Patient Diagnosis Alzheimer’s Disease with Multi-Stage Features Fusion Network and Structural MRI
Abstract
1. Introduction
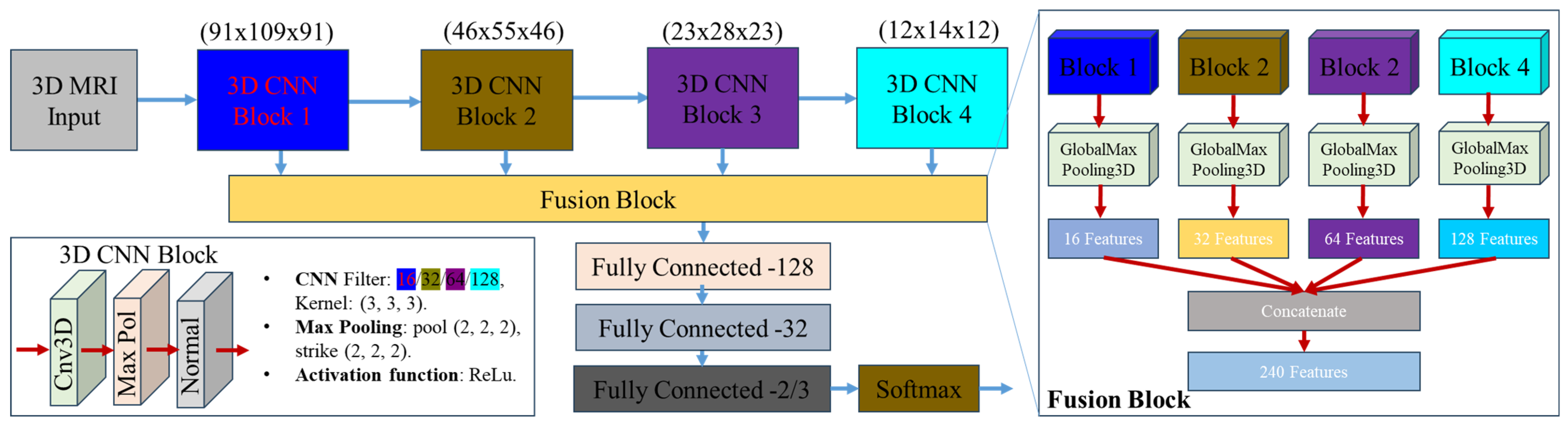
- We proposed a multi-stage feature fusion-based 3D CNN model that fuses feature extraction across multiple blocks of a CNN to avoid information loss and improve classification accuracy [32].
- Optimize model parameters to achieve high computational efficiency at low cost.
- Compare our proposed approach with other 3D CNN models applied in AD classification [33].
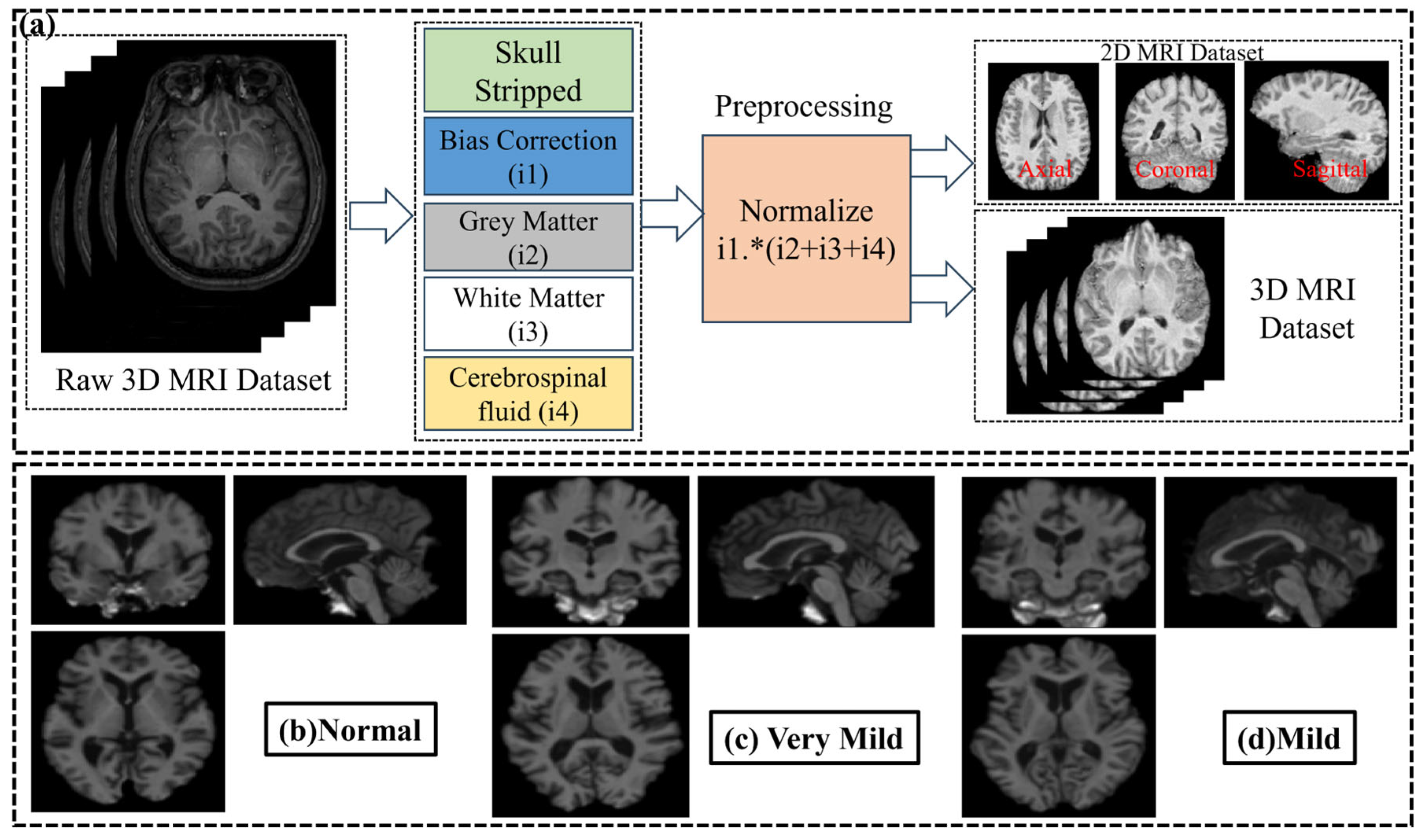
- Preprocessing stage for structural MRI data to remove noise and reconstruct image with four feature image (i.e., white matter, grey matter, cerebrospinal fluid, and bias correction) to improve image quality and reduce computational complexity [34].
