1. Good and Bad Practices in Data Management Projects
This paper introduces a flexible archival and data management model that integrates recent developments across data-grid, cloud, edge, and fog computing technologies. Designed to meet the requirements of large-scale astronomical projects, the model emphasizes resilience, performance, and sustainability while avoiding typical single points of failure (SPOFs), which often arise from short-sighted political management decisions and suboptimal In-Kind Contribution (IKC) allocations.
A recurring issue in large scientific collaborations is the allocation of leadership roles based not on technical expertise, but on political convenience or financial leverage. This leads to fragmented and inefficient work organization, particularly in core areas such as data handling and archiving. It is common to see simple tasks unnecessarily divided among multiple groups, each with distinct visions and leadership, making coordination and integration difficult. In response, project leaders often “descope” activities, reducing group autonomy in favor of hierarchical control. While this may streamline decision-making, it suppresses innovation and undermines project agility.
A particularly harmful trend is the political fragmentation of archive design, where medium- to long-term data management is split across loosely defined entities without real architectural boundaries. Such divisions introduce complexity and delay, especially when multiple groups interact with a shared infrastructure. Leadership may be assigned to individuals with little or no technical background, and the final decision-making authority may reside with administrative bodies rather than developers. This practice results in systems driven by political compromise rather than technological soundness.
Effective archive development must begin with robust planning. As outlined in
Figure 1, project management strategies should reflect the project’s timeline and goals. For short-term implementations, use-case generalization and rapid prototyping are essential to test technological feasibility. For long-term projects, more detailed planning, including thorough documentation of use cases, requirements, and interfaces, should be established early on. However, premature commitment to specific technologies should be avoided, as rapid technological evolution can render early choices obsolete.
The system design phase consolidates all use cases and validated requirements into an integrated solution based on proven technologies. This is followed by code development, pre-production testing, and final deployment. A major constraint, particularly in scientific archiving, is budgetary: long-term maintenance costs are often underestimated or ignored. As a result, hardware acquisition frequently follows funding availability rather than design logic. To overcome this, a virtualized, service-based model is adopted, allowing for the decoupling of hardware from software layers.
This approach enables the implementation of Archive as a Service (AaaS), which builds upon the established paradigms of Infrastructure as a Service (IaaS), Platform as a Service (PaaS), and Software as a Service (SaaS), as illustrated in
Figure 2.
The archive model distinguishes two main user roles:
Data Producers, who supply content at various levels, and
Data Consumers, who access and possibly process the data (
Figure 3). While users may act in both roles, each must interact with the system through standardized, role-specific interfaces.
A good approach in data archival and handling systems is to adopt the Open Archival Information System (OAIS) reference model [
1], which logically separates user interaction from core system operations. The core functions of the archive—Ingest, Search/Browse, and Retrieve/Distribute—are built upon two foundational components: a repository and a database. The choice of technologies in these areas is dictated by the archive’s system topology and performance goals.
The key challenge in developing an effective data handling and archival system is to prioritize the intended use cases over political and personal considerations before implementing any technological solutions. Specifically, once the most common challenges are abstracted, as outlined in the OAIS model, it becomes possible to identify the most widely adopted technologies and approaches. This helps in evaluating which are suitable for our needs and which are not. By doing so, the resulting system will become an intelligent and adaptive framework that facilitates both efficient data access and its medium- to long-term preservation.
In the following sections, we will explore archive topologies, database selection, and data access interfaces as critical factors in designing scalable, user-centered archive and data handling systems. Finally, the versatile and efficient architecture of CTAARCHS will be introduced.
2. Storage Topology: Centralized vs. Distributed Approaches
One of the key design challenges in developing an astronomical archive and data handling system is to understand the data topology, so the first thing to understand is the storage architecture and the related storage service deployments. The decision between a centralized or distributed model depends on the archive’s use cases, particularly when access is required across geographically distributed locations. There is no universal solution—each approach has strengths and limitations based on scalability, resilience, access latency, cost, and administrative complexity [
2].
As summarized in
Table 1, centralized architectures offer simplicity, streamlined security, and ease of management, making them suitable for small-scale or local deployments. However, they pose greater risks of failure and limited scalability. In contrast, distributed architectures support high availability (HA), redundancy, and better performance across dispersed users, though they require more sophisticated orchestration and monitoring.
The choice ultimately depends on system scale, geographic distribution, and acceptable complexity. The model presented here allows flexible configuration—from a single-node centralized instance to a distributed system with multiple nodes in a “leader + followers” configuration (i.e., primary + secondaries) or fully redundant HA configurations, ensuring no single point of failure (no SPOF) [
3].
Before discussing the details of our work, it is appropriate to make a general narrative highlighting specific aspects addressed in different approaches and computing paradigms; this basic overview helps us to design an end-to-end solution that could be adaptable to various projects, each using different methodologies and paradigms, ensuring compatibility within tools and parts required by currently in-production experiments [
4].
Historically,
data-grid computing, see references, was the dominant model in research environments, where computing and storage were distributed across tiered datacenters connected by middleware for data orchestration. While effective in some contexts, its hierarchical structure limited scalability and flexibility. Over past decades, the data-grid model has largely been complemented and/or sometimes replaced by
cloud computing, which enables horizontal scaling, service-based architecture, and global accessibility. Cloud systems offer improved resource outsourcing, built-in redundancy, and disaster recovery, making them better suited to handle complex, large-scale datasets with minimal management overhead [
5].
However, widespread data sharing via cloud platforms raises serious security and privacy concerns [
6], making robust access control and encryption critical challenges [
7].
More recently, computing paradigms have been supplemented by
edge computing, where data processing occurs closer to data sources—often at the sensor or device level. This reduces network congestion, minimizes latency, and enables real-time applications [
8]. Edge computing is particularly valuable in time-sensitive use cases, where immediate processing and decision-making are required. Enhancing this model with
edge intelligence—that is, applying AI and machine learning algorithms locally—enables automated decisions based on complex, use-case-specific criteria. This adds significant value where human intervention must be minimized [
9].
At a broader level, this leads to fog computing, a form of fine-grained distributed processing that extends computing and storage further toward the network edge. By integrating IoT devices and localized data sources, fog architectures process large volumes of unstructured data near their origin, which is essential for real-time analytics [
10].
Even if a lot of scientific computing is historically and technically incapable of embracing the edge (or fog) paradigm since data can not be analyzed at the edge, there is no doubt that these solutions are going to represent the near future. Given the limited computational capacity typical at the edge, adaptive AI algorithms play a critical role in optimizing performance. These systems can identify semantic patterns, adapt compression techniques, and reduce computational loads, enabling efficient data analysis and visualization. The use of optimized low-latency databases becomes essential in transforming raw data into science-ready outputs quickly and interactively.
Note: Although these models raise legitimate concerns about environmental impact—particularly related to the power demands of AI training and edge infrastructure—this paper does not address sustainability [11]. It is misleading to discuss energy use without a comprehensive life-cycle analysis of the hardware and algorithms involved. The sustainability of AI and edge computing should not be reduced to superficial claims but rather evaluated within a systemic framework, which is beyond the scope of this discussion [12].
3. Selecting the Appropriate Database Architecture for Archival Systems
The database lies at the heart of any archive and data management system, making its selection a critical component of the overall design. However, there is no universally optimal solution—the appropriate database choice depends on multiple factors, including the storage use case, system topology, data access patterns, geographical distribution of users, and desired services deployments.
In distributed storage environments, relying on a centralized database for file cataloging introduces significant risks. It creates a single point of failure (SPOF) and becomes a performance bottleneck under concurrent, geographically dispersed queries. This undermines the redundancy and resilience typically sought in distributed systems.
