Generation of Synthetic Hyperspectral Image Cube for Mapping Soil Organic Carbon Using Proximal Remote Sensing †
Abstract
1. Introduction
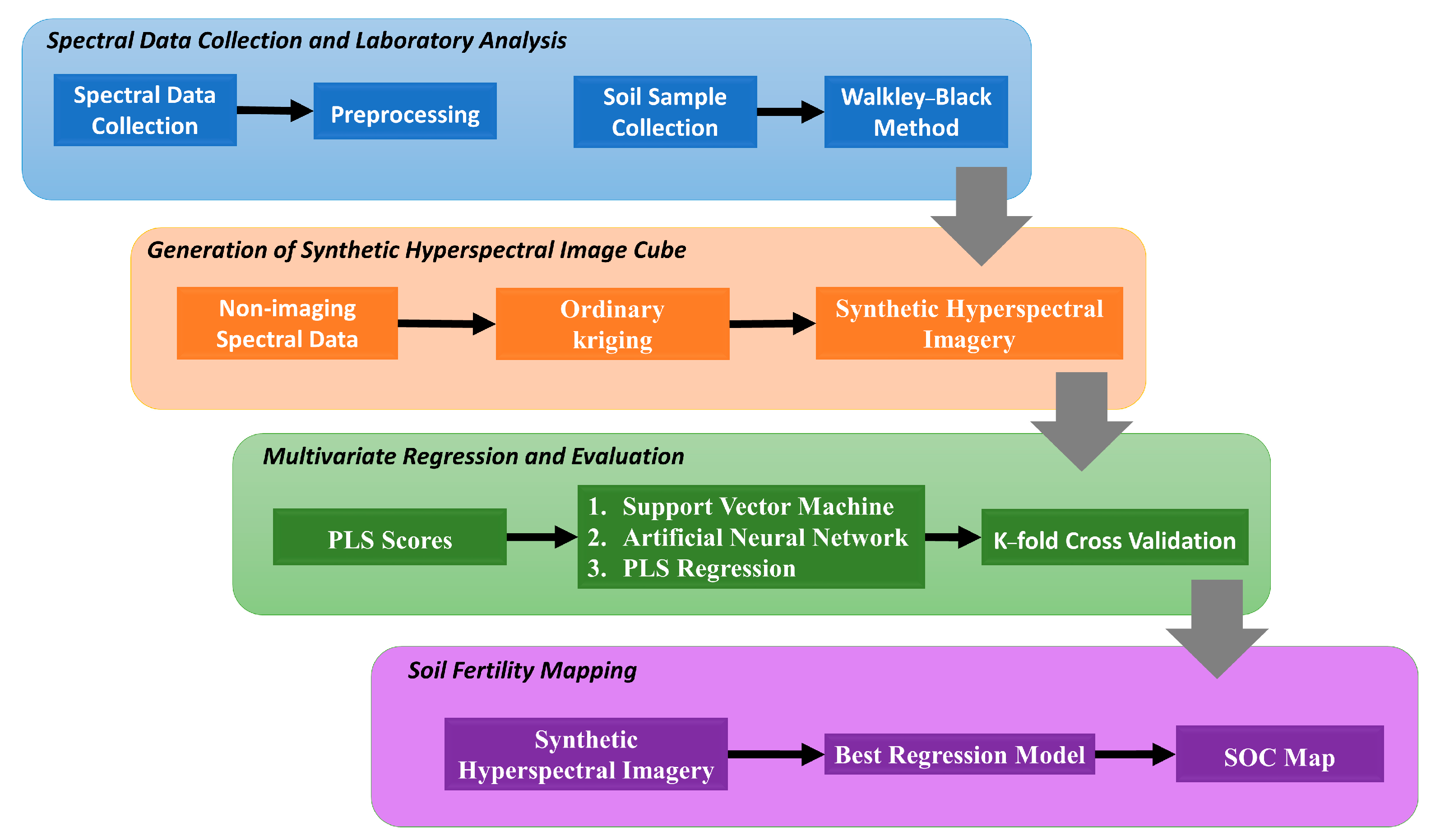
2. Materials and Methods
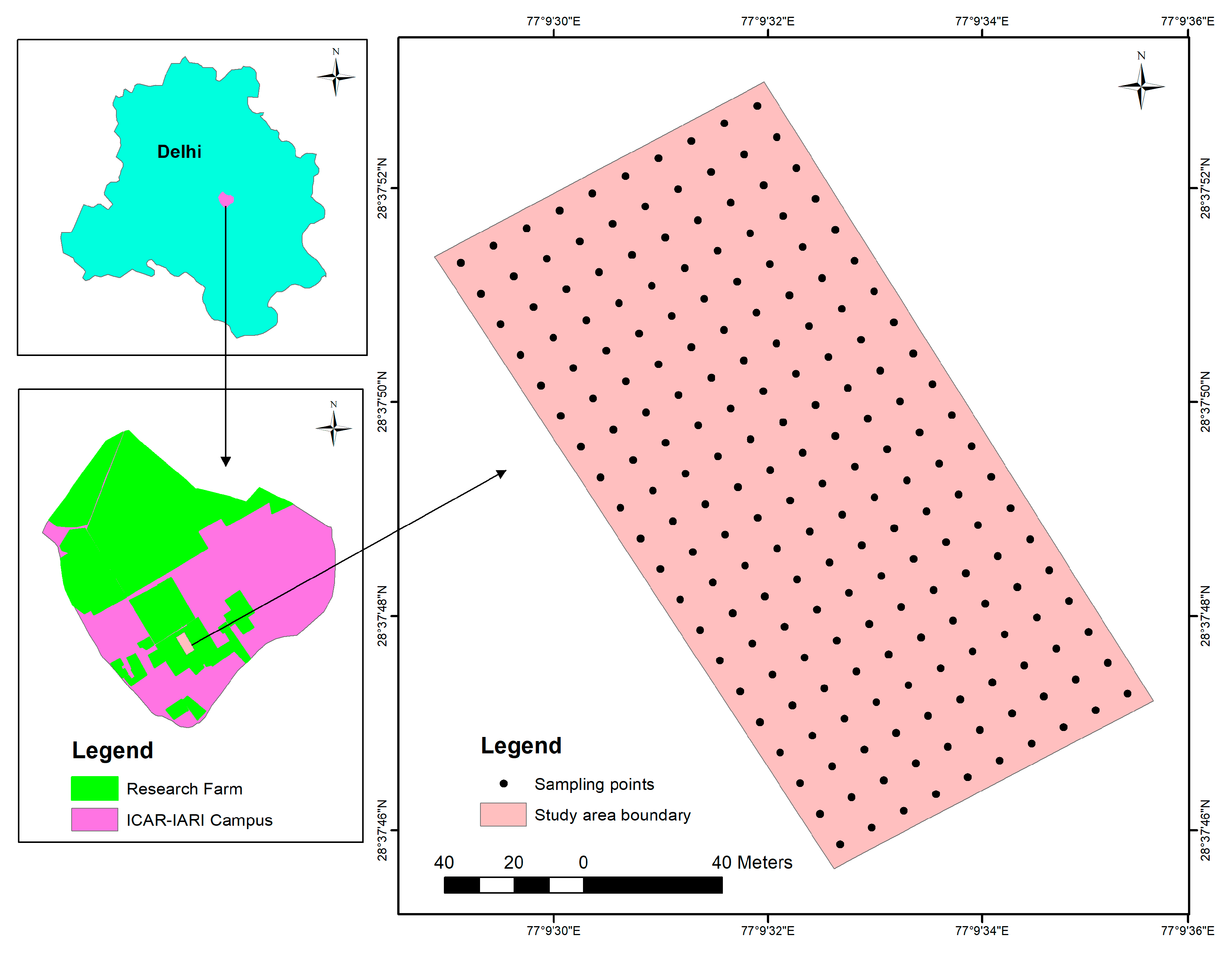
2.1. Study Area and Spectral Data Collection
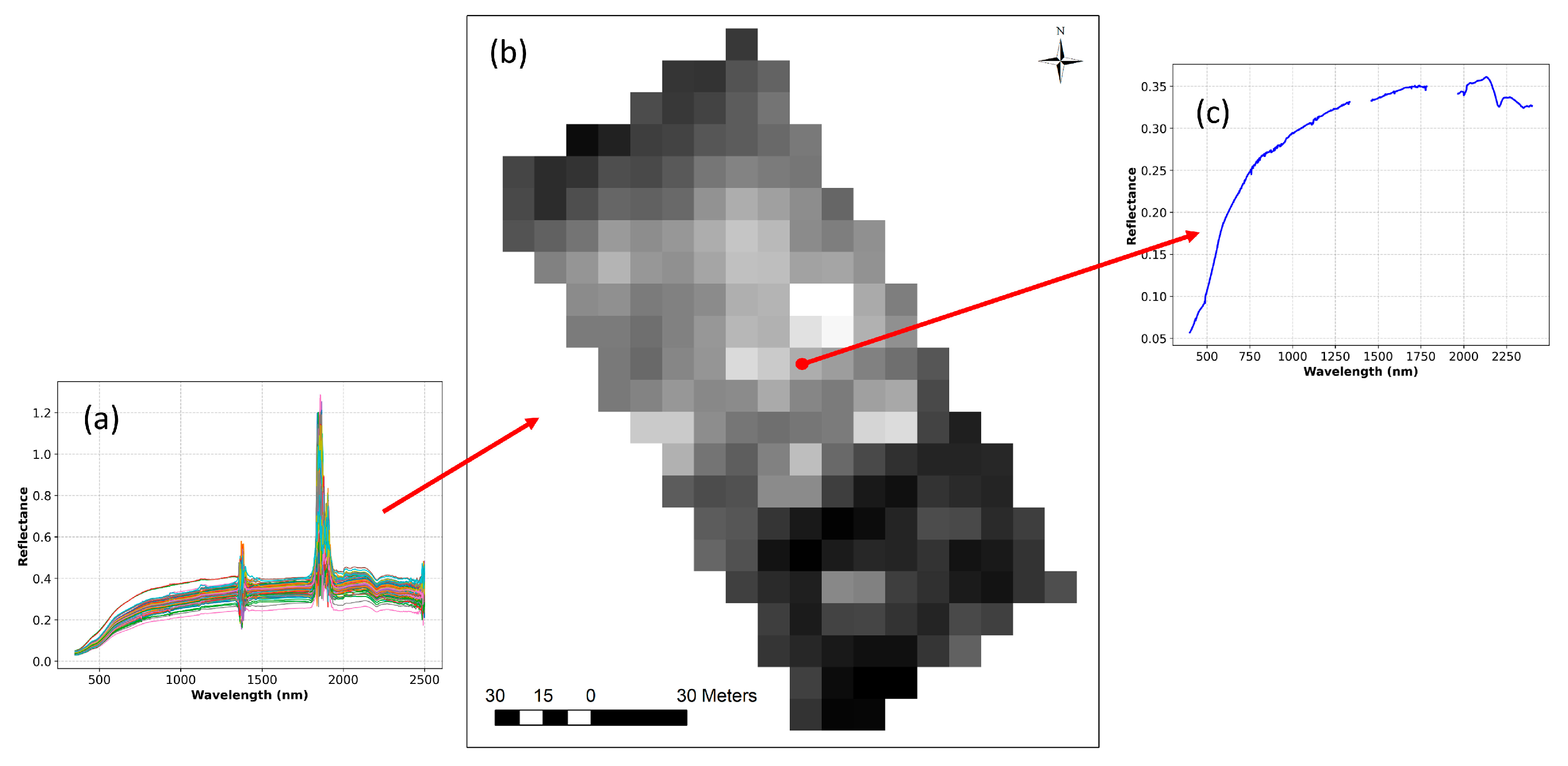
2.2. Generation of Synthetic Hyperspectral Image
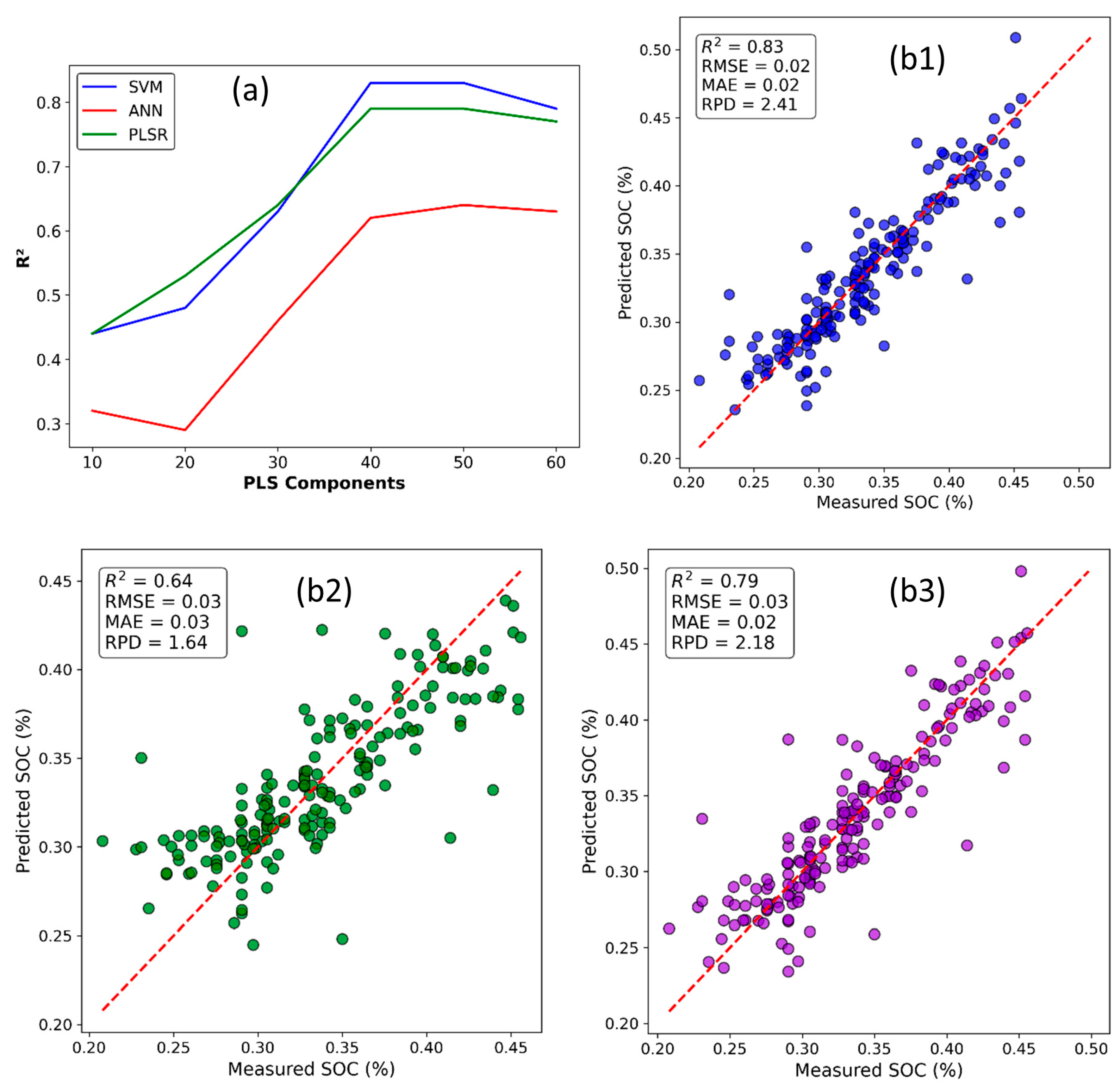
2.3. Optimizing PLS Scores and Machine Learning Regression
3. Results
3.1. Descriptive Statistics of Measured SOC
3.2. Synthetic Hyperspectral Imagery
3.3. Model Evaluation and SOC Mapping
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.; Huang, T.; Liu, J.; Lin, Z.; Li, S.; Wang, R.; Ge, Y. Soil PH Value, Organic Matter and Macronutrients Contents Prediction Using Optical Diffuse Reflectance Spectroscopy. Comput. Electron. Agric. 2015, 111, 69–77. [Google Scholar] [CrossRef]