2. Materials and Methods
2.1. The Open-Access Series of Imaging Studies (OASIS) Dataset
2.2. Data Preprocessing
2.3. Proposed Architecture of Classification Model Based on 3D CNN
2.4. Evaluation Metrics
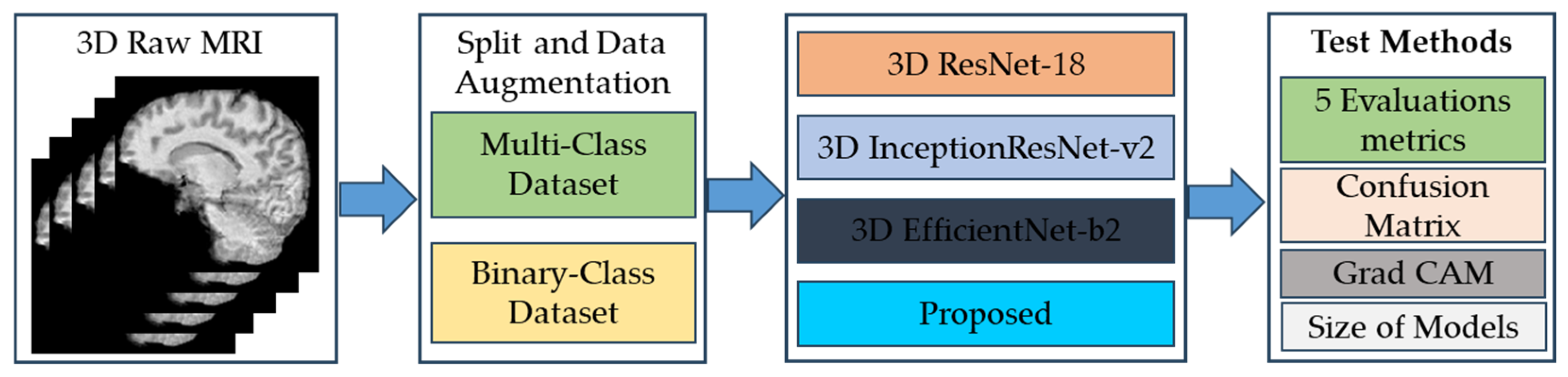
3. Results
3.1. Preparation of Dataset for Training Proposed Classification Model
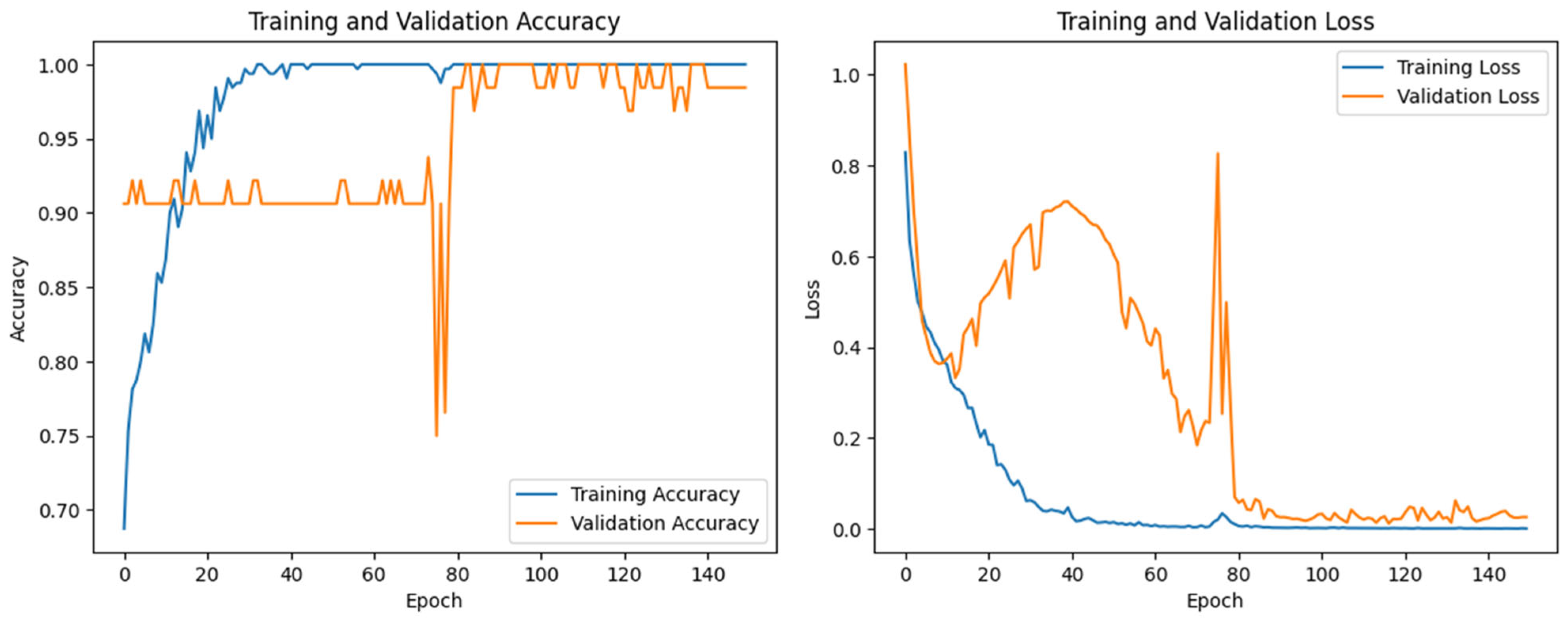
3.2. Evaluation Performance of Proposed Model
3.3. Comparison of Performance of Proposed Model with Three Different Classification 3D CNN Models Based on Transfer Learning
3.3.1. Comparison Evaluation of Metrics After Training
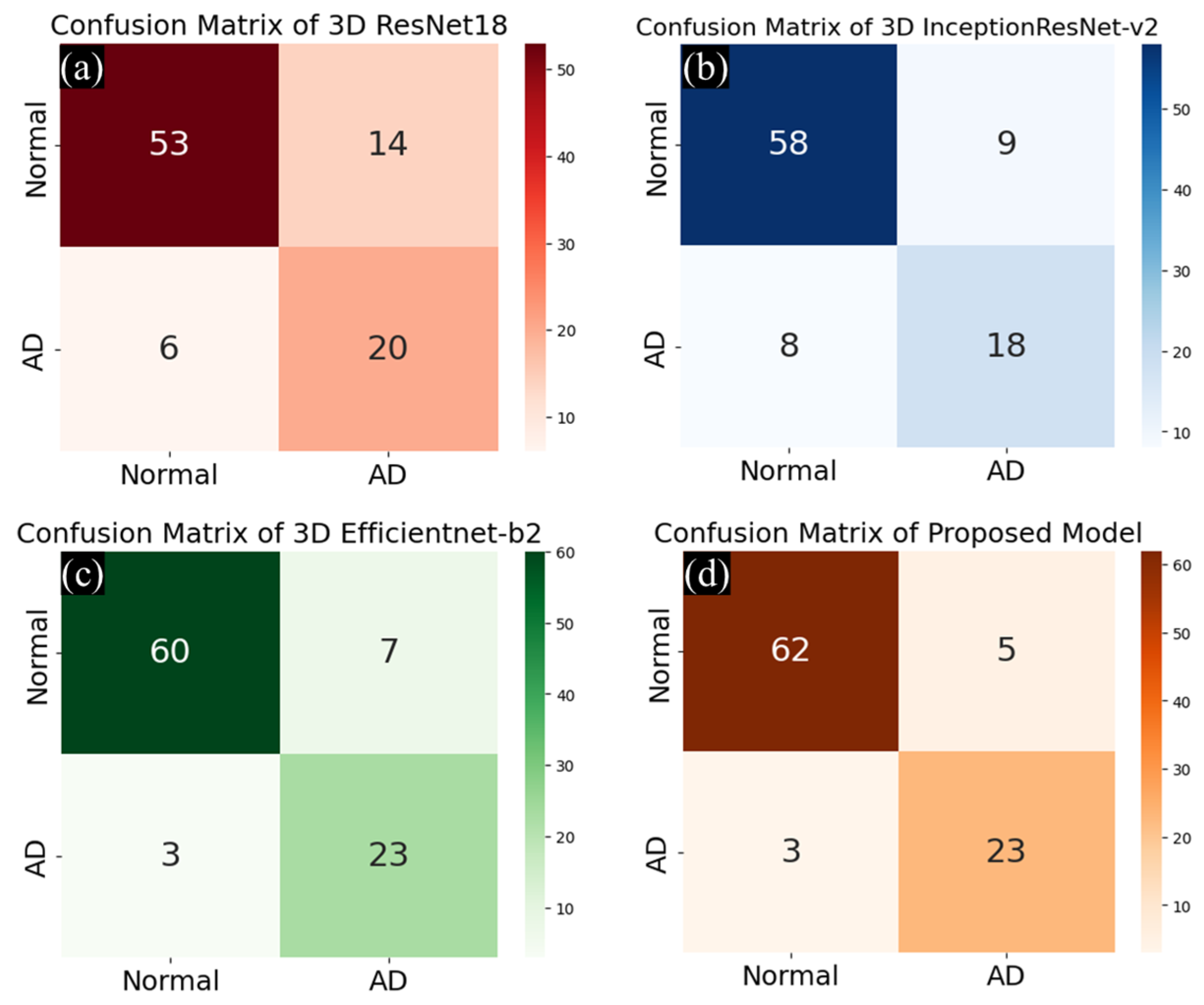
3.3.2. Comparison of Performance of Multi-Class and Binary Classification with Test Set
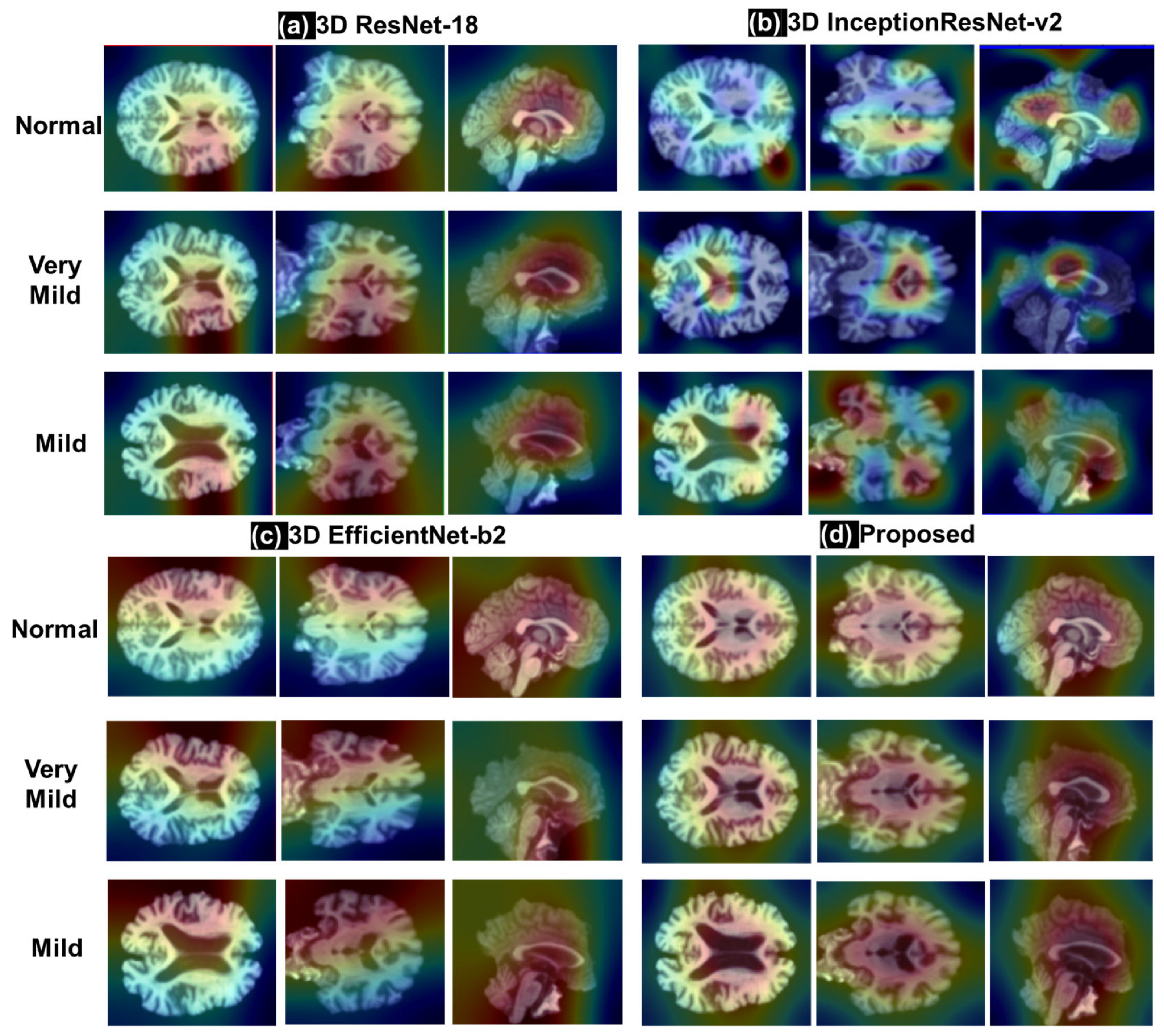
3.3.3. Comparison of Activation Maps with Grad Cam Method
3.3.4. Comparison of Model Sizes for Proposed Model and Three Transfer Learning Models Based on 3D CNN
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, H.; Pedrycz, W.; Hirota, K.; Yan, F. A multiview-slice feature fusion network for early diagnosis of Alzheimer’s disease with structural MRI images. Inf. Fusion 2025, 119, 103010. [Google Scholar] [CrossRef]
- Rahim, N.; El-Sappagh, S.; Ali, S.; Muhammad, K.; Del Ser, J.; Abuhmed, T. Prediction of Alzheimer’s progression based on multimodal Deep-Learning-based fusion and visual Explainability of time-series data. Inf. Fusion 2023, 92, 363–388. [Google Scholar] [CrossRef]
- Fu, J.; Ferreira, D.; Smedby, Ö.; Moreno, R. Decomposing the effect of normal aging and Alzheimer’s disease in brain morphological changes via learned aging templates. Sci. Rep. 2025, 15, 11813. [Google Scholar] [CrossRef]
- Cheng, J.; Wang, H.; Wei, S.; Mei, J.; Liu, F.; Zhang, G. Alzheimer’s disease prediction algorithm based on de-correlation constraint and multi-modal feature interaction. Comput. Biol. Med. 2024, 170, 108000. [Google Scholar] [CrossRef]
- Raza, H.A.; Ansari, S.U.; Javed, K.; Hanif, M.; Mian Qaisar, S.; Haider, U.; Pławiak, P.; Maab, I. A proficient approach for the classification of Alzheimer’s disease using a hybridization of machine learning and deep learning. Sci. Rep. 2024, 14, 30925. [Google Scholar] [CrossRef]
- Liu, S.; Masurkar, A.V.; Rusinek, H.; Chen, J.; Zhang, B.; Zhu, W.; Razavian, N. Generalizable deep learning model for early Alzheimer’s disease detection from structural MRIs. Sci. Rep. 2022, 12, 17106. [Google Scholar] [CrossRef]
