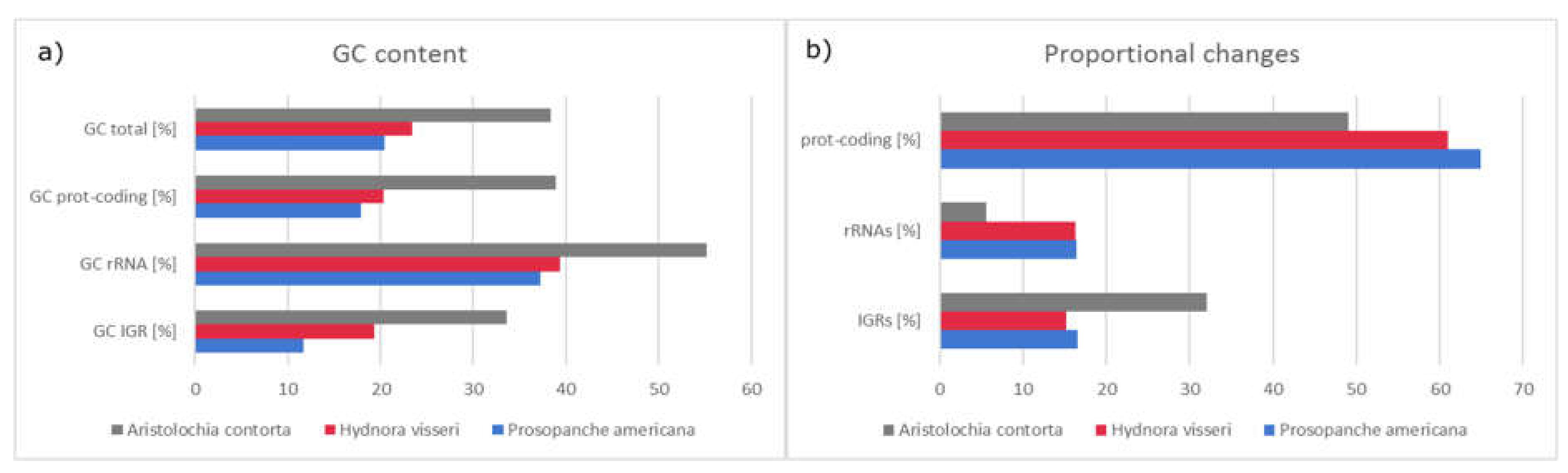
2.2. Comparison of the Plastomes of the Holoparasitic Hydnoraceae
2.3. Plastome Comparison to Aristolochia
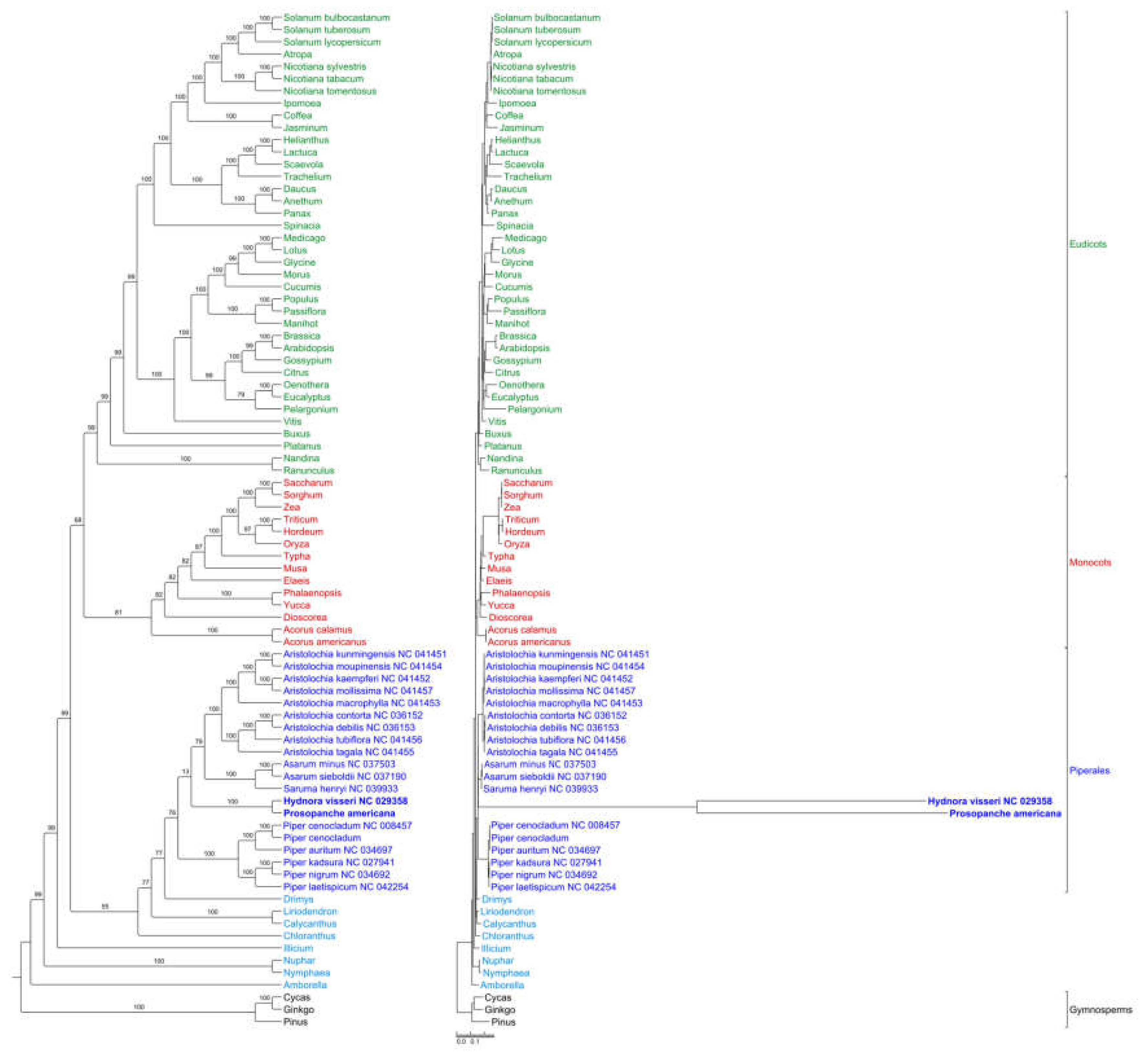
2.4. Phylogenetic Placement of Hydnoraceae
3. Discussion
4. Materials and Methods
4.1. Plant Material, DNA Extraction, Library Preparation and Sequencing
4.2. Assembly and Identification of Scaffolds with Plastid Information
4.3. Gene Annotation and Circularization
4.4. Plastome Comparisons
4.5. Phylogenetic Placement of Prosopanche Americana
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

References
- Palmer, J.D. Comparative organization of chloroplast genomes. Annu. Rev. Genet. 1985, 19, 325–354. [Google Scholar] [CrossRef]
- Ohyama, K.; Fukuzawa, H.; Kohchi, T.; Shirai, H.; Sano, T.; Sano, S.; Umesono, K.; Shiki, Y.; Takeuchi, Y.; Chang, Z.; et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 1986, 322, 572. [Google Scholar] [CrossRef]
- Wicke, S.; Müller, K.F.; de Pamphilis, C.W.; Quandt, D.; Wickett, N.J.; Zhang, Y.; Renner, S.S.; Schneeweiss, G.M. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 2013, 25, 3711–3725. [Google Scholar] [CrossRef] [PubMed]
- Krause, K. Piecing together the puzzle of parasitic plant plastome evolution. Planta 2011, 234, 647. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Naumann, J. Molecular evolution of plastid genomes in parasitic flowering plants. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2018; Volume 85, pp. 315–347. [Google Scholar]
- Barkman, T.J.; McNeal, J.R.; Lim, S.H.; Coat, G.; Croom, H.B.; Young, N.D.; Claude, W.D. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol. Biol. 2007, 7, 248. [Google Scholar] [CrossRef]
- Schneider, A.C.; Braukmann, T.; Banerjee, A.; Stefanovic, S. Convergent plastome evolution and gene loss in holoparasitic Lennoaceae (Boraginales). Genome Biol. Evol. 2018, 10, 2663–2670. [Google Scholar] [CrossRef]
- Wu, C.S.; Wang, T.J.; Wu, C.W.; Wang, Y.N.; Chaw, S.M. Plastome evolution in the sole hemiparasitic genus laurel dodder (Cassytha) and insights into the plastid phylogenomics of Lauraceae. Genome Biol. Evol. 2017, 9, 2604–2614. [Google Scholar] [CrossRef]