- Jiang, R.; Sui, Y.; Zhang, X.; Lin, N.; Zheng, X.; Li, B.; Zhang, L.; Li, X.; Yu, H. Estimation of Soil Organic Carbon by Combining Hyperspectral and Radar Remote Sensing to Reduce Coupling Effects of Soil Surface Moisture and Roughness. Geoderma 2024, 444, 116874. [Google Scholar] [CrossRef]
- Padarian, J.; Minasny, B.; McBratney, A.B. Using Deep Learning to Predict Soil Properties from Regional Spectral Data. Geoderma Reg. 2019, 16, e00198. [Google Scholar] [CrossRef]
- Das, B.; Chakraborty, D.; Singh, V.K.; Das, D.; Sahoo, R.N.; Aggarwal, P.; Murgaokar, D.; Mondal, B.P. Partial Least Square Regression Based Machine Learning Models for Soil Organic Carbon Prediction Using Visible–near Infrared Spectroscopy. Geoderma Reg. 2023, 33, e00628. [Google Scholar] [CrossRef]
- Walkley, A.; Black, I.A. An Examination of the Degtjareff Method for Determining Soil Organic Matter, and a Proposed Modification of the Chromic Acid Titration Method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Pham, T.G.; Kappas, M.; Van Huynh, C.; Nguyen, L.H.K. Application of Ordinary Kriging and Regression Kriging Method for Soil Properties Mapping in Hilly Region of Central Vietnam. ISPRS Int. J. Geo-Inf. 2019, 8, 147. [Google Scholar] [CrossRef]