- Golovanevsky, M.; Eickhoff, C.; Singh, R. Multimodal attention-based deep learning for Alzheimer’s disease diagnosis. J. Am. Med. Inform. Assoc. 2022, 29, 2014–2022. [Google Scholar] [CrossRef]
- Kim, J.S.; Han, J.W.; Bae, J.B.; Moon, D.G.; Shin, J.; Kong, J.E.; Lee, H.; Yang, H.W.; Lim, E.; Kim, J.Y.; et al. Deep learning-based diagnosis of Alzheimer’s disease using brain magnetic resonance images: An empirical study. Sci. Rep. 2022, 12, 18007. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.; Lin, L.; Sun, S.; Wu, S. Three-round learning strategy based on 3D deep convolutional GANs for Alzheimer’s disease staging. Sci. Rep. 2023, 13, 5750. [Google Scholar] [CrossRef] [PubMed]
- Alinsaif, S.; Lang, J. Alzheimer’s Disease Neuroimaging Initiative, 3D shearlet-based descriptors combined with deep features for the classification of Alzheimer’s disease based on MRI data. Comput. Biol. Med. 2021, 138, 104879. [Google Scholar] [CrossRef]
- Guan, H.; Wang, C.; Tao, D. MRI-based Alzheimer’s disease prediction via distilling the knowledge in multi-modal data. Neuroimage 2021, 244, 118586. [Google Scholar] [CrossRef]
- Jytzler, J.A.; Lysdahlgaard, S. Radiomics evaluation for the early detection of Alzheimer’s dementia using T1-weighted MRI. Radiography 2024, 30, 1427–1433. [Google Scholar] [CrossRef] [PubMed]
- Ur Rahman, J.; Hanif, M.; Ur Rehman, O.; Haider, U.; Mian Qaisar, S.; Pławiak, P. Stages prediction of Alzheimer’s disease with shallow 2D and 3D CNNs from intelligently selected neuroimaging data. Sci. Rep. 2025, 15, 9238. [Google Scholar] [CrossRef]
- Chen, K.; Weng, Y.; Hosseini, A.A.; Dening, T.; Zuo, G.; Zhang, Y. A comparative study of GNN and MLP based machine learning for the diagnosis of Alzheimer’s Disease involving data synthesis. Neural Netw. 2024, 169, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Z.; Zhang, Q.; Yin, Z.; Lu, Z.; Li, Y. A new weakly supervised deep neural network for recognizing Alzheimer’s disease. Comput. Biol. Med. 2023, 163, 107079. [Google Scholar] [CrossRef] [PubMed]
- Bloch, L.; Friedrich, C.M. Alzheimer’s Disease Neuroimaging Initiative, Systematic comparison of 3D Deep learning and classical machine learning explanations for Alzheimer’s Disease detection. Comput. Biol. Med. 2024, 170, 108029. [Google Scholar] [CrossRef]
- Park, C.; Jung, W.; Suk, H.I. Deep joint learning of pathological region localization and Alzheimer’s disease diagnosis. Sci. Rep. 2023, 13, 11664. [Google Scholar] [CrossRef]
