Probing the Antitumor Mechanism of Solanum nigrum L. Aqueous Extract against Human Breast Cancer MCF7 Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Aqueous Extract of S. nigrum L. (AESN)
2.3. Cell Culture
2.4. Cytotoxicity Assay
2.5. Wound Healing Migration Assay
2.6. Hexokinase Activity Assay
2.7. Pyruvate Kinase Assay
2.8. JC-1-Mitochondrial Membrane Potential Assay
2.9. Real-Time RT-PCR
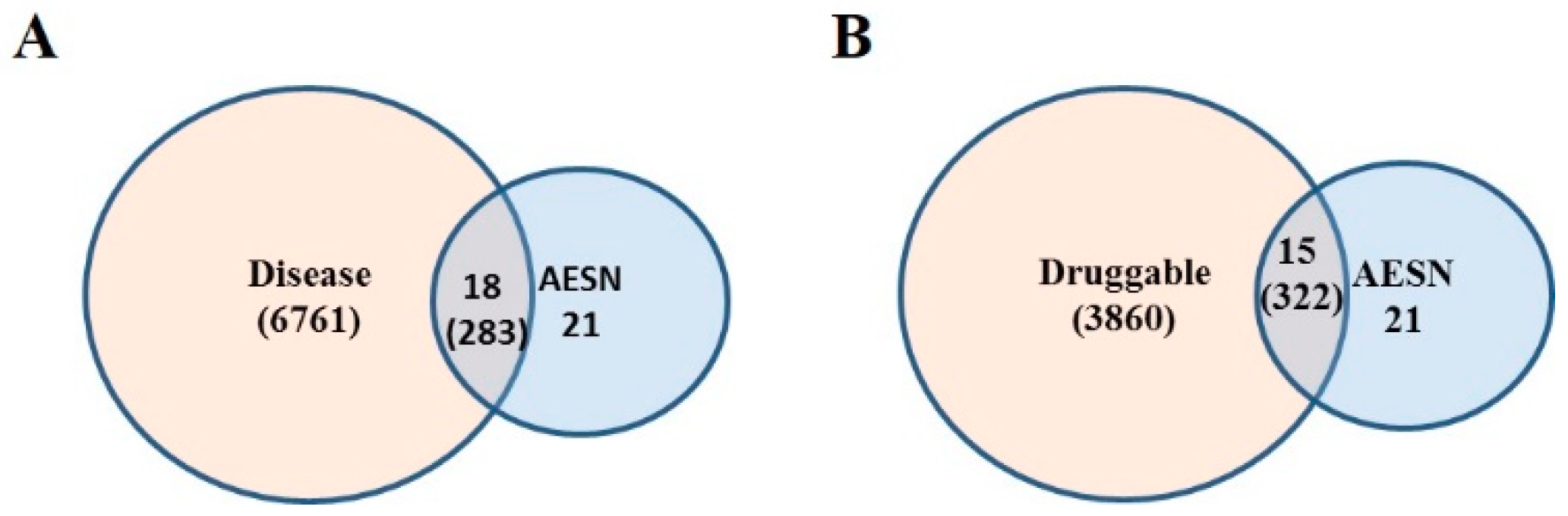
2.10. Druggable Genome
2.11. Disease Gene Set
2.12. Functional Disease Ontology (FunDO) Analysis
2.13. Statistical Analysis
3. Results
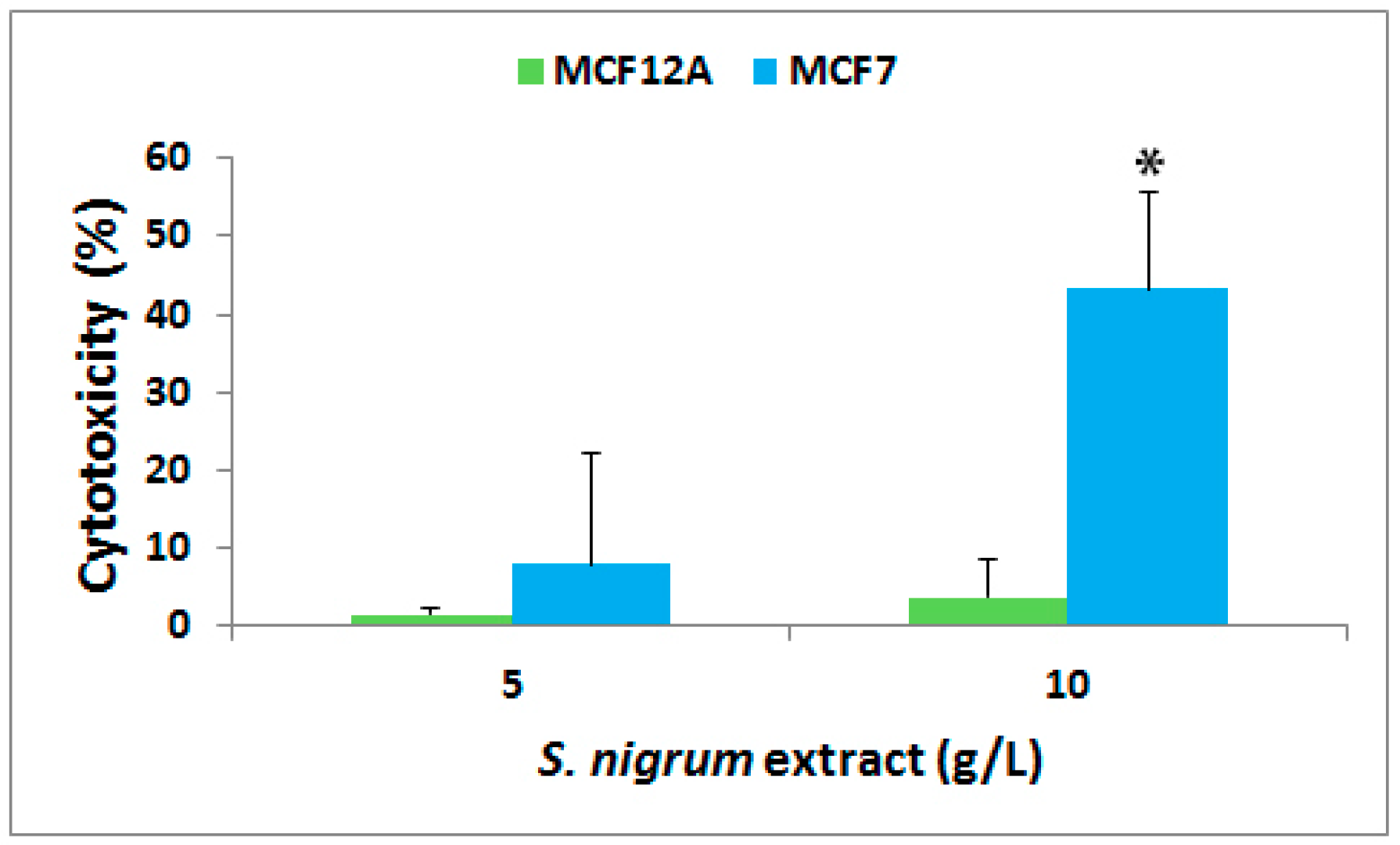
3.1. Effect on Cytotoxicity
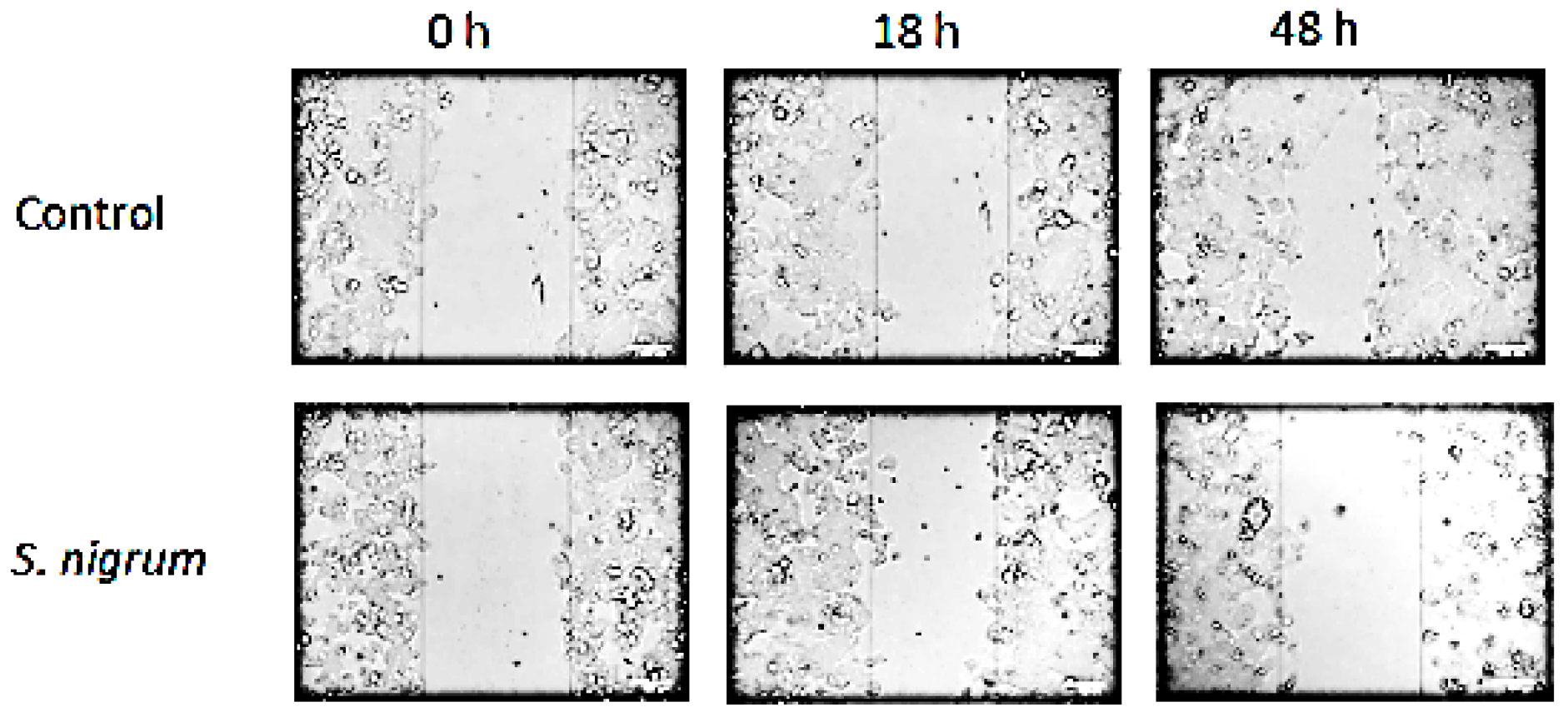
3.2. Effect on Cell Migration
3.3. Effect on Cell Glycolysis
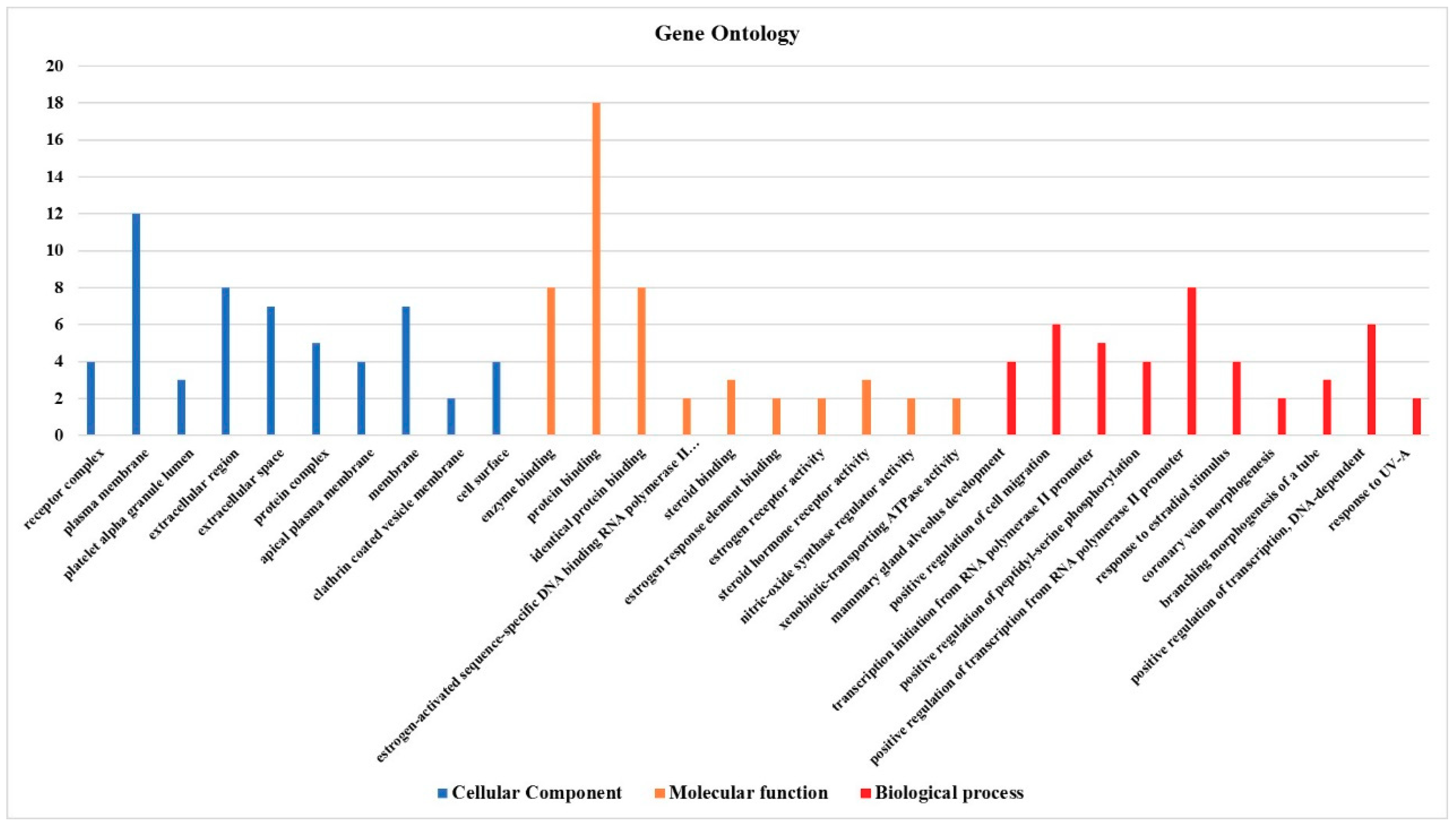
3.4. Effect on the Expression of a Panel of 89-Cancer-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.K.; Buchholz, T.A.; Aggarwal, B.B. Chemosensitization and radiosensitization of tumors by plant polyphenols. Antioxid. Redox Signal. 2005, 7, 1630–1647. [Google Scholar] [CrossRef] [PubMed]
- Konkimalla, V.B.; Efferth, T. Evidence-based Chinese medicine for cancer therapy. J. Ethnopharmacol. 2008, 116, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.S.; Apaya, M.K.; Shyur, L.F. Herbal medicine and acupuncture for breast cancer palliative care and adjuvant therapy. Evid. Based Complement. Alternat. Med. 2013, 2013, 437948. [Google Scholar] [CrossRef]
- Li, S.; Luo, X. Compendium of Materia Medica: (Bencao Gangmu); Foreign Languages Press: Beijing, China, 2003. [Google Scholar]
- Ling, B.; Michel, D.; Sakharkar, M.K.; Yang, J. Evaluating the cytotoxic effects of the water extracts of four anticancer herbs against human malignant melanoma cells. Drug Des. Devel. Ther. 2016, 10, 3563–3572. [Google Scholar] [CrossRef]