- Bellot, S.; Cusimano, N.; Luo, S.; Sun, G.; Zarre, S.; Gröger, A.; Temsch, E.; Renner, S.S. Assembled plastid and mitochondrial genomes, as well as nuclear genes, place the parasite family Cynomoriaceae in the Saxifragales. Genome Biol. Evol. 2016, 8, 2214–2230. [Google Scholar] [CrossRef]
- Naumann, J.; Der, J.P.; Wafula, E.K.; Jones, S.S.; Wagner, S.T.; Honaas, L.A.; Ralph, P.E.; Bolin, J.F.; Maass, E.; Neinhuis, C.; et al. Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae). Genome Biol. Evol. 2016, 8, 345–363. [Google Scholar] [CrossRef]
- Roquet, C.; Coissac, É.; Cruaud, C.; Boleda, M.; Boyer, F.; Alberti, A.; Gielly, L.; Taberlet, P.; Thuiller, W.; Van Es, J.; et al. Understanding the evolution of holoparasitic plants: The complete plastid genome of the holoparasite Cytinus hypocistis (Cytinaceae). Ann. Bot. 2016, 118, 885–896. [Google Scholar] [CrossRef]
- Bellot, S.; Renner, S.S. The plastomes of two species in the endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biol. Evol. 2015, 8, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Petersen, G.; Cuenca, A.; Seberg, O. Plastome evolution in hemiparasitic mistletoes. Genome Biol. Evol. 2015, 7, 2520–2532. [Google Scholar] [CrossRef] [PubMed]
- Molina, J.; Hazzouri, K.M.; Nickrent, D.; Geisler, M.; Meyer, R.S.; Pentony, M.M.; Flowers, J.M.; Pelser, P.; Barcelona, J.; Inovejas, S.A.; et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Mol. Biol. Evol. 2014, 31, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Funk, H.T.; Berg, S.; Krupinska, K.; Maier, U.G.; Krause, K. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 2007, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.W.; Lam, V.K.; Merckx, V.S. Plastomes on the edge: The evolutionary breakdown of mycoheterotroph plastid genomes. New Phytol. 2017, 214, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Barrett, C.F.; Davis, J.I. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am. J. Bot. 2012, 99, 1513–1523. [Google Scholar] [CrossRef]