- Jenber Belay, A.; Walle, Y.M.; Haile, M.B. Deep Ensemble learning and quantum machine learning approach for Alzheimer’s disease detection. Sci. Rep. 2024, 14, 14196. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Luo, S.; Chiong, R. Introducing Transfer Learning to 3D ResNet-18 for Alzheimer’s Disease Detection on MRI Images. In Proceedings of the 35th International Conference on Image and Vision Computing New Zealand (IVCNZ), Wellington, New Zealand, 25–27 November 2020; pp. 1–6. [Google Scholar]
- Cao, G.; Zhang, M.; Wang, Y.; Zhang, J.; Han, Y.; Xu, X.; Huang, J.; Kang, G. End-to-end automatic pathology localization for Alzheimer’s disease diagnosis using structural MRI. Comput. Biol. Med. 2023, 163, 107110. [Google Scholar]
- Hussain, M.Z.; Shahzad, T.; Mehmood, S.; Akram, K.; Khan, M.A.; Tariq, M.U.; Ahmed, A. A fine-tuned convolutional neural network model for accurate Alzheimer’s disease classification. Sci. Rep. 2025, 15, 11616. [Google Scholar] [CrossRef]
- Zhang, Y.; Peng, S.; Xue, Z.; Zhao, G.; Li, Q.; Zhu, Z.; Gao, Y.; Kong, L. Alzheimer’s Disease Neuroimaging Initiative, AMSF: Attention-based multi-view slice fusion for early diagnosis of Alzheimer’s disease. PeerJ Comput. Sci. 2023, 9, e1706. [Google Scholar] [CrossRef]
- Priyadharshini, S.; Ramkumar, K.; Vairavasundaram, S.; Narasimhan, K.; Venkatesh, S.; Madhavasarma, P.; Kotecha, K. Bio-inspired feature selection for early diagnosis of Parkinson’s disease through optimization of deep 3D nested learning. Sci. Rep. 2024, 14, 23394. [Google Scholar] [CrossRef]
- Parmar, H.; Nutter, B.; Long, R.; Antani, S.; Mitra, S. Spatiotemporal feature extraction and classification of Alzheimer’s disease using deep learning 3D-CNN for fMRI data. J. Med. Imaging 2020, 7, 056001. [Google Scholar] [CrossRef]
- Pan, D.; Zeng, A.; Jia, L.; Huang, Y.; Frizzell, T.; Son, X.; Initia, A. Early Detection of Alzheimer’s Disease Using Magnetic Resonance Imaging: A Novel Approach Combining Convolutional Neural Networks and Ensemble Learning. Front. Neurosci. 2020, 14, 259. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.; Chung, Y.-C.; Kim, K.W.; Kim, W.-S.; Oh, I.-S. Classification and Visualization of Alzheimer’s Disease using Volumetric Convolutional Neural Network and Transfer Learning. Sci. Rep. 2019, 9, 18150. [Google Scholar] [CrossRef] [PubMed]