- Uen, W.C.; Lee, B.H.; Shi, Y.C.; Wu, S.C.; Tai, C.J.; Tai, C.J. Inhibition of aqueous extracts of Solanum nigrum (AESN) on oral cancer through regulation of mitochondrial fission. J. Tradit. Complement. Med. 2018, 8, 220–225. [Google Scholar] [CrossRef]
- Huang, H.C.; Syu, K.Y.; Lin, J.K. Chemical composition of Solanum nigrum linn extract and induction of autophagy by leaf water extract and its major flavonoids in AU565 breast cancer cells. J. Agric. Food Chem. 2010, 58, 8699–8708. [Google Scholar] [CrossRef]
- Li, J.; Li, Q.; Feng, T.; Li, K. Aqueous extract of Solanum nigrum inhibit growth of cervical carcinoma (U14) via modulating immune response of tumor bearing mice and inducing apoptosis of tumor cells. Fitoterapia 2008, 79, 548–556. [Google Scholar] [CrossRef]
- Lin, H.M.; Tseng, H.C.; Wang, C.J.; Chyau, C.C.; Liao, K.K.; Peng, P.L.; Chou, F.P. Induction of autophagy and apoptosis by the extract of Solanum nigrum Linn in HepG2 cells. J. Agric. Food Chem. 2007, 55, 3620–3628. [Google Scholar] [CrossRef]
- Tai, C.J.; Wang, C.K.; Chang, Y.J.; Lin, C.S.; Tai, C.J. Aqueous extract of Solanum nigrum leaf activates autophagic cell death and enhances docetaxel-induced cytotoxicity in human endometrial carcinoma cells. Evid. Based Complement. Alternat. Med. 2012, 2012, 859185. [Google Scholar] [CrossRef]
- Tai, C.J.; Wang, C.K.; Tai, C.J.; Lin, Y.F.; Lin, C.S.; Jian, J.Y.; Chang, Y.J.; Chang, C.C. Aqueous extract of Solanum nigrum leaves induces autophagy and enhances cytotoxicity of cisplatin, doxorubicin, docetaxel, and 5-fluorouracil in human colorectal carcinoma cells. Evid. Based Complement. Alternat. Med. 2013, 2013, 514719. [Google Scholar] [PubMed]