Conversely, centralized database architectures are well-suited for smaller or geographically constrained archives, where high availability can be ensured through network and service redundancy. These systems benefit from ACID-compliant transactions—atomicity, consistency, isolation, and durability—which are essential in contexts requiring strong data integrity, such as financial systems.
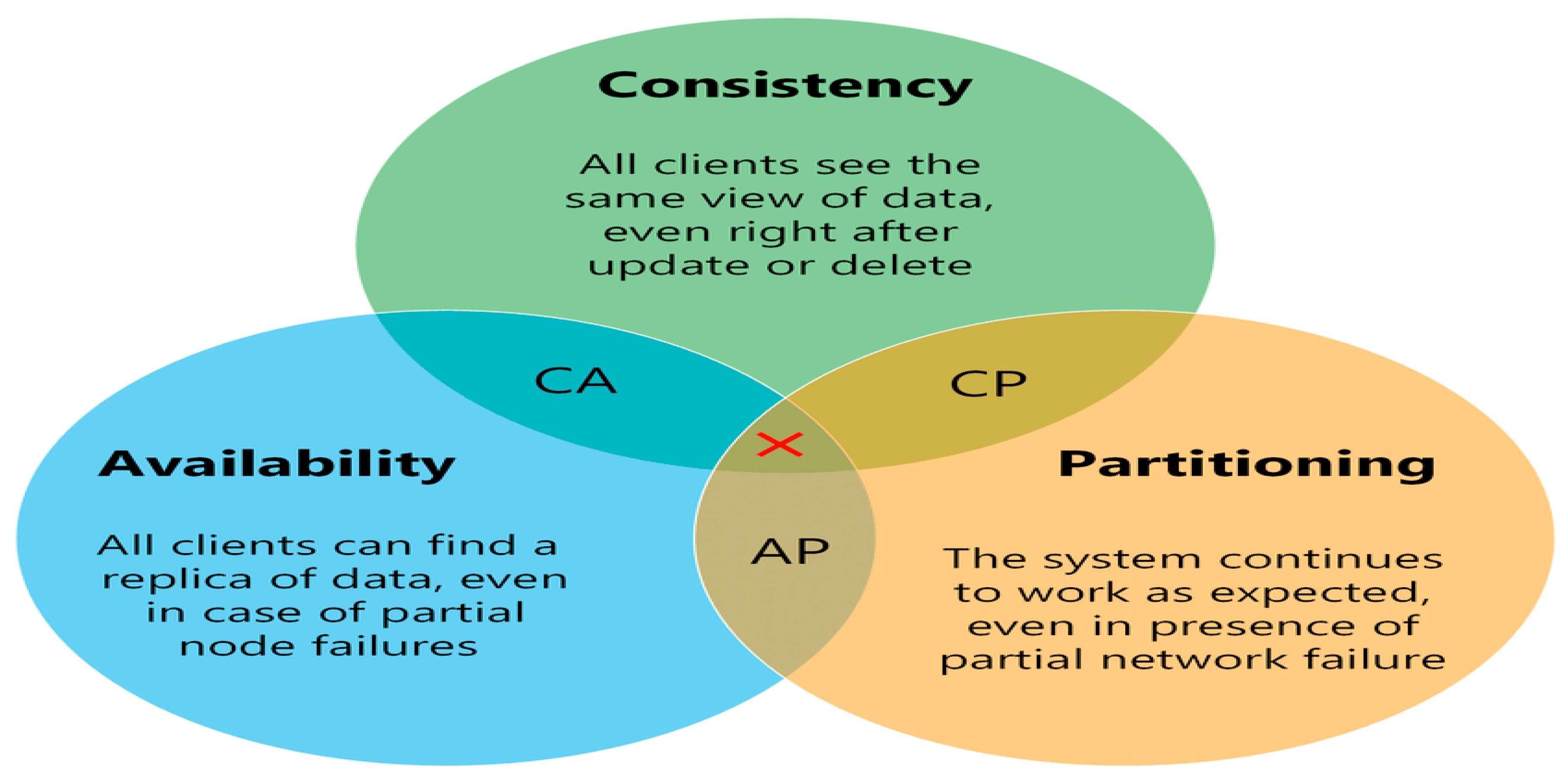
However, distributed databases cannot fully guarantee ACID properties and instead operate under the
CAP Theorem (Brewer’s Theorem), which states that a distributed system can only simultaneously satisfy two of the following: consistency, availability, and partition tolerance. Trade-offs among these properties must be carefully evaluated depending on the archive’s performance and reliability needs (see
Table 2 and
Figure 4).
In summary, the choice between centralized and distributed database architectures must align with the system’s scale, access requirements, and fault-tolerance goals. The database model must not only support efficient data access but also integrate seamlessly into the broader storage and computing infrastructure.
In distributed databases, data are replicated across multiple nodes. When network partitions occur—isolating one or more nodes—the system must prioritize among consistency, availability, and partition tolerance (the CAP Theorem).
Prioritizing consistency may halt reads or writes to prevent divergence, sacrificing availability.
Prioritizing availability ensures responsiveness but may serve outdated or inconsistent data.
Prioritizing partition tolerance allows continued operation despite communication failures, though it may compromise either consistency or availability.
Many systems dynamically balance these trade-offs based on application needs. For archival systems, using pre-assigned physical file names and a Write Once, Read Many (WORM) model minimizes consistency concerns. Once written, immutable data simplifies coherence across nodes. This permits a focus on availability and partition tolerance (AP), ensuring the system remains operational and responsive—even if some nodes are unreachable.
Partition tolerance is often the most critical factor in large-scale or globally distributed environments, as network disruptions are inevitable. Ensuring only a single version of any file exists and is replicated guarantees that if a file is accessible, it is valid and consistent system-wide.
Another key factor in choosing a database system is balancing data scalability with the complexity of the data model and queries. As illustrated in
Figure 5, certain database families are inherently unsuited to large-scale data. For instance, relational databases (SQL), while efficient for smaller datasets and simpler queries, struggle when dealing with high-complexity joins or terabyte-scale tables. At this point, only three options remain:
Simplify the data model or queries.
Scale up the hardware infrastructure.
Migrate to a different database family—such as a document-oriented (NoSQL) system.
In practice, restructuring or hardware upgrades often cause service interruptions, particularly when the database was not properly designed from the outset. This underscores the importance of selecting the appropriate architecture early in the project lifecycle.
Databases can broadly be categorized into two groups: Relational DBMS (RDBMS) and Not Only SQL (NoSQL) systems [
13]. A comparative summary is provided in
Table 3.
4. Polyglot Persistence in Modern Archive Systems
For this archival model, we focus on the versatility, schema-less nature, and aggregation capabilities of document-oriented databases. Their architecture supports scalability through replication, sharding, and clustering, depending on performance demands and availability requirements. Strategies for scaling read/write capacity and ensuring high availability are summarized in
Table 4.
If data size exceeds single-server capacity, two strategies are available: scaling up infrastructure or scaling out via clustering. Similarly, read performance can be improved through replication and caching, while write scalability benefits from partitioning and sharding. To mitigate SPOFs and ensure service resilience, especially in geographically distributed collaborations, combining clustering with cross-site replication is essential. Inter-datacenter distances of several hundred kilometers are generally sufficient to safeguard against regional failures and enable disaster recovery.
A key principle here is polyglot persistence, which leverages multiple database types, each tailored to a specific data class, written as follows:
Relational databases (e.g., PostgreSQL, MariaDB) for structured data like observation proposals.
Document-oriented databases (e.g., MongoDB) for semi-structured metadata.
Column stores (e.g., Cassandra) for streaming telemetry.
Key–value stores (e.g., Voldemort) for fast-access logs.
Graph databases (e.g., Neo4j, Cosmos DB) for user interaction mapping.
Array or functional query languages for analytical pipelines.