- Balboni, E.; Nocetti, L.; Carbone, C.; Dinsdale, N.; Genovese, M.; Guidi, G.; Malagoli, M.; Chiari, A.; Namburete, A.I.L.; Jenkinson, M.; et al. The impact of transfer learning on 3D deep learning convolutional neural network segmentation of the hippocampus in mild cognitive impairment and Alzheimer disease subjects. Hum. Brain Mapp. 2022, 43, 3427–3438. [Google Scholar] [CrossRef] [PubMed]
- Rogeau, A.; Hives, F.; Bordier, C.; Lahousse, H.; Roca, V.; Lebouvier, T.; Pasquier, F.; Huglo, D.; Semah, F.; Lopes, R. A 3D convolutional neural network to classify subjects as Alzheimer’s disease, frontotemporal dementia or healthy controls using brain 18F-FDG PET. Neuroimage 2024, 288, 120530. [Google Scholar] [CrossRef]
- Alp, S.; Akan, T.; Bhuiyan, M.S.; Disbrow, E.A.; Conrad, S.A.; Vanchiere, J.A.; Kevil, C.G.; Bhuiyan, M.A.N. Joint transformer architecture in brain 3D MRI classification: Its application in Alzheimer’s disease classification. Sci. Rep. 2024, 14, 8996. [Google Scholar] [CrossRef]
- Zhang, J.; Zheng, B.; Gao, A.; Feng, X.; Liang, D.; Long, X. A 3D densely connected convolution neural network with connection-wise attention mechanism for Alzheimer’s disease classification. Magn. Reson. Imaging 2021, 78, 119–126. [Google Scholar] [CrossRef]
- Zhang, X.; Han, L.; Zhu, W.; Sun, L.; Zhang, D. An Explainable 3D Residual Self-Attention Deep Neural Network for Joint Atrophy Localization and Alzheimer’s Disease Diagnosis Using Structural MRI. IEEE J. Biomed. Health Inform. 2022, 26, 5289–5297. [Google Scholar] [CrossRef]
- Muhammad, G.; Shamim Hossain, M. COVID-19 and Non-COVID-19 Classification using Multi-layers Fusion From Lung Ultrasound Images. Inf. Fusion 2021, 72, 80–88. [Google Scholar] [CrossRef]
- Solovyev, R.; Kalinin, A.A.; Gabruseva, T. 3D convolutional neural networks for stalled brain capillary detection. Comput. Biol. Med. 2022, 141, 105089. [Google Scholar] [CrossRef]
- Friston, K.J.; Holmes, A.P.; Worsley, K.J.; Poline, J.-P.; Frith, C.D.; Frackowiak, R.S.J. Statistical Parametric Maps in Functional Imaging: A General Linear Approach. Hum. Brain Mapp. 1994, 2, 189–210. [Google Scholar] [CrossRef]
- Marcus, D.; Wang, T.H.; Parker, J.; Csernansky, J.G.; Morris, J.C.; Buckner, R.L. Open Access Series of Imaging Studies (OASIS): Cross-Sectional MRI Data in Young, Middle Aged, Nondemented, and Demented Older Adults. J. Cogn. Neurosci. 2007, 19, 1498–1507. [Google Scholar] [CrossRef]