- Fu, R.; Wang, X.; Hu, Y.; Du, H.; Dong, B.; Ao, S.; Zhang, L.; Sun, Z.; Zhang, L.; Lv, G.; et al. Solamargine inhibits gastric cancer progression by regulating the expression of lncNEAT1_2 via the MAPK signaling pathway. Int. J. Oncol. 2019, 54, 1545–1554. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zhang, X.; Chen, J.; Tang, X.; Wang, M.; Zhang, L.; Guo, Z.; Shen, W. Fe3O4-solamargine induces apoptosis and inhibits metastasis of pancreatic cancer cells. Int. J. Oncol. 2019, 54, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tang, Q.; Xiao, Q.; Yang, L.; Hann, S.S. Targeting EP 4 downstream c-Jun through ERK 1/2-mediated reduction of DNMT 1 reveals novel mechanism of solamargine-inhibited growth of lung cancer cells. J. Cell Mol. Med. 2017, 21, 222–233. [Google Scholar] [CrossRef]
- Al Sinani, S.S.; Eltayeb, E.A.; Coomber, B.L.; Adham, S.A. Solamargine triggers cellular necrosis selectively in different types of human melanoma cancer cells through extrinsic lysosomal mitochondrial death pathway. Cancer Cell Int. 2016, 16, 11. [Google Scholar] [CrossRef]
- Xiang, S.; Zhang, Q.; Tang, Q.; Zheng, F.; Wu, J.; Yang, L.; Hann, S.S. Activation of AMPKα mediates additive effects of solamargine and metformin on suppressing MUC1 expression in castration-resistant prostate cancer cells. Sci. Rep. 2016, 6, 36721. [Google Scholar] [CrossRef]
- Sani, I.K.; Marashi, S.H.; Kalalinia, F. Solamargine inhibits migration and invasion of human hepatocellular carcinoma cells through down-regulation of matrix metalloproteinases 2 and 9 expression and activity. Toxicol. In Vitro 2015, 29, 893–900. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, Z.; Xu, T.; An, Z.; Chen, W.; Wang, X.; Huang, M.; Zhu, F. Solamargine derived from Solanum nigrum induces apoptosis of human cholangiocarcinoma QBC939 cells. Oncol. Lett. 2018, 15, 6329–6335. [Google Scholar]
