1. Introduction
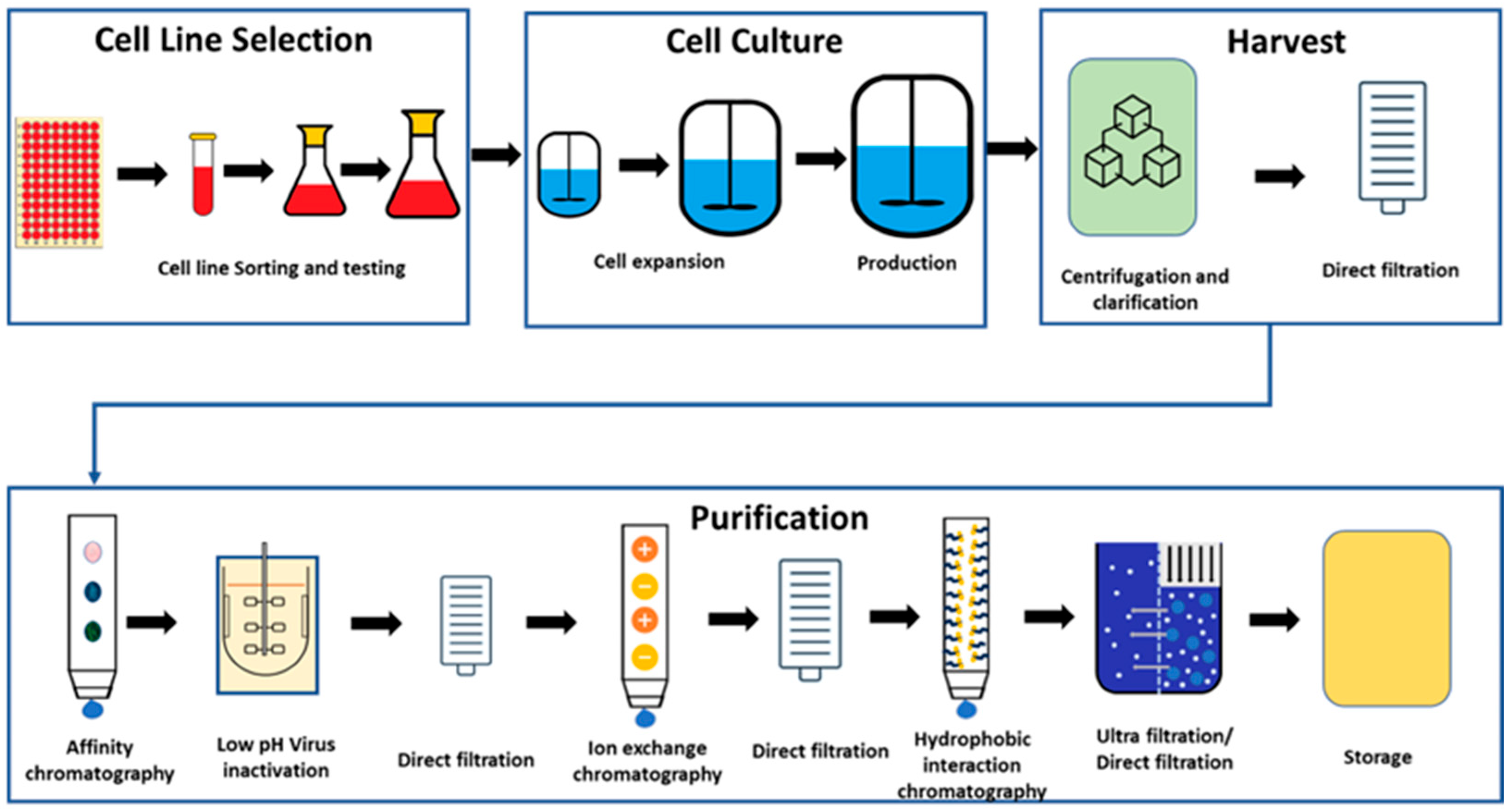
Biopharmaceutical manufacturing across several modalities, such as monoclonal antibodies (mAbs), antibody drug conjugates (ADCs), vaccines, and cellular and gene therapies (C>s) represent a major part of the emerging pharmaceutical portfolio for a variety of medical conditions, such as autoimmune diseases, cancer, and infectious diseases. Monoclonal antibodies (mAbs) have emerged as top-grossing pharmaceutical products, with substantial sales and significant approvals between 2020 and 2022 [
1,
2]. Manufacturing mAbs is a complex, regulated process aimed at ensuring product quality, safety, and efficacy, involving cell culture, harvest, purification, formulation, and packaging stages. The key objective is to maximize yield (titre) and minimize impurities (including host cell proteins) while complying with regularities guidelines. Such processes are sensitive to various chemical and physical factors, including dissolved oxygen (DO) levels, pH, temperature, glucose concentration, nitrogen, and carbon dioxide flow rates, along with mixing speeds. These variables significantly impact cell growth, metabolism, protein concentration, and cell viability. Given the extensive and intricate nature of the mAb manufacturing process, a monitoring platform is essential to confirm robustness and process consistency [
3,
4].