- Salami, F.; Bozorgi-Amiri, A.; Hassan, G.M.; Tavakkoli-Moghaddam, R.; Datta, A. Designing a clinical decision support system for Alzheimer’s diagnosis on OASIS-3 data set. Biomed. Signal Process. Control 2022, 74, 103527. [Google Scholar] [CrossRef]
- Mishra, S.K.; Kumar, D.; Kumar, G.; Kumar, S. Multi-Classification of Brain MRI Using Efficientnet. In Proceedings of the 2022 International Conference for Advancement in Technology (ICONAT), Goa, India, 21–22 January 2022; pp. 1–6. [Google Scholar]
- Matharaarachchi, S.; Domaratzki, M.; Muthukumarana, S. Enhancing SMOTE for imbalanced data with abnormal minority instances. Mach. Learn. Appl. 2024, 18, 100597. [Google Scholar] [CrossRef]
- Chaudhary, P.K.; Pachori, R.B. Automatic diagnosis of glaucoma using two-dimensional Fourier-Bessel series expansion based empirical wavelet transform. Biomed. Signal Process. Control 2021, 64, 102237. [Google Scholar] [CrossRef]
- Fernando, K.R.M.; Tsokos, C.P. Dynamically Weighted Balanced Loss: Class Imbalanced Learning and Confidence Calibration of Deep Neural Networks. IEEE Trans. Neural Netw. Learn. Syst. 2022, 33, 2940–2951. [Google Scholar] [CrossRef] [PubMed]
- Bouguerra, O.; Attallah, B.; Brik, Y. MRI-based brain tumor ensemble classification using two stage score level fusion and CNN models. Egypt. Inform. J. 2024, 28, 100565. [Google Scholar] [CrossRef]
- Wang, W.R.; Pan, B.; Ai, Y.; Li, G.H.; Fu, Y.L.; Liu, Y.J. ParaCM-PNet: A CNN-tokenized MLP combined parallel dual pyramid network for prostate and prostate cancer segmentation in MRI. Comput. Biol. Med. 2024, 170, 107999. [Google Scholar] [CrossRef]


| Reference | Year | Dataset | Image Type | Method | Classification Type | Accuracy (%) | No. of Layers/No. of Parameters |
|---|---|---|---|---|---|---|---|
| Priyadharshini et al. [23] | 2024 | PPMI database | MRI | 3D CNN | Multi-class | 97 | 23 layers |
| Parmar et al. [24] | 2020 | ADNI | fMRI | 3D CNN | Multi-class | 93 | 8 layers |
| Pan et al. [25] | 2020 | ADNI | MR | 2D CNN | Multi-class | 84 | 8 layers |
| Oh et al. [26] | 2020 | ADNI | MRI | Convolutional autoencoder (CAE)-based | Multi-class | 86.6 | 371 K |
| Balboni et al. [27] | 2022 | ADNI | MRI | Spatial warping network segmentation (SWANS) 3D- CNN | Multi-class | 90 | 10 layers |
| Rogeau et al. [28] | 2024 | ADNI | MRI | 3D CNN | Multi-class | 89.8 | 3D VGG16 |