- Wang, H.C.; Wu, D.H.; Chang, Y.C.; Li, Y.J.; Wang, C.J. Solanum nigrum Linn. water extract inhibits metastasis in mouse melanoma cells in vitro and in vivo. J. Agric. Food Chem. 2010, 58, 11913–11923. [Google Scholar] [CrossRef]
- Zhao, Z.; Jia, Q.; Wu, M.S.; Xie, X.; Wang, Y.; Song, G.; Zou, C.Y.; Tang, Q.; Lu, J.; Huang, G.; et al. Degalactotigonin, a Natural Compound from Solanum nigrum L., Inhibits Growth and Metastasis of Osteosarcoma through GSK3beta Inactivation-Mediated Repression of the Hedgehog/Gli1 Pathway. Clin. Cancer Res. 2018, 24, 130–144. [Google Scholar] [CrossRef]
- Lai, Y.J.; Tai, C.J.; Wang, C.W.; Choong, C.Y.; Lee, B.H.; Shi, Y.C.; Tai, C.J. Anti-Cancer Activity of Solanum nigrum (AESN) through Suppression of Mitochondrial Function and Epithelial-Mesenchymal Transition (EMT) in Breast Cancer Cells. Molecules 2016, 21, 553. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.O.; Kim, J.; Lim, J.C.; Chung, Y.; Chung, G.H.; Lee, J.C. Ripe fruit of Solanum nigrum L. inhibits cell growth and induces apoptosis in MCF-7 cells. Food Chem. Toxicol. 2003, 41, 1421–1428. [Google Scholar] [CrossRef]
- Heo, K.S.; Lee, S.J.; Ko, J.H.; Lim, K.; Lim, K.T. Glycoprotein isolated from Solanum nigrum L. inhibits the DNA-binding activities of NF-kappaB and AP-1, and increases the production of nitric oxide in TPA-stimulated MCF-7 cells. Toxicol. In Vitro 2004, 18, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Tuan