This modular approach allows independent scaling of archive components and optimization of performance and cost. The main drawback lies in the complexity of managing diverse technologies and the associated manpower and training costs.
5. Polyglot Persistence in a Data Lake Scenario
In modern observatories, archives manage more than just raw scientific data. A data lake approach is adopted to incorporate a wide range of heterogeneous data products—proposals, schedules, weather station outputs, logs, alarms, analytics, and system monitoring.
Different database systems are better suited for handling different types:
Relational databases for structured data.
Object storage for unstructured or large datasets (e.g., images, videos, documents).
NOSQL databases for semi-structured data that does not fit into a rigid schema.
Graph databases for analyzing complex relationships and social semantic analytics.
Polyglot persistence ensures that each data type is managed by the most appropriate database and storage technology, enabling long-term flexibility and integration across services.
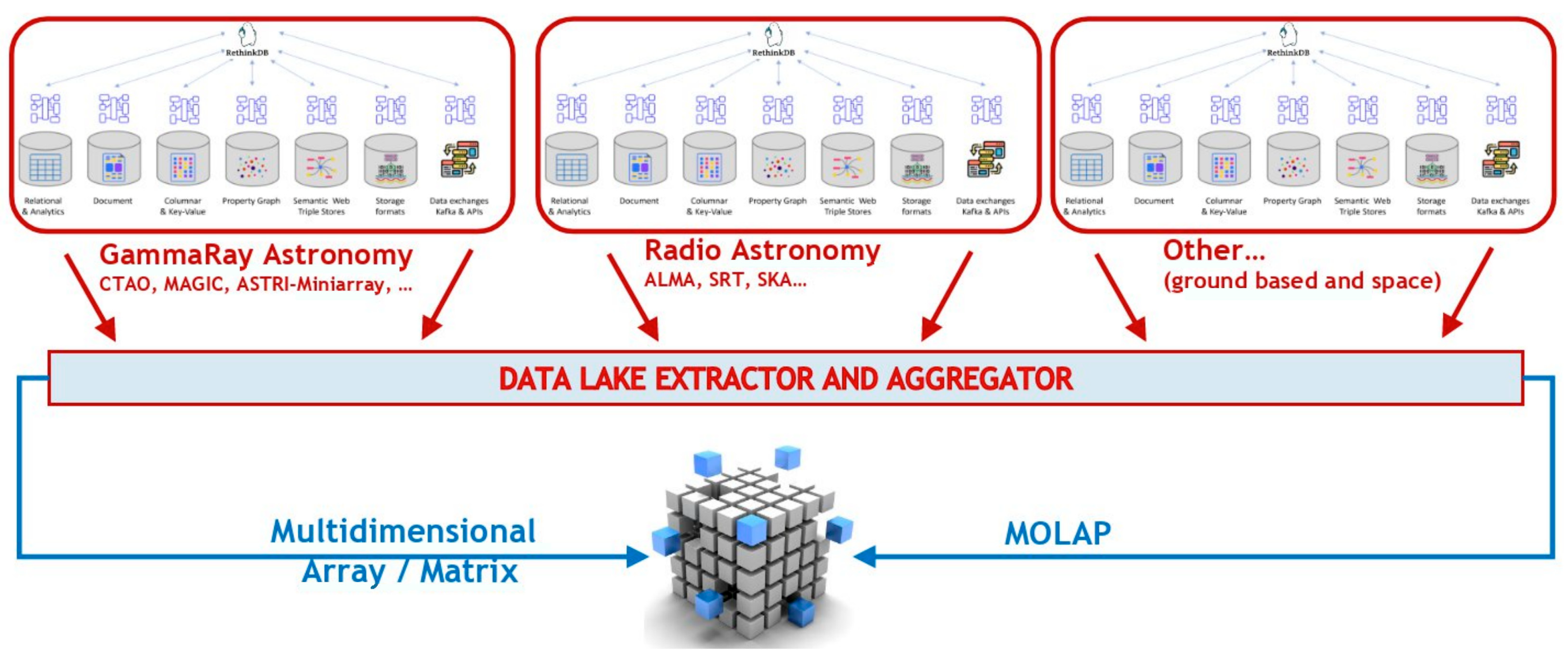
Figure 6 shows a generic case study of the archives commonly managed within an astronomical observatory facility.
Polyglot persistence relies on a unified access layer—a middleware abstraction that enables seamless querying, handling, and processing of heterogeneous datasets across diverse storage backends. This layer simplifies interaction with various database systems and protocols within a distributed archive.
Different data types are best served by specialized database technologies, such as the following:
Structured proposal data can be easily managed by a relational DBMS (e.g., MariaDB, PostgreSQL).
Logs and alarms require high-throughput so a key–value stores (e.g., Voldemort) can well fit.
JSON-based scientific metadata can rely on a document-oriented DBs (e.g., MongoDB).
Streaming telemetry and event data may need column–family databases (e.g., Cassandra) approach
Tracking accesses and users interactions could be managed by graph databases (e.g., Neo4j, Azure Cosmos DB).
Data analytics/pipelines can be easily stored by an array or functional query systems approach.
By matching each data type to the most suitable database family, this model enables
independent scaling of archive components and
optimized performance. Object storage handles large unstructured datasets efficiently, while NoSQL systems provide high responsiveness for semi-structured content. However, this flexibility comes at the cost of increased operational complexity and a steep learning curve for different technologies. In
Figure 7 it is possible to see a high-level description of how different database technologies, used for various types of data within a common observatory infrastructure, could be linked and aggregated into a unified service. The core aggregator is a document oriented distributed dB (in the image is indicated ReThinkDB). A common approach to a data lake scenario can be easily implemented once all these types of data are aggregated into a single cluster: the final work is a matter of implementing a suitable “extractor service” for what it is needed.
Extending this model, a
multi-observatory abstraction layer can integrate science-ready data products from multiple facilities into a unified archive, enabling
MOLAP-based multiwavelength research with consistent access to distributed, heterogeneous datasets, optimized and standardized by Virtual Observatory standards (see
Figure 7).
6. Distributed Strategy for a Petascale Astronomical Observatory
Consider a distributed observatory composed of mountaintop telescope arrays, multiple observing sites, and geographically dispersed data centers. Managing tens of petabytes of data annually and enabling broad scientific access—potentially to proprietary datasets—requires an archive system that is scalable, efficient, and responsive (see
Figure 8).
A data cloud paradigm must be adapted to a research context rather than a commercial one. In an astronomical observatory, the only form of “payment” is through the submission of an observing proposal. If the proposal is highly rated by the Time Allocation Committee, it is prioritized in the observing schedule. This means that the resulting data from the observatory is directly linked to the specific proposal, and the associated scientific data (i.e., high-level data produced after standard reduction) must be accessible only to the principal investigator (PI) and the co-investigators (co-PIs) of that proposal.
This PIship-data archival can be managed quite simply if the database that provides access to the data also stores the proposal metadata at the time of acquisition and carries it through the entire data reduction pipeline. The size of the files, whether large or small, is not considered to have commercial value—only the observational data collected during the allocated time is relevant and must be reduced at different levels to reach a scientific readiness when they are provided to the PI according to the observatory policies.
As widely described for this task database technology is central in any archive solution so taking into account such a distributed scenario, where data are generated on-site and transferred to off-site facilities for long-term storage and processing, the database architecture must mirror the data’s geographical organization. A well-designed geographical topology reduces latency between clients and databases. Geographical proximity helps ensure fast response times, which is particularly important for real-time applications or those with large traffic volumes (see references). To reach such versatile potential geographical distribution, with its flexibility and scalability characteristics, a document-oriented, schema-less database is optimal.