To optimize and predict outcomes in large-scale biopharmaceutical manufacturing, mechanistic and semi-mechanistic mathematical models are employed, though they are complex and need rigorous efforts to build and test [
5]. Advancements in data collection, storage, and processing have led to the evolution of data-driven methods, data engineering, and data analytics [
6,
7,
8]. Multivariate Statistical Process Monitoring (MSPM) is a prominent data-driven analytics approach, offering advantages over traditional univariate methods by accounting for variable correlations to detect process drifts, deviations, and anomalies or atypicality. Early detection of process atypicality allows for potential for preventative intervention and batch saving [
9]. MSPM can be a powerful component in understanding the holistic process as it provides a comprehensive view of the manufacturing process in a multivariate approach and can be used for dimensionality reduction and visualization by transforming variables into uncorrelated components [
10,
11]. Common data-driven approaches like principal component analysis (PCA) and partial least square (PLS) have attracted significant interest from both academia and industry [
10]. PCA is a multivariate statistical technique used for dimensionality reduction and data visualization with the primary objective of transforming the original variables into a new set of uncorrelated principal components [
11]. In the context of multivariate process monitoring, PCA is often employed to analyze and monitor the variation in multiple variables and identify abnormal patterns in a test dataset that may indicate issues in the manufacturing process. The use of PCA model diagnostics like residuals and Hotelling’s T
2 is critical for detecting such deviations from the normal operating conditions represented in the historical data used to train the model [
11,
12].
Despite its efficiency in handling static data, PCA struggles with dynamic or time series data. Unfolding methods, such as batch-wise and variable-wise unfolding, address the challenges when time series data are available. Batch-wise unfolding enables the detailed analyses of temporal behaviour by segmenting time data, though it grapples with irregular or no-periodic batch structures like those characterizing bioreactor stages. Variable-wise unfolding averages variability across time points, suitable for static variables but challenging for dynamic ones, necessitating alternative modelling approaches. The PCA is efficient to handle the static data but lacks the capability to model the dynamic/time series data. When time series data are available for real-time process monitoring, batch-wise unfolding and variable-wise unfolding are often used. For dynamic variables, process batch trajectory modelling approaches like dynamic PCA and PLS, which consider maturity index and assumption-free batch process modelling, can be used to track the evolution of key process variables over time during a batch production cycle.
Implementing Multivariate Statistical Process Monitoring (MSPM) offers significant advantages and has demonstrated benefits for process monitoring in several manufacturing modalities [
13,
14]. MSPM can help identify process atypicality by detecting interactions among variables; for example, a pH increase alongside uncontrolled CO
2 overlay can negatively impact cell metabolism and productivity. Timely detection of such atypicality allows for corrective actions in a preventative manner, leveraging the MSPM model outcomes. Early detection of variations can help reduce process waste and enhance operational efficiency and productivity. In essence, MSPM aids predictive maintenance by analyzing equipment sensor correlations, predicting equipment health and maintenance needs, and preventing downtime in manufacturing.
One of the biggest challenges for the development of the process monitoring models, particularly for expensive biologicals, is the limited availability of at-scale manufacturing process data. Often, suitable data are available for only a few batches, making it difficult to generate robust MSPM models [
15,
16,
17]. In biopharmaceutical manufacturing, “low N” refers to situations where there is a small sample size or limited data availability for analysis. Low-N data scenarios often arise under several conditions: introducing a new product with limited production history at a manufacturing site, meeting only clinical or early commercial demand, transferring an established product from another site, or changing the setup of an established product process [
18]. The impact of the low-N scenario on the MSPM model development is significant due to several statistical constraints. PCA scores cannot be assumed to be normally distributed if process variables are not normally distributed, and it is not possible to apply the central-limit theorem (CLT). Consequently, the assumption that residuals’ statistics are Gaussian does not necessarily hold since the results from CLT are not applicable. Under the low-N scenario, the variability on the control limits derived for Hotelling’s T
2 and residuals Q statistic can be quite large, introducing challenges in accurately defining thresholds and interpreting the result of the monitored processes. This necessitates alternative approaches to ensure reliability. To address the challenges posed by the low-N scenario in MSPM model development, a potential solution involves leveraging the existing data to generate an arbitrary number of in silico data points to augment the dataset. This approach aims at improving the coverage of data across normal operations. By augmenting the real dataset with in silico data, it becomes feasible to build robust MSPM models following the same robust strategy typically applied in high-N scenarios. This combined use of real and in silico data enhances the statistical reliability and generalization capabilities of the models, allowing for better defined control limits and more accurate process monitoring. The objective of this study is to demonstrate the use of in silico data along with real batch data for developing MSPM models for mAb manufacturing to monitor the bioreactor process for low-N data scenarios. The result of this study displays that this approach bridges the gap created by insufficient data, ensuring comprehensive and effective MSPM model development for the manufacturing of biologics.