Anh, H.L.; Tran, P.T.; Thao, D.T.; Trang, D.T.; Dang, N.H.; Van Cuong, P.; Kiem, P.V.; Minh, C.V.; Lee, J.H. Degalactotigonin, a Steroidal Glycoside from Solanum nigrum, Induces Apoptosis and Cell Cycle Arrest via Inhibiting the EGFR Signaling Pathways in Pancreatic Cancer Cells. BioMed Res. Int. 2018, 2018, 3120972. [Google Scholar] [CrossRef]
- Shen, K.H.; Liao, A.C.; Hung, J.H.; Lee, W.J.; Hu, K.C.; Lin, P.T.; Liao, R.F.; Chen, P.S. α-Solanine inhibits invasion of human prostate cancer cell by suppressing epithelial-mesenchymal transition and MMPs expression. Molecules 2014, 19, 11896–11914. [Google Scholar] [CrossRef]
- Weihua, Z.; Saji, S.; Makinen, S.; Cheng, G.; Jensen, E.V.; Warner, M.; Gustafsson, J.A. Estrogen receptor (ER) beta, a modulator of ERalpha in the uterus. Proc. Natl. Acad. Sci. USA 2000, 97, 5936–5941. [Google Scholar] [CrossRef]
- Christie, E.L.; Pattnaik, S.; Beach, J.; Copeland, A.; Rashoo, N.; Fereday, S.; Hendley, J.; Alsop, K.; Brady, S.L.; Lamb, G.; et al. Multiple ABCB1 transcriptional fusions in drug resistant high-grade serous ovarian and breast cancer. Nat. Commun. 2019, 10, 1295. [Google Scholar] [CrossRef]
- Gilroy, D.W.; Colville-Nash, P.R. New insights into the role of COX 2 in inflammation. J. Mol. Med. (BERL) 2000, 78, 121–129. [Google Scholar] [CrossRef]
- Goos, J.A.; Hiemstra, A.C.; Coupe, V.M.; Diosdado, B.; Kooijman, W.; Delis-Van Diemen, P.M.; Karga, C.; Belien, J.A.; Menke-van der Houven van Oordt, C.W.; Geldof, A.A.; et al. PET group. Epidermal growth factor receptor (EGFR) and prostaglandin-endoperoxide synthase 2 (PTGS2) are prognostic biomarkers for patients with resected colorectal cancer liver metastases. Br. J. Cancer 2014, 111, 749–755. [Google Scholar] [CrossRef]