- Lu, P.; Niu, Z.; Li, L. Prediction of Soil Organic Carbon by Hyperspectral Remote Sensing Imagery. In Proceedings of the 2012 Third Global Congress on Intelligent Systems, Wuhan, China, 6–8 November 2012; IEEE: Orlando, FL, USA; pp. 291–293. [Google Scholar]
- Sahoo, R.N.; Rejith, R.G.; Gakhar, S.; Verrelst, J.; Ranjan, R.; Kondraju, T.; Meena, M.C.; Mukherjee, J.; Dass, A.; Kumar, S.; et al. Estimation of Wheat Biophysical Variables through UAV Hyperspectral Remote Sensing Using Machine Learning and Radiative Transfer Models. Comput. Electron. Agric. 2024, 221, 108942. [Google Scholar] [CrossRef]
- Sahoo, R.N.; Gakhar, S.; Rejith, R.G.; Ranjan, R.; Meena, M.C.; Dey, A.; Mukherjee, J.; Dhakar, R.; Arya, S.; Daas, A.; et al. Unmanned Aerial Vehicle (UAV)–Based Imaging Spectroscopy for Predicting Wheat Leaf Nitrogen. Photogramm. Eng. Remote Sens. 2023, 89, 107–116. [Google Scholar] [CrossRef]