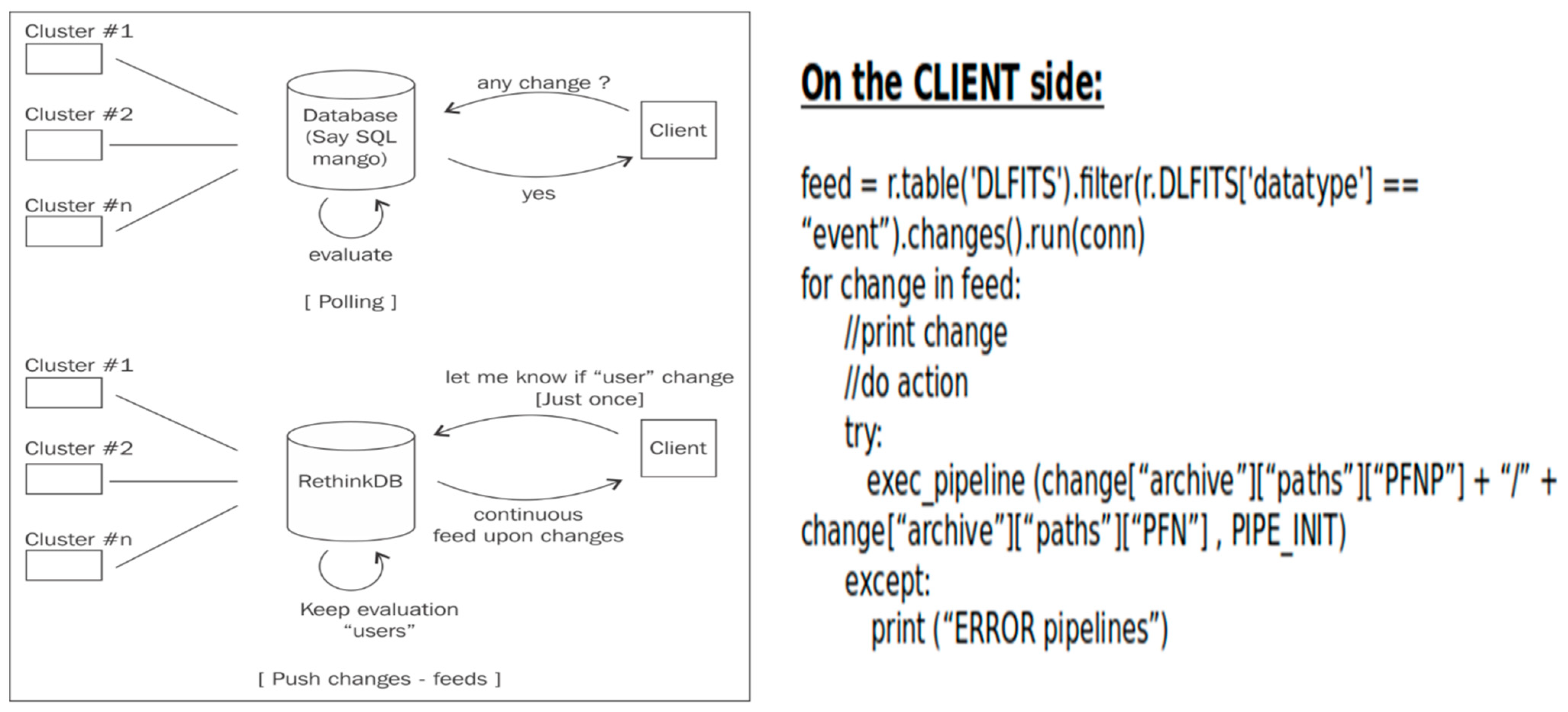
The key point in the database selection is the need to represent the intelligent part of the data handling system, at least ensuring the automation of standard actions to be triggered on the client side. To perform such an automation, it is possible to implement a standard “polling” mechanism or a “change-feed” mechanism.
Several open-source document-oriented databases were evaluated for this. While MongoDB and Couchbase were considered, RethinkDB was selected due to its native “change-feed” mechanism, which enables real-time triggers for any database event. This functionality supports near-automated archival operations, reducing human intervention and eliminating the need for resource-heavy polling systems (
Figure 9). Only Azure Cosmos DB offers similar changefeed support, but RethinkDB provided a more lightweight, open-source alternative with low complexity and ease of deployment.
In particular, the change-feed mechanism allows any client to perform an operation when it is triggered by the database: an action can be executed each time there is a change in the result of predefined query (i.e., execute on the client node a standard reduction when new data are present in the DB collection and related calibrations are available). The alternative triggering mechanism is the standard “polling strategy” that executes the same query several times; it compares with the result of a previous query, and if there are differences then it triggers an action (+ sleep + redo!).
Other available database solutions, including the proprietary relational ones, do not have the change-feed functionality integrated, and to develop similar functions, it is necessary to put in place a standard “polling” mechanism (see
Figure 9) that consumes a lot of resources and performs several “not-needed” queries and consequent I/O traffic. The change-feed has to be preferred over the polling mechanism for real-time applications.
In principle, when an HA database is distributed and follows the storage distribution, the data management and archival system can be easily customized and adapted to every use case. For this reason a recommended configuration for DB distribution involves deploying at least two RethinkDB instances per data center, ensuring local availability, distributed processing, and high resilience (
Figure 10). This configuration is what is explained as edge computing applied to database operations; here, database nodes follow the physical data geographical distribution in order to allow change-feed executions on the single node/datacenter even in the case of network failure, producing output results to be synchronized within the cluster as soon as the network connection is again available.
7. FAIR Principles and VO Integration in Polyglot Persistence
In modern polyglot persistence/data lake environments handling heterogeneous data types, the
FAIR principles—Findable, Accessible, Interoperable, and Reusable—serve as foundational guidelines for enabling data discoverability and reuse. These principles, combined with the
Open Archives Initiative (OAI), support metadata standardization and cross-repository interoperability [
14].
To ensure scientific data are interoperable and accessible at the final stage, adherence to
Virtual Observatory (VO) standards is essential. These standards, defined by the International
Virtual Observatory Alliance (IVOA), require metadata to be exposed via TAP services and formatted as VO-Tables. This enables seamless integration with VO tools for accessing and analyzing high-level science products such as multi-wavelength catalogs, spectra, and images. Execution workflows are brokered via standardized APIs (e.g., OpenAPI, REST) and submitted to local resource managers such as Slurm, as shown in
Figure 11.
Note: In this paper, depending on the context, we use the VO notation both for the Virtual Observatory (for public data access) and the Virtual Organization (for managing access rights and group policies).
8. CTAARCHS Implementation
8.1. Modular Design and Data Transfer Workflow
CTAARCHS provides flexible access to its archive functionalities through the following modular access interfaces:
Command-Line Interface (CLI): executable Python (up to v.3.10) scripts with standardized input/output.
Python Library: core actions encapsulated in run_action () functions, enabling seamless integration into external applications.
REST API: web-based access via HTTP methods (POST, GET, PUT/PATCH, DELETE), allowing CRUD operations through scripts or clients (e.g., CURL, Requests).
Containerized Deployment: distributed as a software container (AMASLIB_IO) to ensure platform compatibility and ease of deployment in Kubernetes (K8s) environments.
8.2. On-Site–Off-Site Data Transfer System
In typical observatory setups, raw data are generated on-site and archived off-site. To facilitate this, CTAARCHS implements a dedicated Data Transfer System (DTS) with optimized bandwidth, error handling, and transfer resumption via client-server architecture and different communication protocols (RPC, gRPC, HTTP/REST, message–queue, etc.).
The on-site storage is treated as a passive element, exposed only to authorized services via secure authentication protocols. This avoids performance bottlenecks and long-term maintenance overhead. Data management and archiving responsibilities reside with off-site data centers, integrated into a broader grid/cloud/edge/fog infrastructure, each with its own virtual organization (VO) (see
Figure 12 for architecture).