| Alp et al. [29] | 2024 | ADNI | MRI | Vision Transformer (ViT) | Multi-class | 99 | x |
| Zhang et al. [30] | 2021 | ADNI | MRI | 2D CNN | Multi-class | 78.79 | Transfer learing with ResNet and DenseNet |
| Zhang et al. [31] | 2021 | ADNI | MRI | 3D CNN | Multi-class | 95.6 | Transfer learing with 3D-ResAttNet34 |
| Kang et al. [9] | 2023 | ADNI | MRI | 3D Deep Convolutional Generative Adversarial Networks (DCGANs) | Multi-class | 92.8 | Transfer learing with 3D ResNet |
| Categories | CDR = 0 | CDR = 0.5 | CDR = 1.0 | CDR = 2.0 |
|---|---|---|---|---|
| Age (year) | 43.8 ± 23.72 | 76.21 ± 7.14 | 77.75 ± 6.68 | 82.0 ± 4.0 |
| Male | 127 | 48 | 9 | 1 |
| Female | 209 | 52 | 19 | 1 |
| Total | 336 | 100 | 28 | 2 |
| Normal | AD | |
|---|---|---|
| Train/Validation | 269 | 102 |
| Test | 67 | 26 |
| CDR = 0 (Normal) | CDR = 0.5 (Very Mild) | CDR = 1.0 (Mild) | |
|---|---|---|---|
| Train/Validation | 269 | 80 | 16 |
| Test | 67 | 20 | 10 |
| Hyper-Parameters | Value |
|---|---|
| Epoch | 30 |
| k-folds | 5 |
| Batch size | 32 |
| Learning rate | 0.0001 |
| Data augmentation | Rotation angles = (−20, −10, −5, 5, 10, 20) |
| Optimizer | Adam |
| Loss function | Cross-entropy |
| Metrics | Train_accuracy, train_loss, val_accuaracy, val_loss |
| Parameter | Cross-Validation | Average (Mean ± SD) (95% CI) | ||||
|---|---|---|---|---|---|---|
| k = 1 | k = 2 | k = 3 | k = 4 | k = 5 | ||
| F1-score | 0.9278 | 0.9255 | 0.9407 | 0.9742 | 0.9927 | 0.9522 ± 0.0267 (0.9151–0.9892) |
| Accuracy | 0.9265 | 0.9355 | 0.9523 | 0.9886 | 0.9901 | 0.9586 ± 0.0264 (0.9219–0.9953) |
| Specificity | 0.9321 | 0.9269 | 0.9412 | 0.9623 | 0.9923 | 0.9510 ± 0.0239 (0.9177–0.9842) |
| Sensitivity | 0.9123 | 0.9117 | 0.9320 | 0.9615 | 0.9957 | 0.9426 ± 0.0321 (0.8980–0.9872) |
| Precision | 0.9256 | 0.9360 | 0.9307 | 0.9709 | 0.9961 | 0.9515 ± 0.0272 (0.9141–0.9897) |
| Parameter | Cross-Validation | Average (Mean ± SD) (95% CI) | ||||
|---|---|---|---|---|---|---|
| k = 1 | k = 2 | k = 3 | k = 4 | k = 5 | ||
| F1-score | 0.9155 | 0.9156 | 0.8904 | 0.9604 | 0.9521 | 0.9268 ± 0.0259 (0.8909–0.9627) |
| Accuracy | 0.9236 | 0.9298 | 0.8896 | 0.9769 | 0.9601 | 0.9360 ± 0.0303 (0.8939–0.9781) |
| Specificity | 0.9228 | 0.9305 | 0.8936 | 0.9556 | 0.9678 | 0.9341 ± 0.0260 (0.8980–0.9701) |
| Sensitivity | 0.9286 | 0.9288 | 0.8821 | 0.9774 | 0.9551 | 0.9344 ± 0.0319 (0.8902–0.9786) |
| Precision | 0.9144 | 0.9159 | 0.8845 | 0.9654 | 0.9668 | 0.9294 ± 0.0320 (0.8850–0.9738) |