- Rejith, R.G.; Sundararajan, M.; Gnanappazham, L.; Seenipandi, K.; Ramaswamy, S. GIS-Based Machine Learning Algorithms for Mapping Beach Placer Deposits in the Southwest Coast of India Using Landsat-8 OLI Images. J. Appl. Remote Sens. 2021, 16, 012011. [Google Scholar] [CrossRef]
- Das, B.; Manohara, K.K.; Mahajan, G.R.; Sahoo, R.N. Spectroscopy Based Novel Spectral Indices, PCA- and PLSR-Coupled Machine Learning Models for Salinity Stress Phenotyping of Rice. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 229, 117983. [Google Scholar] [CrossRef]
- Viscarra Rossel, R.A.; Walvoort, D.J.J.; McBratney, A.B.; Janik, L.J.; Skjemstad, J.O. Visible, near Infrared, Mid Infrared or Combined Diffuse Reflectance Spectroscopy for Simultaneous Assessment of Various Soil Properties. Geoderma 2006, 131, 59–75. [Google Scholar] [CrossRef]
- Mondal, B.P.; Sahoo, R.N.; Das, B.; Ahmed, N.; Bandyopadhyay, K.K.; Mukherjee, J.; Arora, A.; Moursy, A.R.A. Comparison of Multivariate Machine Learning Models for Major Soil Nutrients Prediction Using Laboratory-Based and Airborne (AVIRIS-NG) Visible near-Infrared Spectroscopy. Eur. J. Agron. 2025, 170, 127726. [Google Scholar] [CrossRef]