8.2.1. Prerequisites
To enable automated data transfer from observatory sites to archival facilities, the following prerequisites must be met:
- A.
Remote Access to On-Site Storage: On-site storage must be remotely accessible via secure, standardized protocols (e.g., HTTPS or XRootD), with appropriate ports opened between datacenters. This can be achieved through object storage systems or secure web-accessible file directories.
- B.
File Monitoring and Triggering: On-site storage must monitor a designated _new_data/ directory to detect new files and trigger transfer actions. A lightweight Python watchdog script can monitor for symbolic links—created upon file completion—and initiate transfer, then remove or relocate the link upon success.
- C.
Off-Site Download Mechanism: Off-site datacenters must run an RPC service hosting the Aria2c downloader. Aria2c supports high-throughput parallel downloads, chunking, resume capability, and integrity verification via checksums. A web UI provides real-time monitoring and automatic retries.
Note: While tools such as GridFTP and GFAL2 remain valid alternatives, the use of CERN FTS is generally discouraged due to its complexity, its reliance on site-specific RSE configuration, and its historically high failure rates. For example, in Tier-0 transfers of LHC experiments using FTS 2.0, failure rates ranged from 5% to 15%, averaging around 10% for simulation chains—primarily due to network timeouts (see ATLAS Tier-0 exercise 2019). FTS 3.0 improved the situation by introducing better monitoring and enhanced retry mechanisms, leading to reduced failure rates.
For this use case, Aria2c offers better control, higher reliability, and simpler integration. In our tests, Aria2c achieved a success rate between 99% and 100%, depending on the number of retry attempts and network availability. The tests were conducted using bulk transfers of real scientific data collected over several years from different projects, as well as simulation data generated by various algorithms. The current dataset consists of approximately 1.2 million files (ranging from 1 to 2.5 GB each), totaling around 2.2 PB of data.
8.2.2. Typical Workflow
- (1)
Data Generation: Telescope systems write data to local storage; upon completion, a symbolic link is placed in _totransfer/.
- (2)
Trigger Detection: A local Python client monitors the directory and detects new links.
- (3)
Transfer Initialization:
- (a)
The symbolic link is resolved to a URI.
- (b)
The target off-site datacenter is selected based on policy rules (e.g., time-based, data level, or project ID).
- (c)
The client invokes a command to the off-site Aria2c service, initiating parallel downloads.
- (d)
Transfer progress is tracked, and completion is confirmed via status queries.
- (e)
Upon success, the symbolic link is removed.
- (4)
Post-Transfer Actions: Additional use cases, such as replication or data ingestion, can be triggered automatically on the off-site side.
8.3. Dataset Ingestion
The ingestion process must adhere to the
Open Archival Information System (OAIS) model, which requires that only verified and validated data products be archived. This mandates a structured, pre-ingestion validation phase, where data integrity and metadata completeness are confirmed before registration, and for ingesting datasets, minimal
Data Product Acceptable Requirements (DPAR) are applicable (i.e., checksum, fits header format, and content verified). These verification steps cannot be postponed to an on-the-fly registration since the file catalog can be updated only when the data product is ready to be registered/stored, even for temporary data (see
Figure 13).
8.3.1. Prerequisites
- A.
The _toingest/storage-pool directory must be POSIX-accessible, even if hosted on object storage.
- B.
The Python environment must include the fitsio (or astropy), json, rucio, and rethinkdb libraries.
- C.
The external storage endpoints called Remote Storage Elements (RSEs) must be accessible via standard A&A protocols (e.g., IAM tokens or legacy credentials).
- D.
A write-enabled RethinkDB node must be reachable on the local network.
8.3.2. Typical Workflow
- (1)
Data Staging: Data products from Data Producers (pipelines, simulations, or DTS) are placed in _toingest/.
- (2)
SIP Creation: A software information package (SIP) is generated, including checksums to verify file integrity.
- (3)
Metadata Validation: FITS headers are parsed and validated to ensure required metadata fields are present, correctly typed, and semantically consistent.
- (4)
Storage Upload:
- (a)
Files are uploaded to an Object Storage path (e.g., dCache FS) using RUCIO or equivalent tools like StoRM for common Storage Resource Manager in DataGrid implementations, see references [
15].
- (b)
If already present in the storage, only a move to a final archive path is needed.
- (c)
Upload status is monitored; once confirmed, metadata (e.g., scope, dataset, RSE) is added to a corresponding JSON record.
Alternative: Use gfal2 to upload directly, guided by storage protocol settings in the ReThinkDB StoragePool collection.
- (5)
Database Registration: Finalized JSON is ingested into the RethinkDB archive, changing file status to “ingested” and completing the Archive Information Package (AIP) creation.
- (6)
Trigger Replication: Upon new entry detection (via RethinkDB’s changefeed), the MAKE_REPLICA process is automatically launched.
8.4. Replica Management in CTAARCHS: Automation and Policy Enforcement
As part of the data ingestion process (point no. 6), automated replication ensures compliance with redundancy and long-term preservation policies. Triggered via a change-feed from the ReThinkDB file catalog, the replication logic references a DATA_POLICY_REPLICATION table to determine the required number of copies per data type and storage level. This shared DB collection is the interface to collect all Retention Policies and Custodial Rules to be assigned to data products.
If no policy rule is found, the data product is assumed to be for temporary processing only. Policies define replication support types (e.g., hot, cold, or hot + cold) and preservation intent. This mechanism fulfills key archival use cases such as tracking preservation state and monitoring physical data locations across distributed storage resources.
8.4.1. Replication Status Levels
Ingested: one off-site catalog record exists.
Archived: at least one replica stored across another RSE.
Preserved: includes a backup on cold storage.
Each replication rule specifies the data type, number of required replicas, and preferred storage configuration. The following is an example:
Any record of the DATA_POLICY_REPLICATION table is called “Replication Rule”, the following is an example:
{ “ruleid”: “1”, “rulename”: “AMAS_dl0-raw”, “datatype”: “dl0.raw”, “replica_lev”: “2”, “rule”: “preserve”, “supports”: “hot + cold”, “timeseries”: [ {“RSE1”: “jan-mar” }, {“RSE2”: “apr-jun” },{“RSE3”: “jul-sep” },{“RSE4”: “oct-dec” } ] }
{ “ruleid”: “1”, “rulename”: “AMAS_dl0-fits”, “datatype”: “dl0.fits”, “replica_lev”: “3”, “rule”: “preserve”, “supports”: “any”}
{ “ruleid”: “2”, “rulename”: “AMAS_dl1-fits”, “datatype”: “dl1[a-c].fits”, “replica_lev”: “1”, “rule”: “ingest”, “supports”: “any”}
{ “ruleid”: “3”, “rulename”: “AMAS_dl3-fits”, “datatype”: “dl3.fits”, “replica_lev”: “3”, “rule”: “ingest”, “supports”: “any”}
A generic UML of the Make_Replica is shown in
Figure 14.
8.4.2. Prerequisites
- A.
All target RSEs must be reachable over secure protocols (e.g., HTTPS, xrootd), and relevant ports must be open across data centers.
- B.
The ReThinkDB cluster must support read/write access from local clients.
- C.
Each off-site RSE must run an
ARIA2c daemon for parallel downloads and transfer monitoring, see
Appendix A.
8.4.3. Typical Workflow
- (0)
Data coming from Data Producers generates a change in the DB catalog.
- (1)
Ingestion completion updates the file catalog, triggering the replication process via the change-feed.
- (2)
The client fetches the file’s URI (2a), matches it against the replication policy (2b), and evaluates eligible RSEs based on latency, throughput, and availability (2c).
- (3)
It initiates parallel data transfers using ARIA2c RPC (3a) and monitors each transfer (3c).
- (4)
On success, the checksum is verified, a new replica record is added to the file’s JSON metadata, and the replica count is updated.