- Wicke, S.; Müller, K.F.; dePamphilis, C.W.; Quandt, D.; Bellot, S.; Schneeweiss, G.M. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc. Natl. Acad. Sci. USA 2016, 113, 9045–9050. [Google Scholar] [CrossRef]
- Wicke, S. Genomic evolution in Orobanchaceae. In Parasitic Orobanchaceae; Springer: Berlin/Heidelberg, Germany, 2013; pp. 267–286. [Google Scholar]
- Westwood, J.H.; Yoder, J.I.; Timko, M.P.; dePamphilis, C.W. The evolution of parasitism in plants. Trends Plant Sci. 2010, 15, 227–235. [Google Scholar] [CrossRef]
- Arias-Agudelo, L.M.; González, F.; Isaza, J.P.; Alzate, J.F.; Pabón-Mora, N. Plastome reduction and gene content in New World Pilostyles (Apodanthaceae) unveils high similarities to African and Australian congeners. Mol. Phylogenet. Evol. 2019, 135, 193–202. [Google Scholar] [CrossRef]
- Su, H.J.; Barkman, T.J.; Hao, W.; Jones, S.S.; Naumann, J.; Skippington, E.; Wafula, E.K.; Hu, J.M.; Palmer, J.D.; dePamphilis, C.W. Novel genetic code and record-setting AT-richness in the highly reduced plastid genome of the holoparasitic plant Balanophora. Proc. Natl. Acad. Sci. USA 2019, 116, 934–943. [Google Scholar] [CrossRef]
- Lavin, M.; Doyle, J.J.; Palmer, J.D. Evolutionary significance of the loss of the chloroplast-DNA inverted repeat in the Leguminosae subfamily Papilionoideae. Evolution 1990, 44, 390–402. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Osorio, B.; Aldrich, J.; Thompson, W.F. Chloroplast DNA evolution among legumes: Loss of a large inverted repeat occurred prior to other sequence rearrangements. Curr. Genet. 1987, 11, 275–286. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; Depamphilis, C.W.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Guo, W.; Gupta, S.; Fan, W.; Mower, J.P. Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016, 209, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Maréchal, A.; Brisson, N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010, 186, 299–317. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Thompson, W.F. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 1982, 29, 537–550. [Google Scholar] [CrossRef]