- Kuwano, T.; Nakao, S.; Yamamoto, H.; Tsuneyoshi, M.; Yamamoto, T.; Kuwano, M.; Ono, M. Cyclooxygenase 2 is a key enzyme for inflammatory cytokine-induced angiogenesis. FASEB J. 2004, 18, 300–310. [Google Scholar] [CrossRef]
- Zaidi, S.K.; Hoda, M.N.; Tabrez, S.; Ansari, S.A.; Jafri, M.A.; Shahnawaz Khan, M.; Hasan, S.; Alqahtani, M.H.; Mohammed Abuzenadah, A.; Banu, N. Protective Effect of Solanum nigrum Leaves Extract on Immobilization Stress Induced Changes in Rat’s Brain. Evid. Based Complement. Alternat. Med. 2014, 2014, 912450. [Google Scholar] [CrossRef] [PubMed]
- Eckel, R.H.; Kahn, S.E.; Ferrannini, E.; Goldfine, A.B.; Nathan, D.M.; Schwartz, M.W.; Smith, R.J.; Smith, S.R. Obesity and type 2 diabetes: What can be unified and what needs to be individualized? J. Clin. Endocrinol. Metab. 2011, 96, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Twig, G.; Afek, A.; Derazne, E.; Tzur, D.; Cukierman-Yaffe, T.; Gerstein, H.C.; Tirosh, A. Diabetes risk among overweight and obese metabolically healthy young adults. Diabetes Care 2014, 37, 2989–2995. [Google Scholar] [CrossRef] [PubMed]
- Rugo, H.; Shtivelman, E.; Perez, A.; Vogel, C.; Franco, S.; Tan Chiu, E.; Melisko, M.; Tagliaferri, M.; Cohen, I.; Shoemaker, M.; et al. Phase I trial and antitumor effects of BZL101 for patients with advanced breast cancer. Breast Cancer Res. Treat. 2007, 105, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Perez, A.T.; Arun, B.; Tripathy, D.; Tagliaferri, M.A.; Shaw, H.S.; Kimmick, G.G.; Cohen, I.; Shtivelman, E.; Caygill, K.A.; Grady, D.; et al. A phase 1B dose escalation trial of Scutellaria barbata (BZL101) for patients with metastatic breast cancer. Breast Cancer Res. Treat. 