8.5. Dataset Search
Once a data product is ingested—regardless of its archival status (“ingested”, “archived”, or “preserved”)—its metadata becomes searchable through the
ReThinkDB catalog. This enables external users to retrieve dataset identifiers and associated replica information (see
Figure 15).
8.5.1. Prerequisite
- A.
Read-only access to the ReThinkDB cluster must be available from at least one node in the local network.
8.5.2. Typical Workflow
- (1)
A user submits a query via the archive interface, specifying metadata fields of interest.
- (2)
The interface maps the request to searchable metadata intervals.
- (3)
It then queries the ReThinkDB cluster through a local node.
- (4)
The database returns a list of matching data products in JSON format, including URIs and identifiers.
- (5)
This list is delivered to the user for potential retrieval.
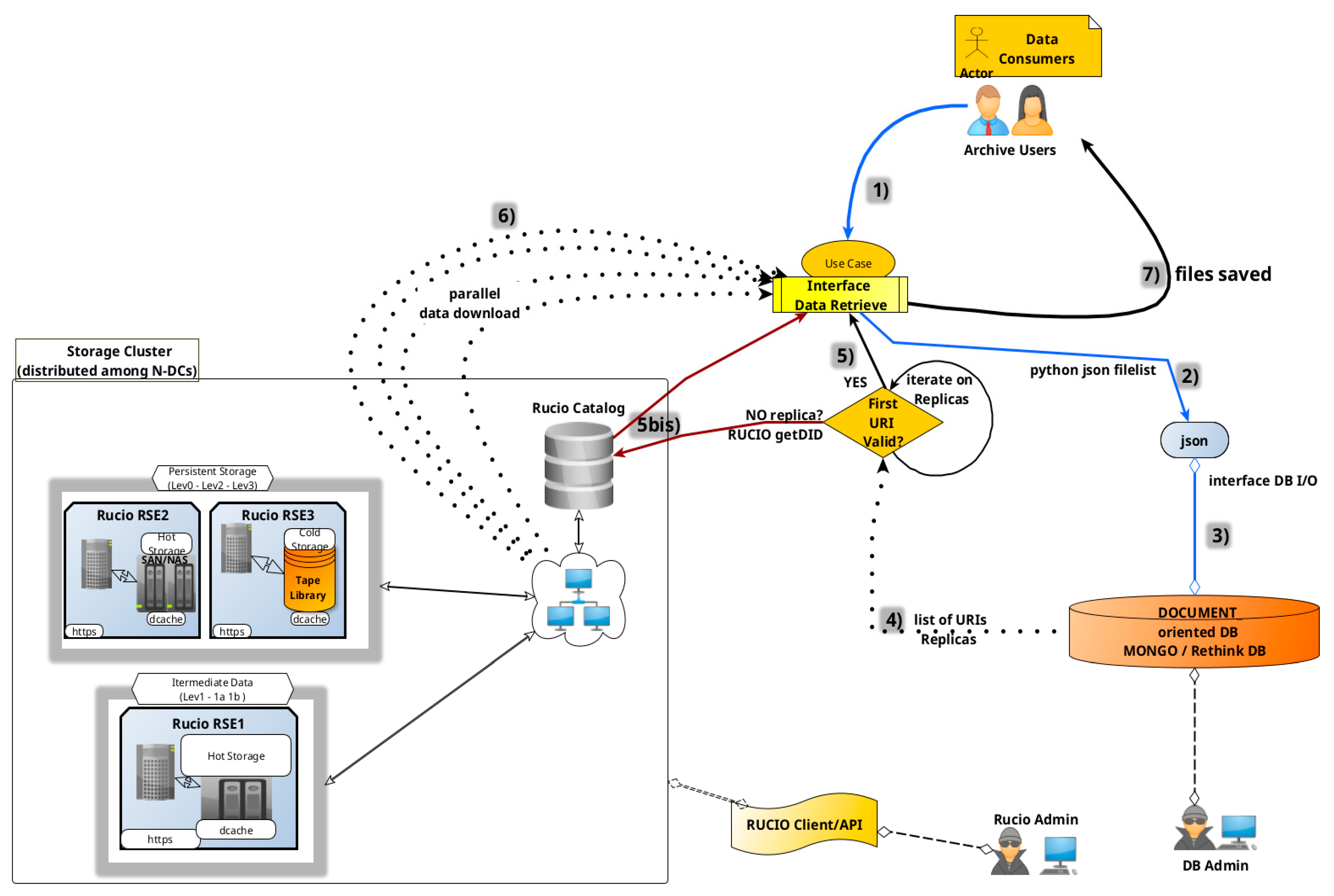
8.6. Dataset Retrieval
Once a dataset is ingested into the archive, regardless of its status (ingested, archived, or preserved), external users can query the RethinkDB metadata catalog to retrieve corresponding datasets and their available replicas. This process involves querying the catalog for metadata, translating the request into predefined searchable metadata intervals, and executing the query via a local node connection. The database returns a JSON file list containing URIs and identifiers of data products matching the query criteria, which are then provided to the user, see
Figure 16 for a generic workflow.
8.6.1. Prerequisites
- A.
Remote Storage Elements (RSEs) must be accessible across data centers via secure protocols (e.g., HTTPS, XRootD), with required ports open. Resources may be object storage pools or directories exposed via HTTPS with encryption and authentication.
- B.
The RethinkDB cluster must be accessible in read-write mode from at least one node within the local network.
8.6.2. Typical Workflow
- (1)
A Data Consumer provides a JSON list of requested data products to the retrieval interface.
- (2)
The system queries the local RethinkDB node.
- (3)
The database returns a list of replica URIs for each product.
- (4)
The interface verifies the existence of each replica.
- (5)
Valid URIs are downloaded in parallel;
(5bis) if no URI from the replica list is available, the system calls the RUCIO catalog to obtain a DID (filename + scope) and find it in the RUCIO catalog.
- (6)
The parallel download starts for any available URI.
- (7)
Retrieved files are stored in a user-specified local or remote directory.
8.7. Search and Retrieve Integration/Concatenation
Search and Retrieve are often combined as a single use case, chaining Python methods to locate metadata and then download the associated data products efficiently, see
Figure 17.
The search.py utility interfaces with the RethinkDB cluster to locate data products based on metadata queries. Depending on the execution context, results may point to internal POSIX paths, external URIs, or any remote-storage-based identifiers (i.e., in RUCIO we have RSE + LFN + SCOPE).
The advanced AMAS Search Interface exposes a REST API via a dedicated web server, supporting fast and complex metadata-based queries across distributed data centers. Users can execute searches from any location or pipeline stage, provided they have network access, the AMAS-REST domain is public, and it can be browsed without authorization (see GITlab CTAARCHS references).
A typical query can be executed with a simple curl command, specifying key–value filters such as date, run number, or filename (see Figure 18).
echo $LIST
{
“files”: [
“… more URIs …”],
“nfiles”: 9
}
The dataset search returns a JSON-formatted file list containing URIs pointing to RSE storage locations. Access typically requires user authentication.
The retrieve.py interface reads this list (e.g., from STDIN) and then downloads the corresponding files to a user-specified directory. It connects to the local RethinkDB node using read-only credentials to fetch replica metadata.
To optimize performance, the system dynamically selects the most efficient replica for each file using a “down-cost” algorithm. This decision is based on the following site-specific parameters:
Cost(i): estimated retrieval cost from site I;
Latency(i): time to initiate transfer;
FileSize: total size of the file;
Throughput(i): nominal data rate;
Workload(i): current system load (0 = idle, 1 = saturated);
Distance(i): network or geographic distance.
These parameters are used to minimize download time and network usage. Workload reflects real-time system strain, while throughput, latency, and distance help assess the optimal retrieval path—especially important in geographically distributed storage systems or under regulatory constraints. Distance could be affected by latency or used explicitly if needed for geo-pinning or regulatory concerns.