| k = 1 | k = 2 | k = 3 | k = 4 | k = 5 | t-Test | ||
|---|---|---|---|---|---|---|---|
| Acc of Binary Classification | ResNet-18 | 0.7621 | 0.8022 | 0.8629 | 0.8782 | 0.8817 | t-statistic: 9.2936 p-value: 0.0007 |
| Our | 0.8721 | 0.9021 | 0.9384 | 0.9432 | 0.9496 | ||
| Acc of Multi-class Classification | ResNet-18 | 0.7522 | 0.7715 | 0.8099 | 0.8267 | 0.8398 | t-statistic: 8.5168 p-value: 0.001 |
| Our | 0.8012 | 0.8823 | 0.9015 | 0.9218 | 0.9312 |
| Task | Evaluation Metrics | Model | |||
|---|---|---|---|---|---|
| 3D ResNet18 | 3D InceptionResNet-v2 | 3D Efficientnet-b2 | Proposed | ||
| Multi-class Classification | Train Acc | 0.9364 | 0.9921 | 0.9736 | 0.9860 |
| Validation Acc | 0.8416 | 0.8098 | 0.9057 | 0.9360 | |
| Train Loss | 0.0261 | 0.0056 | 0.0098 | 0.0174 | |
| Validation Loss | 0.1651 | 0.1827 | 0.1398 | 0.1152 | |
| Binary Classification | Train Acc | 0.9447 | 0.9949 | 0.9952 | 0.9886 |
| Validation Acc | 0.8851 | 0.8604 | 0.9155 | 0.9586 | |
| Train Loss | 0.0056 | 0.0016 | 0.0029 | 0.0101 | |
| Validation Loss | 0.1125 | 0.1230 | 0.0879 | 0.0771 | |
| Model | Trainable Params | Total Params |
|---|---|---|
| 3D ResNet-18 | 33,236,548 (126.79 MB) | 33,244,486 (126.82 MB) |
| 3D InceptionResNet-v2 | 67,654,115 (258.08 MB) | 67,714,659 (258.31 MB) |
| 3D Efficientnet-b2 | 9,898,577 (37.76 MB) | 9,980,865 (38.07 MB) |
| Proposed | 1,387,929 (5.28 MB) | 1,386,489 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, T.M.T.; Bui, N.T. Patient Diagnosis Alzheimer’s Disease with Multi-Stage Features Fusion Network and Structural MRI. J. Dement. Alzheimer's Dis. 2025, 2, 35. https://doi.org/10.3390/jdad2040035
Nguyen TMT, Bui NT. Patient Diagnosis Alzheimer’s Disease with Multi-Stage Features Fusion Network and Structural MRI. Journal of Dementia and Alzheimer's Disease. 2025; 2(4):35. https://doi.org/10.3390/jdad2040035
Chicago/Turabian StyleNguyen, Thi My Tien, and Ngoc Thang Bui. 2025. "Patient Diagnosis Alzheimer’s Disease with Multi-Stage Features Fusion Network and Structural MRI" Journal of Dementia and Alzheimer's Disease 2, no. 4: 35. https://doi.org/10.3390/jdad2040035
APA StyleNguyen, T. M. T., & Bui, N. T. (2025). Patient Diagnosis Alzheimer’s Disease with Multi-Stage Features Fusion Network and Structural MRI. Journal of Dementia and Alzheimer's Disease, 2(4), 35. https://doi.org/10.3390/jdad2040035