- Naumann, J.; Salomo, K.; Der, J.P.; Wafula, E.K.; Bolin, J.F.; Maass, E.; Frenzke, L.; Samain, M.S.; Neinhuis, C.; dePamphilis, C.W.; et al. Single-copy nuclear genes place haustorial Hydnoraceae within Piperales and reveal a Cretaceous origin of multiple parasitic angiosperm lineages. PLoS ONE 2013, 8, e79204. [Google Scholar] [CrossRef]
- Bolin, J.F.; Lupton, D.; Musselman, L.J. Hydnora arabica (Aristolochiaceae), a new species from the Arabian Peninsula and a key to Hydnora. Phytotaxa 2018, 338, 99. [Google Scholar] [CrossRef]
- Funez, L.A.; Ribeiro-Nardes, W.; Kossmann, T.; Peroni, N.; Drechsler-Santos, E.R. Prosopanche demogorgoni: A new species of Prosopanche (Aristolochiaceae: Hydnoroideae) from southern Brazil. Phytotaxa 2019, 422, 93–100. [Google Scholar] [CrossRef]
- Musselman, L.J.; Visser, J.H. Taxonomy and natural history of Hydnora (Hydnoraceae). Aliso J. Syst. Evol. Bot. 1989, 12, 317–326. [Google Scholar] [CrossRef]
- Nickrent, D.L.; Blarer, A.; Qiu, Y.L.; Soltis, D.E.; Soltis, P.S.; Zanis, M. Molecular data place Hydnoraceae with Aristolochiaceae. Am. J. Bot. 2002, 89, 1809–1817. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, X.; Cui, Y.; Sun, W.; Li, Y.; Wang, Y.; Song, J.; Yao, H. Molecular structure and phylogenetic analyses of complete chloroplast genomes of two Aristolochia medicinal species. Int. J. Mol. Sci. 2017, 18, 1839. [Google Scholar] [CrossRef] [PubMed]
- CLC. Genomics Workbench. Available online: https://www.qiagenbioinformatics.com (accessed on 15 February 2020).
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2014, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Beck, N.; Lang, B. MFannot, Organelle Genome Annotation Websever; Université de Montréal: Montréal, QC, Canada, 2010. [Google Scholar]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Search and Contextual Analysis of Transfer RNA Genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Geneious 11.1.5. Available online: https://www.geneious.com (accessed on 22 January 2020).