- Xu, S.; Wang, M.; Shi, X. Hyperspectral Imaging for High-Resolution Mapping of Soil Carbon Fractions in Intact Paddy Soil Profiles with Multivariate Techniques and Variable Selection. Geoderma 2020, 370, 114358. [Google Scholar] [CrossRef]
- Yang, J.; Dong, H.; Zhang, H.; Wang, J.; Dai, H.; Zhou, L.; Liang, T.; Zhang, Y. Enhancing the Estimation of Low-Spectral-Sensitivity Soil Properties: A Case Study of Active Organic Carbon Using Vis-NIR Hyperspectral Data. Int. J. Remote Sens. 2025, 46, 4941–4958. [Google Scholar] [CrossRef]
- Casa, R.; Pignatti, S.; Pascucci, S.; Castaldi, F.; Marrone, L. Estimation of Agronomic Soil Properties from Multitemporal PRISMA Satellite Imaging Spectroscopy. In Precision Agriculture ’23; Brill|Wageningen Academic: Leiden, The Netherlands, 2023; pp. 839–845. [Google Scholar]
- Gao, Y.; Cui, L.; Lei, B.; Zhai, Y.; Shi, T.; Wang, J.; Chen, Y.; He, H.; Wu, G. Estimating Soil Organic Carbon Content with Visible–Near-Infrared (Vis-NIR) Spectroscopy. Appl. Spectrosc. 2014, 68, 712–722. [Google Scholar] [CrossRef]