3. Results and Discussion
Model-based real-time process monitoring and process control are key areas of interest for a number of scientific fields, such as (bio)chemical processing, pharmaceutical manufacturing, and energy and gas production [
17,
24,
25]. In this work, we have focused on developing real-time process monitoring capability based on multivariate statistical modelling approaches to monitor the performance of upstream bioreactors for the production of monoclonal antibodies (mAbs). Through the implementation of MSPM methods, we aim to track multiple variables simultaneously, offering a comprehensive view of system performance. This approach enables us to take preventative actions in response to any potential excursions, ensuring optimal system operation.
3.1. Evaluation of In Silico Batches and Comparison with Real Data
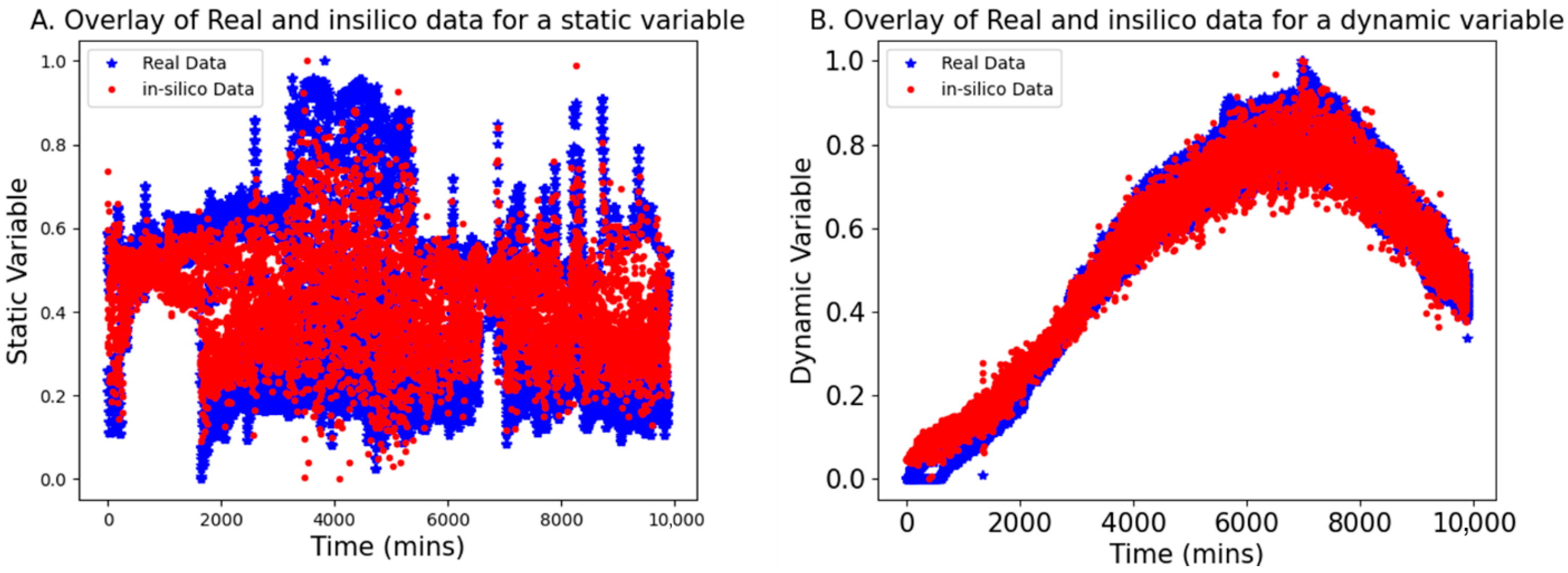
This section presents the results of using bGen to generate in silico datasets for low-N scenarios, applied to both static and dynamic variable examples. The bGen applies Gaussian process state-space models to build in silico datasets from the few historical batch datasets. In
Figure 3, the red points represent the data points generated in silico, while the blue points display the real low-N data points. The two real batch data have a sampling frequency of 2 min, while in silico batches are generated with a sampling frequency of 50 min. The bounds for the in silico data were defined based on the bounds of the real data.