2010, 120, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Salmenperä, L.; Suominen, T.; Vertio, H. Physicians’ attitudes towards the use of complementary therapies (CTs) by cancer patients in Finland. Eur. J. Cancer Care (Engl.) 2003, 12, 358–364. [Google Scholar] [CrossRef]
- Mohd Mujar, N.M.; Dahlui, M.; Emran, N.A.; Abdul Hadi, I.; Wai, Y.Y.; Arulanantham, S.; Hooi, C.C.; Mohd Taib, N.A. Complementary and alternative medicine (CAM) use and delays in presentation and diagnosis of breast cancer patients in public hospitals in Malaysia. PLoS ONE 2017, 12, e0176394. [Google Scholar] [CrossRef]
- Johnson, S.B.; Park, H.S.; Gross, C.P.; Yu, J.B. Complementary Medicine, Refusal of Conventional Cancer Therapy, and Survival among Patients with Curable Cancers. JAMA Oncol. 2018, 4, 1375–1381. [Google Scholar] [CrossRef]

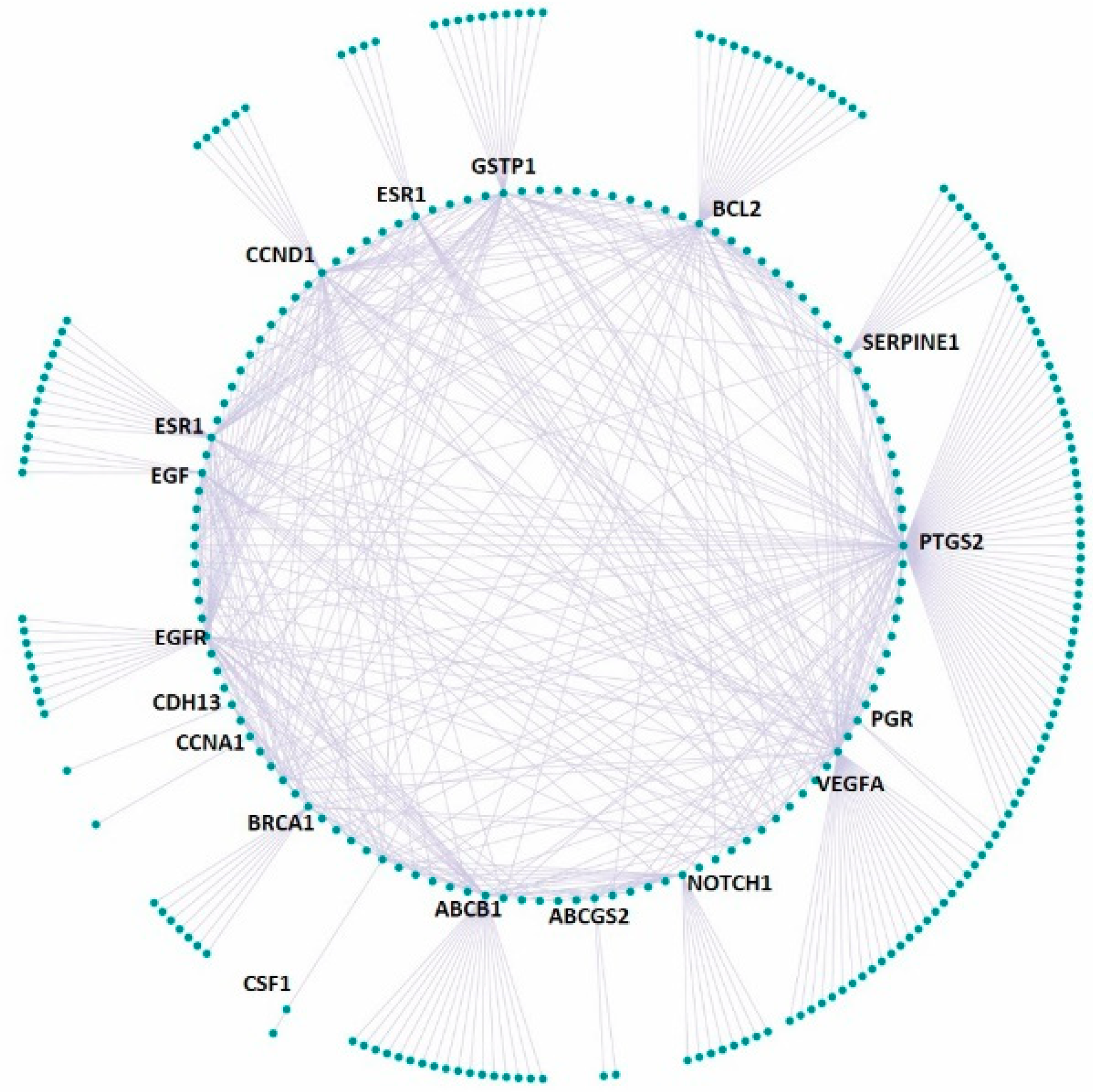
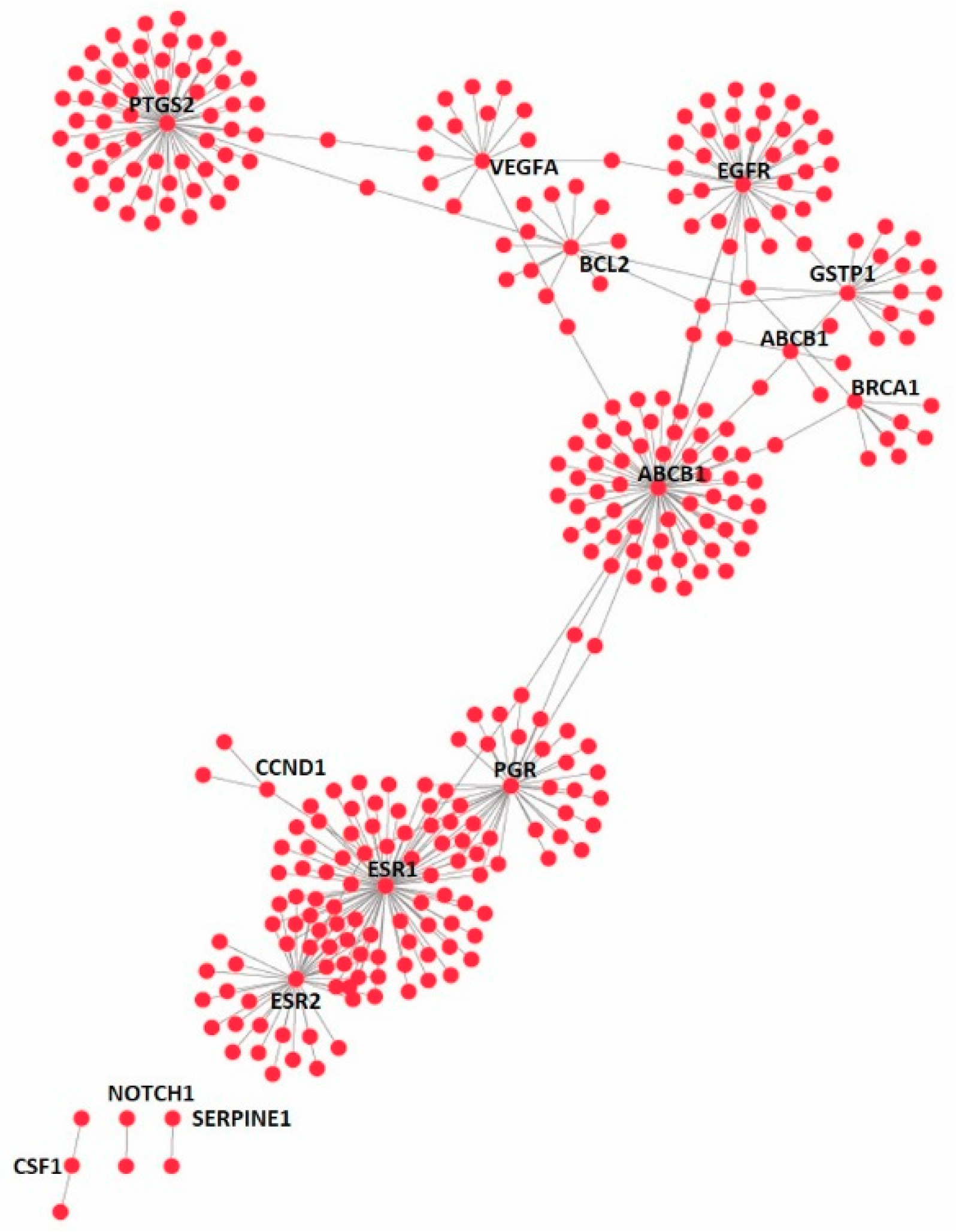

| Gene Name | Gene Name | Gene Name | Gene Name | ||||
|---|---|---|---|---|---|---|---|
| ABCB1 | −2.5 | CST6 | −1.0 | KRT8 | −1.7 | SERPINE1 | 3.1 |
| ABCG2 | 2.5 | CTNNB1 | −1.3 | MAPK1 | 1.3 | SFN | 1.4 |
| ADAM23 | 3.9 | CTSD | 1.0 | MAPK3 | 1.1 | SFRP1 | −1.0 |
| AKT1 | 1.1 | EGF | 2.8 | MAPK8 | 1.4 | SLC39A6 | −1.6 |
| APC | 1.3 | EGFR | 2.0 | MGMT | −1.1 | SLIT2 | −1.3 |
| AR | −1.2 | ERBB2 | −1.3 | MKI67 | 1.7 | SSNAI2 | 1.4 |
| ATM | 1.2 | ESR1 | −4.3 | MLH1 | 1.1 | SRC | 1.6 |
| BAD | 1.3 | ESR2 | 2.3 | MMP2 | −1.0 | TFF3 | −1.3 |
| BCL2 | −2.9 | FOXA1 | −1.1 | MMP9 | 1.1 | TGFB1 | −1.1 |
| BIRC5 | −1.3 | GATA3 | −1.6 | MUC1 | −1.3 | THBS1 | −1.9 |
| BRCA1 | 2.1 | GLI1 | −1.4 | MYC | −1.0 | TP53 | 1.1 |
| BRCA2 | 1.9 | GRB7 | 1.0 | NME1 | 1.0 | TP73 | −1.3 |