- Stenberg, B.; Viscarra Rossel, R.A.; Mouazen, A.M.; Wetterlind, J. Visible and Near Infrared Spectroscopy in Soil Science. Adv. Agron. 2010, 107, 163–215. [Google Scholar]
- Wenjun, J.; Zhou, S.; Jingyi, H.; Shuo, L. In Situ Measurement of Some Soil Properties in Paddy Soil Using Visible and Near-Infrared Spectroscopy. PLoS ONE 2014, 9, e105708. [Google Scholar] [CrossRef] [PubMed]
- Vasques, G.M.; Grunwald, S.; Sickman, J.O. Comparison of Multivariate Methods for Inferential Modeling of Soil Carbon Using Visible/near-Infrared Spectra. Geoderma 2008, 146, 14–25. [Google Scholar] [CrossRef]
- Xu, S.; Zhao, Y.; Wang, M.; Shi, X. Comparison of Multivariate Methods for Estimating Selected Soil Properties from Intact Soil Cores of Paddy Fields by Vis–NIR Spectroscopy. Geoderma 2018, 310, 29–43. [Google Scholar] [CrossRef]
- Odebiri, O.; Odindi, J.; Mutanga, O. Basic and Deep Learning Models in Remote Sensing of Soil Organic Carbon Estimation: A Brief Review. Int. J. Appl. Earth Obs. Geoinf. 2021, 102, 102389. [Google Scholar] [CrossRef]