The optimal replica for download is dynamically selected by computing the retrieval cost (Costi) in real time. The replica with the lowest cost is chosen, and its URI is returned. Final access requires authentication and authorization.
Latency
i is easily measured via network ping; Throughput
i and Distance
i are typically available from infrastructure documentation. Estimating Workload
i, however, is more complex and can be approximated by comparing the MeasuredThroughput
i—from a small test download—to the NominalThroughput
i.
if latency > threshold or throughput < expected × 0.5:
Workload_i = 0.8 # heavy
elif throughput < expected * 0.8:
Workload_i = 0.5 # moderate
else:
Workload_i = 0.1 # low
The computation is performed periodically (hourly) by an agent module on each datacenter, a temporary ranking is assigned to any storage resources, and the result is saved in a dedicated collection of the rethinkDB cluster in order to be queried before starting each data transfer.
The interfaces (see Python implementations in
Appendix A) are readily accessible via the amas-api_1.0.2
docker image and/or podman in order to be run on Containerd/CRI on Kubernetes or HPC environments.
docker load -i amas-api_1.0.2.tar;
docker run -it amas-environment bash;
./venv/bin/python ./search.py
9. Deployment of CTAARCHS at the CIDC and AMAS
Deploying a data center requires careful planning to ensure efficiency, scalability, and security. At the CTA Italy Data Center (CIDC), deployment of the ASTRI and Miniarray Archive System followed a structured strategy aligned with observatory goals and technical constraints. Emphasis was placed on building a secure, scalable infrastructure, minimizing risks while supporting operational demands. Initial phases included logical and physical design—rack layouts, network topology, cooling, and power—supported by the Tier-2 facility at INFN Frascati, where CIDC is currently hosted [
16,
17].
9.1. Hardware Resources
The CTAARCHS implementation is based on the AMAS archive system, supporting the ASTRI-Horn prototype and the nine-telescope ASTRI Miniarray at Teide Observatory, Tenerife. AMAS represents the complete off-site infrastructure for these projects and serves as the technical deployment of CTAARCHS.
Built on the CTAARCHS/AMAS IaaS, the CTA Italy Data Center (CIDC) forms one of four designated off-site data centers for the CTAO Project (see
Section 9.7.1). Hardware requirements for computing and storage are defined annually by each project office and reflected in a procurement plan for 2025–2026.
Software services follow a Continuous Integration/Delivery (CI/CD) model, with the exception of the archive system, which must be accessible from project initiation. Archive deployment is coordinated with collaboration partners and adapted through a virtualized abstraction layer.
The AMAS implementation of CTAARCHS relies on shared hardware located at the following three main sites:
INAF—OAR, Astronomical Observatory of Rome;
INAF—SSDC, ASI Science Data Center;
INFN—LNF, National Laboratoies of Frascati.
In total, AMAS hardware list consists of a federated distributed “hot” storage of 6PB (directly upgradable to 10PB); around 10PB (directly upgradable up to 100PB) of “cold” storage (Fiber Channel Tape Library); an HPC@OAR consisting of about 800 cores (8.8 kHS06) with ~1TB RAM; and a grid HTC@LNF consisting of about 1400 cores (15.4 kHS06) with ~2.5TB of RAM. In SSDC are foreseen only minimal services and resources not listed here for sharing MWL data.
9.2. The Setup
Datacenters can join the CTAARCHS environment by registering to access repositories of Docker containers, virtual machines, and Kubernetes (K8s) orchestration for various services.
The K8s clusters at INAF-OAR and INFN-LNF sites share resources within the ReDB “resource_pools” collection, managing storage, computing, services, and user registrations. The distributed RethinkDB cluster spans multiple sites—OAR (DC1), LNF (DC2), and SSDC (DC3, pending activation)—as illustrated in
Figure 19.
9.3. Users Interfaces
The main users of the common archive and data management system are:
Pipeline/Simulation users—for low-level data products
Science users—for higher-level data products
BDMS users and Admin—for high-level operations on archives
Access type depends on the nature of the task. If specific archive or database management operations are required, the preferred method is through the command-line interface (CLI) from the BDMS or Science Archive machines. This access is POSIX-compliant, and BDMS/Science Admin roles are mapped within the authorization framework (see
Section 9.8).
Bulk database operations are restricted to administrators and can be executed via the ReThinkDB console. Portal access supports both science users (in the principal investigator role) and admins, who may also manage LDAP credentials.
CLI and POSIX access is granted to low-level users such as automated pipelines, enabling operations like bulk search and retrieval, file renaming/removal, and other data management tasks. Authorization for all access types adheres to the A&A policy and its associated services (see
Section 9.8).
Using the Data I/O Interfaces, authorized science users can search, query, and retrieve proprietary data within defined POSIX paths. Proprietary data mapping is handled at the Science Database-level, ensuring secure and structured access.
9.4. Pipeline/Simulation Users Access and Interface
Users access data through a variety of tools and workflows. Simulation and Pipeline users utilize Workload Management Systems (WMS) such as DIRAC or PANDA to execute Directed Acyclic Graphs (DAGs) on grid computing or HPC environments (e.g., Condor, Slurm), interacting with remote object storage systems.
A more recent approach involves a Kubernetes-based computing element service (CES) to orchestrate job queues and manage virtual organizations and authorization mechanisms. However, current WMS platforms like DIRAC and PANDA are not yet compatible with Kubernetes environments.
Simulation users typically write their output directly to Object Storage for asynchronous ingestion. Pipeline users, on the other hand, first query the archive using metadata to locate input datasets, and then process data close to storage to reduce data transfer overhead (this process is detailed in the “Search” use case). Higher-level data products can be generated inside the same data centers where the bulk of the dataset physically resides, using
Airflow’s DAG Editor, which delivers science-ready outputs to the
Science Archive collection (see
Section 9.7).
Note: All I/O operations must strictly adhere to defined Use Cases (UC) without custom modifications. If a WMS cannot conform to these requirements, it should be adapted or replaced. The archive design must remain unchanged: interactions between the archive and WMS must occur exclusively through defined interfaces. Customization is permitted only at the interface level, as the two systems must remain strictly decoupled.
9.5. Unconventionl Challenges
International collaborations face challenges due to political mandates to use pre-existing systems or software developed by IKC and used for other data models and/or scientific scenarios. For instance, the RUCIO Data Management System and/or DIRAC for the Workload Management System impose several limitations on CTAARCHS. These software often become single points of failure (SPOFs) in a no-SPOF infrastructure, forcing inefficient archive adaptations and violating OAIS principles that mandate strict separation between Data Producers, Data Consumers, and archive submodules through standard interfaces. Modifying the archive requirements to adapt to these limitations becomes detrimental to the continuation of a good collaboration.
For example, RUCIO suffers from SPOF in its centralized PostgreSQL catalog and is complex for multi-institutional sharing due to its fixed CERN-centric data model, leading to storage overhead and high operational costs. A natural antagonist of RUCIO is the
OneData, which is a distributed data management system too, designed to integrate diverse storage resources, facilitating seamless data access and sharing across institutions. Differently from RUCIO, OneData offers a storage federation model based on a distributed, document-oriented database model; being based on a distributed, document-oriented DB cluster (i.e., CouchBase), it offers a storage federation that better supports metadata management, open data, and collaboration, aligning with Open Science goals (see
Table 5). Choosing storage federation technology to serve an astronomical observatory community should prioritize technical effectiveness and use case fit over political or economic pressures [
18].
Finally, because of the several points of failure involved with the RUCIO environment it is clear that NO PERSISTENT ARCHIVAL SERVICE can be dependent on potentially unstable archival software without the possibility of having a “plan-B” ready and usable.