| CCNA1 | 3.8 | GSTP1 | 3.0 | NOTCH1 | 2.0 | TWIST1 | −1.4 |
| CCND1 | −2.3 | HIC1 | 1.7 | NR3C1 | 1.3 | VEGFA | 3.0 |
| CCND2 | 1.6 | ID1 | 1.3 | PGR | −3.5 | XBP1 | −1.6 |
| CCNE1 | 1.1 | IGF1 | −1.5 | PLAU | 1.5 | B2M | −1.1 |
| CDH1 | −1.4 | IGF1R | −1.2 | PRDM2 | 1.2 | HPRT1 | 1.2 |
| CDH13 | −2.4 | IGFBP3 | 1.2 | PTEN | 1.2 | RPL13A | −1.0 |
| CDK2 | −1.1 | IL6 | 2.3 | PTGS2 | −2.3 | GAPDH | 1.1 |
| CDKN1A | 1.5 | JUN | 1.9 | PYCARD | −1.1 | ACTB | −1.1 |
| CDKN1C | 1.0 | KRT18 | −1.4 | RARB | −1.1 | ||
| CDKN2A | −1.0 | KRT19 | −2.1 | RASSF1 | 1.4 | ||
| CSF1 | 3.4 | KRT5 | −1.2 | RB1 | −1.0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ling, B.; Xiao, S.; Yang, J.; Wei, Y.; Sakharkar, M.K.; Yang, J. Probing the Antitumor Mechanism of Solanum nigrum L. Aqueous Extract against Human Breast Cancer MCF7 Cells. Bioengineering 2019, 6, 112. https://doi.org/10.3390/bioengineering6040112
Ling B, Xiao S, Yang J, Wei Y, Sakharkar MK, Yang J. Probing the Antitumor Mechanism of Solanum nigrum L. Aqueous Extract against Human Breast Cancer MCF7 Cells. Bioengineering. 2019; 6(4):112. https://doi.org/10.3390/bioengineering6040112
Chicago/Turabian StyleLing, Binbing, Shujun Xiao, Jinha Yang, Ying Wei, Meena K. Sakharkar, and Jian Yang. 2019. "Probing the Antitumor Mechanism of Solanum nigrum L. Aqueous Extract against Human Breast Cancer MCF7 Cells" Bioengineering 6, no. 4: 112. https://doi.org/10.3390/bioengineering6040112
APA StyleLing, B., Xiao, S., Yang, J., Wei, Y., Sakharkar, M. K., & Yang, J. (2019). Probing the Antitumor Mechanism of Solanum nigrum L. Aqueous Extract against Human Breast Cancer MCF7 Cells. Bioengineering, 6(4), 112. https://doi.org/10.3390/bioengineering6040112