- Engelmann, L.; Bierl, R.; Kirchhoff, M.; Ries, J.B. Application of Mid-Infrared Spectroscopy for Soil Analysis in Calcareous Argania Spinosa Forests in Morocco. Geoderma Reg. 2025, 42, e00964. [Google Scholar] [CrossRef]
- Mondal, B.P.; Sahoo, R.N.; Das, B.; Ahmed, N.; Bandyopadhyay, K.K.; Mukherjee, J.; Arora, A.; Ali Moursy, A.R. Digital Mapping of DTPA Extractable Micronutrients Using Combined AVIRIS-NG Hyperspectral and Multispectral Data of Sentinel-2 and SRTM-DEM. Comput. Electron. Agric. 2025, 239, 110905. [Google Scholar] [CrossRef]
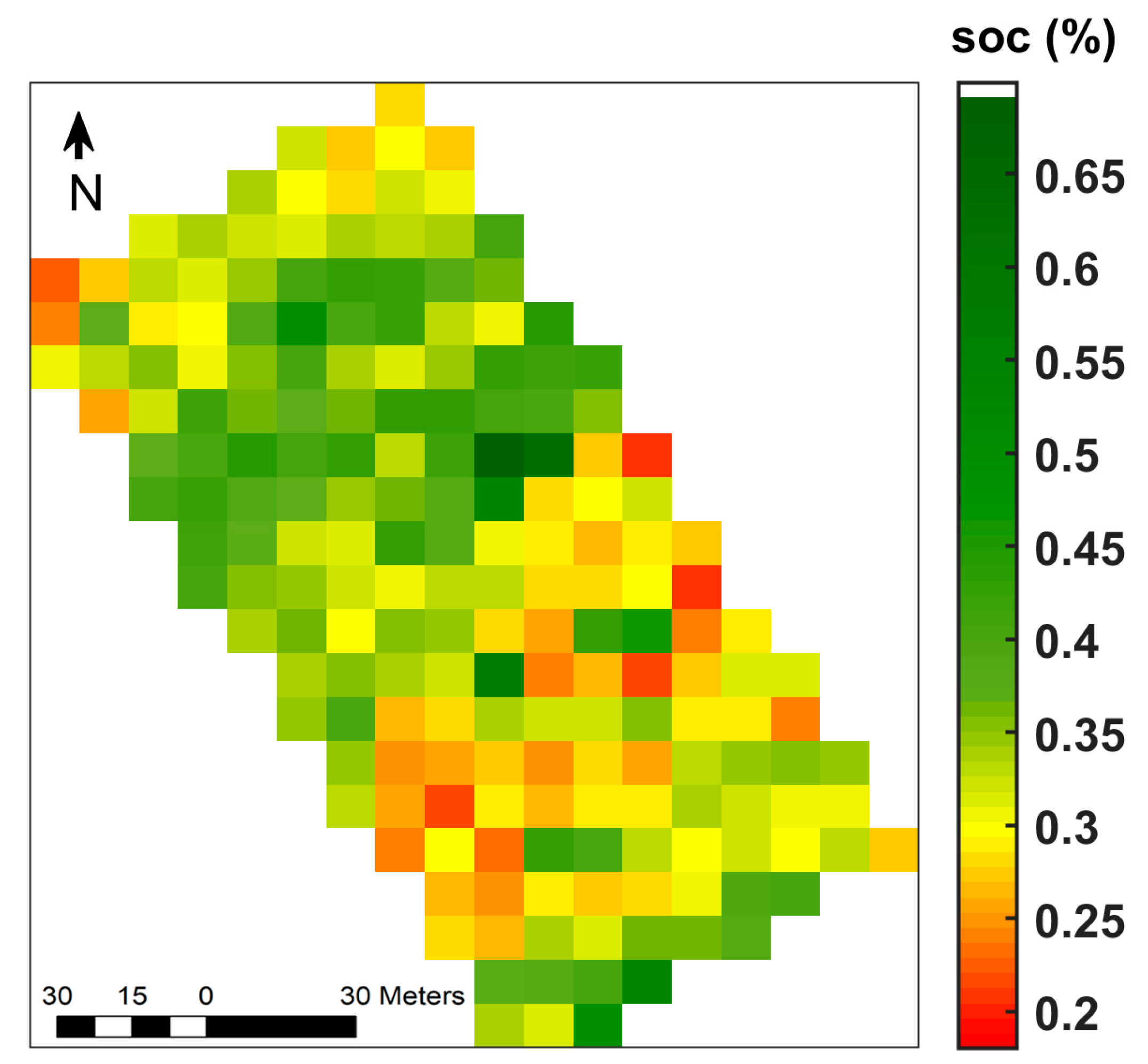
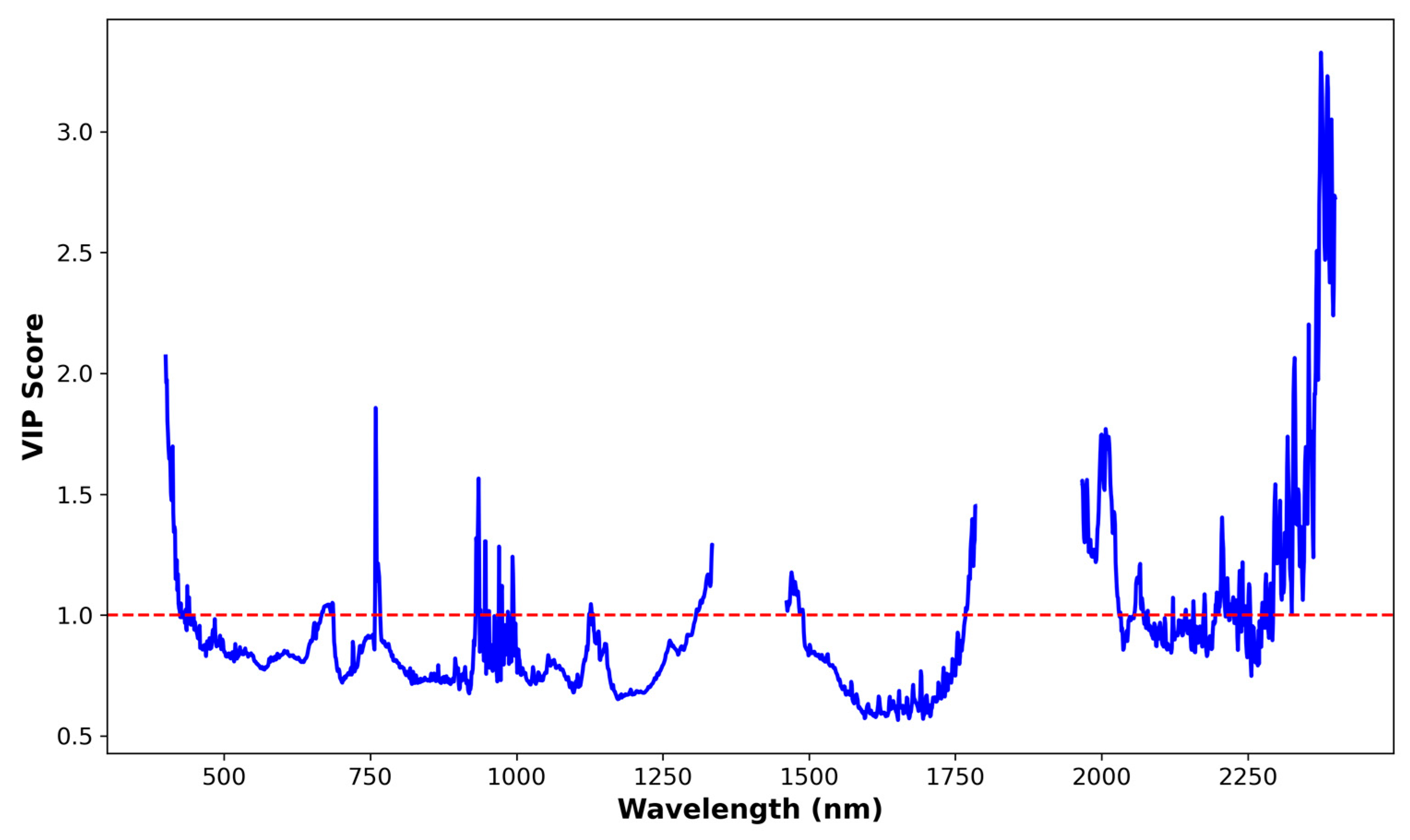
| Minimum | Maximum | Mean | Median | SD 1 | %CV 2 | Skewness | Kurtosis |
|---|---|---|---|---|---|---|---|
| 0.18 | 0.52 | 0.33 | 0.33 | 0.06 | 18.05 | 0.16 | −0.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rejith, R.G.; Sahoo, R.N.; Kondraju, T.; Bhandari, A.; Ranjan, R.; Moursy, A. Generation of Synthetic Hyperspectral Image Cube for Mapping Soil Organic Carbon Using Proximal Remote Sensing. Environ. Earth Sci. Proc. 2025, 36, 3. https://doi.org/10.3390/eesp2025036003
Rejith RG, Sahoo RN, Kondraju T, Bhandari A, Ranjan R, Moursy A. Generation of Synthetic Hyperspectral Image Cube for Mapping Soil Organic Carbon Using Proximal Remote Sensing. Environmental and Earth Sciences Proceedings. 2025; 36(1):3. https://doi.org/10.3390/eesp2025036003
Chicago/Turabian StyleRejith, Rajan G., Rabi N. Sahoo, Tarun Kondraju, Amrita Bhandari, Rajeev Ranjan, and Ali Moursy. 2025. "Generation of Synthetic Hyperspectral Image Cube for Mapping Soil Organic Carbon Using Proximal Remote Sensing" Environmental and Earth Sciences Proceedings 36, no. 1: 3. https://doi.org/10.3390/eesp2025036003
APA StyleRejith, R. G., Sahoo, R. N., Kondraju, T., Bhandari, A., Ranjan, R., & Moursy, A. (2025). Generation of Synthetic Hyperspectral Image Cube for Mapping Soil Organic Carbon Using Proximal Remote Sensing. Environmental and Earth Sciences Proceedings, 36(1), 3. https://doi.org/10.3390/eesp2025036003