So we need to deprecate the wide use of RUCIO as a central storage system for a good archive, and we auspicate to relegate it only as a marginal common interface because it is optimized for different storage element protocols.
Throughout this work, the term RSE (Remote Storage Element) is used generically to denote any remote storage resource accessible via standard protocols, independent of the RUCIO framework.
9.6. Database and Data Model Interfaces
Intermediate and end users may require direct access to metadata for scientific analysis or simulation output. To support this, a dedicated read-only user role enables querying across all data levels. For FITS files, primary headers are indexed within the data model, allowing advanced search capabilities. A
sample data model and query interface are provided in
Appendix A (see
Section 9.7.1), with customizable code available for tailored use cases.
The code sample is similar to those used for the find and query interface client but can be expressly customized on demand (see
Appendix A for insights).
RethinkDB supports the creation of secondary indexes on metadata fields, enabling faster queries as datasets grow. This feature is simple to implement, with no strict limits on the number of indexes, making it highly effective for optimizing search performance over time.
r.table(“DLFITS”).index_create(“dateobs”).run(conn) #CREATE INDEX
r.table(“DLFITS”).index_wait(“dateobs”).run(conn) #WAIT COMPLETITION
# Query using the index
r.table(“DLFITS”).get_all(“2024-12-06”, index=“dateobs”).run(conn)
9.7. Web Archive Portal for the End-User and Other Interfaces
Science users—primarily researchers accessing high-level data products—interact with the archive via a dedicated web portal. These users are considered as Data Consumer and are planned to retrieve level-3 datasets in read-only mode to conduct analyses or run customizable pipelines. Data dissemination relies on the distributed database, with pipeline execution triggered by change-feed mechanisms monitoring the level-3 collection (see
Figure 20).
9.7.1. Prerequisites
- A.
Python environment must include fitsio (via Astropy), json, rethinkdb, and rucio libraries.
- B.
Bulk and science RSEs must be accessible via supported authentication methods: IAM tokens (preferred), legacy grid certificates (deprecated), or credentials.
- C.
The ReThinkDB cluster must be accessible in read-only mode via at least one local node.
- D.
The Science Database may reside within ReThinkDB or any compatible RDBMS.
9.7.2. Typical Workflow
- (0)
A pipeline processes data and ingests new DL3 products into the archive.
- (1)
Detection of new DL3 entries triggers the get&process action.
- (2)
The associated URI is fetched from the source RSE and transferred to the Science RSE.
- (3)
DL3 metadata are extracted from ReThinkDB and written to the Science DB.
- (4)
Optional automated workflows convert DL3 to DL4 and DL5 products.
Note: Since higher-level science data (DL3–DL5) involve smaller volumes, they may be handled via
lightweight solutions such as local Airflow DAGs and executed on dedicated clusters (see Figure 21).
Figure 21.
Simple processing to pass from DL0 to science data.
Figure 21.
Simple processing to pass from DL0 to science data.
Community LDAP or VPN access enables shared resource usage and supports defining Airflow pipeline steps. The Search and Retrieve Python APIs remain functional but require read-only access to the ReThinkDB cluster. Alternatively, REST-API endpoints can be used to bypass direct database access (see
Figure 22).
The low level processing, for a large amount of data, can be easily shared and distributed among DPPN datacenters, while the science processing can be easily concentrated in one site using a dedicated slurm queue and an AIRFLOW DAG authomatic processing.
The output Science RSE can benefit from a localized access dedicated only to scientific end-users passing through a web portal to browse and access proprietary “proposals” data or through a web gateway facility sharing a user-defined policy repository bucket on a cloud-based storage utility like Min-IO.
Note: Scientific end-user data access cannot rely on complicated grid-based data I/O access like IAM (grid-based certificates/tokens for authentication) required for low-level big data processing. So a cloud-based approach, like amazon-AWS (i.e.,
a customized MinIO facility), gives the end user very simple access customized on a common LDAP authenticaiton (login + password) and permits access to proprietary data products using standard POSIX and REST API access, as well as mounting and sharing local storage areas for analysis and collaboration within research groups [19].
A simple implementation for High Energy Astronomical Archives has been realized for the
ASTRI Project in the
AMAS, ASTRI and Miniarray Archive System, containing the proposal handling system, an observing scheduler and planner a PI-web-based data access for all relevant scientific data levels and if necessary also for logs/alarms, housekeeping, data quality checks, and a quicklook browser as a service to browse data, perform a very simple evaluation on the goodness of a data acquisition, and provide the viewer of plots and graphs. The common usage foresees the end-user accessing through a collaboration VPN access and sharing a local-cluster facility, using find/search queries and data retrieval and using a user-defined namespace local bucket shared on a MinIO infrastructure interface [
20,
21].
The bulk data processing is distributed among three different nodes, and the archive system is distributed too; the science processing instead is principally driven through an AIRFLOW facility running on top of a SLURM HPC queue in the OAR cluster.
As shown, storage and computing resources are built on the
AAAS (Astronomical Archive as a Service) paradigm applied to the astronomical use case; the resulting infrastructure is easily horizontally scalable as well as upgradable without out-of-service when needed. See
Figure 23 for current implementations.
9.8. Authentication and Authorization (A&A) Challenges
Collaboration and interconnection with international resources require a common approach to
Authentication and Authorization (A&A), as the management and usage of resources must be orchestrated across multiple institutions, countries, and potentially varying ICT security levels. CTAARCHS is ready to implement most A&A protocols, ranging from customized LDAP-based accounting to combinations of various A&A services [
22].
Typically, the identification of who is accessing a system, software, or service relies on storing a user ID associated with standard eduGAIN credentials in a credential database. Some collaborations adopt proprietary systems, such as Microsoft Entra ID, to store user credentials and maintain a unified authentication layer.
Once a user is authenticated, their authorization level must be verified to determine whether they are permitted to access a specific platform, resource, or dataset. A common solution is to integrate Grouper as part of a Common Trusted Access Platform architecture, which enables attribute- and role-based authorization and group membership management in an auditable manner. This allows Grouper-enabled services to decentralize the management of authorization for authenticated users.
In scenarios where multiple services work together, authorization levels are often established through SSL grid certificates or via temporary (or persistent) tokens. Using Indigo IAM, services can validate these tokens to grant access to resources. This is especially useful for automated agents and pipelines that need access to storage or computing facilities. In such cases, a specialized mapping mechanism is required to distinguish between public and proprietary data.
Any high-level data produced by a pipeline process must be mapped back to its origin via the proposal database, ensuring access is granted only to the relevant Principal Investigators (PIs and Co-PIs). In CTAARCHS, this mapping is managed through the ReThinkDB Science collection, which links data to the PI table within the proposal handling system, and is ready to integrate PI information with any A&A service as needed. For small-level services, CTAARCHS allows A&A a simple and persistent LDAP-structured database, including a customizable list of user credentials, attributes, and roles.
10. Conclusions and Recommendations
The definition of archive solutions to be adopted in a wide range of scientific collaborations is a crucial step for the success of a project, especially in the astronomical fields where, differently from nuclear and subnuclear particle experiments, the number of end-users is several orders of magnitude greater.
Although political choices can be made and pushed on the basis of pre-existing economic and technological contributions, choosing the best technologies to assemble the most efficient system for the project’s use cases is the most important obstacle to overcome and depends extremely on the project management capabilities of the different teams (working groups) identified to assemble the different modules/packages.
In our CTAARCHS we present a feasible and versatile implementation of all archival and data management ecosystems needed for an astronomical observatory use case, with a set of possibilities or alternate scenarios and equally valid technological choices.
11. Software Resources and Repositories
In this section we summarized a small list of CTAARCH software packages, modules, resources, and repositories, including third-party packages.
11.1. CTAARCH and AMAS
- -
- -
- -
- -
- -