- Peden, J. CodonW Version 1.4. 2. 2005. Available online: http://codonw.sourceforge.net/ (accessed on 15 February 2020).
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Depamphilis, C.W.; Leebens-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef]
- Massoni, J.; Forest, F.; Sauquet, H. Increased sampling of both genes and taxa improves resolution of phylogenetic relationships within Magnoliidae, a large and early-diverging clade of angiosperms. Mol. Phylogenet. Evol. 2014, 70, 84–93. [Google Scholar] [CrossRef]
- Palmer, J.D. Chloroplast DNA exists in two orientations. Nature 1983, 301, 92–93. [Google Scholar] [CrossRef]
- Zhang, H.R.; Zhang, X.C.; Xiang, Q.P. Directed repeats co-occur with few short-dispersed repeats in plastid genome of a Spikemoss, Selaginella vardei (Selaginellaceae, Lycopodiopsida). BMC Genom. 2019, 20, 484. [Google Scholar]
- Xu, Z.; Xin, T.; Bartels, D.; Li, Y.; Gu, W.; Yao, H.; Liu, S.; Yu, H.; Pu, X.; Zhou, J.; et al. Genome analysis of the ancient tracheophyte Selaginella tamariscina reveals evolutionary features relevant to the acquisition of desiccation tolerance. Mol. Plant 2018, 11, 983–994. [Google Scholar] [CrossRef]
- Mower, J.P.; Ma, P.F.; Grewe, F.; Taylor, A.; Michael, T.P.; VanBuren, R.; Qiu, Y.L. Lycophyte plastid genomics: Extreme variation in GC, gene and intron content and multiple inversions between a direct and inverted orientation of the rRNA repeat. New Phytol. 2019, 222, 1061–1075. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, T.; Yuan, X.; Huang, H.; Yao, G.; Mo, X.; Xue, X.; Yan, H. The plastid genome and its implications in barcoding specific-chemotypes of the medicinal herb Pogostemon cablin in China. PLoS ONE 2019, 14, e0215512. [Google Scholar] [CrossRef] [PubMed]
- Schelkunov, M.I.; Shtratnikova, V.Y.; Nuraliev, M.S.; Selosse, M.A.; Penin, A.A.; Logacheva, M.D. Exploring the limits for reduction of plastid genomes: A case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol. Evol. 2015, 7, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2016; Available online: https://rstudio.com/ (accessed on 15 February 2020).
- Harris, R.S. Improved Pairwise Alignmnet of Genomic DNA. Ph.D. Thesis, Penn State University, State College, PA, USA, 2007. [Google Scholar]
- Geneious 8.1. Available online: https://www.geneious.com (accessed on 22 January 2020).
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Stöver, B.C.; Müller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010, 11, 7. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jost, M.; Naumann, J.; Rocamundi, N.; Cocucci, A.A.; Wanke, S. The First Plastid Genome of the Holoparasitic Genus Prosopanche (Hydnoraceae). Plants 2020, 9, 306. https://doi.org/10.3390/plants9030306
Jost M, Naumann J, Rocamundi N, Cocucci AA, Wanke S. The First Plastid Genome of the Holoparasitic Genus Prosopanche (Hydnoraceae). Plants. 2020; 9(3):306. https://doi.org/10.3390/plants9030306
Chicago/Turabian StyleJost, Matthias, Julia Naumann, Nicolás Rocamundi, Andrea A. Cocucci, and Stefan Wanke. 2020. "The First Plastid Genome of the Holoparasitic Genus Prosopanche (Hydnoraceae)" Plants 9, no. 3: 306. https://doi.org/10.3390/plants9030306
APA StyleJost, M., Naumann, J., Rocamundi, N., Cocucci, A. A., & Wanke, S. (2020). The First Plastid Genome of the Holoparasitic Genus Prosopanche (Hydnoraceae). Plants, 9(3), 306. https://doi.org/10.3390/plants9030306