As seen in
Figure 3, the in silico batches are contained within the bounds of the real batches and demonstrate excellent coverage of the gaps between the trajectories of real data. Furthermore, the trajectories of the in silico batches closely resemble those of the real batches. The result confirmed that even with the low-frequency data, the in silico batches completely cover the space bracketed between real data, while retaining the structure and trends of the original batches. This reconstructing of variability in data facilitates the development of robust models for further applications.
3.2. Model Development Results
3.2.1. PCA Model Result
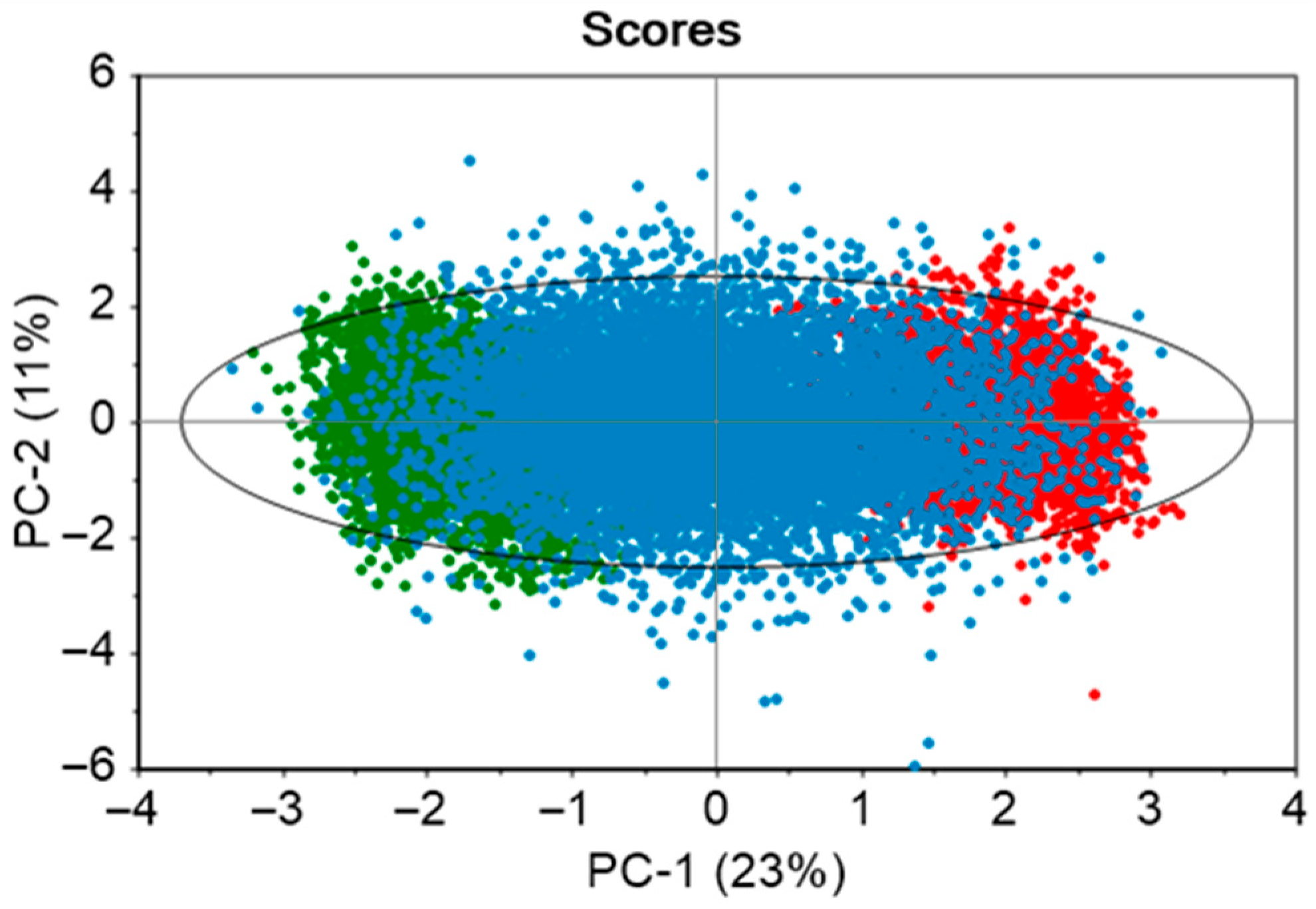
The developed PCA model consisted of seven principal components to explain 83% variance of the ten static variables. The dataset contained two historical batches, and 20 in silico generated batches. From the scores plot in
Figure 4, it is observed that there is an even distribution of the data points across the model space. The real batches (green and red solid points) are clustered on the opposite ends of the ellipse, while the in silico batches (blue solid points) occupy the space between them. This enhances the model’s coverage and its capability to capture the variability within the real batches. The green and red areas indicate process conditions that are slightly distinct from one another.
3.2.2. Trajectory Model Result
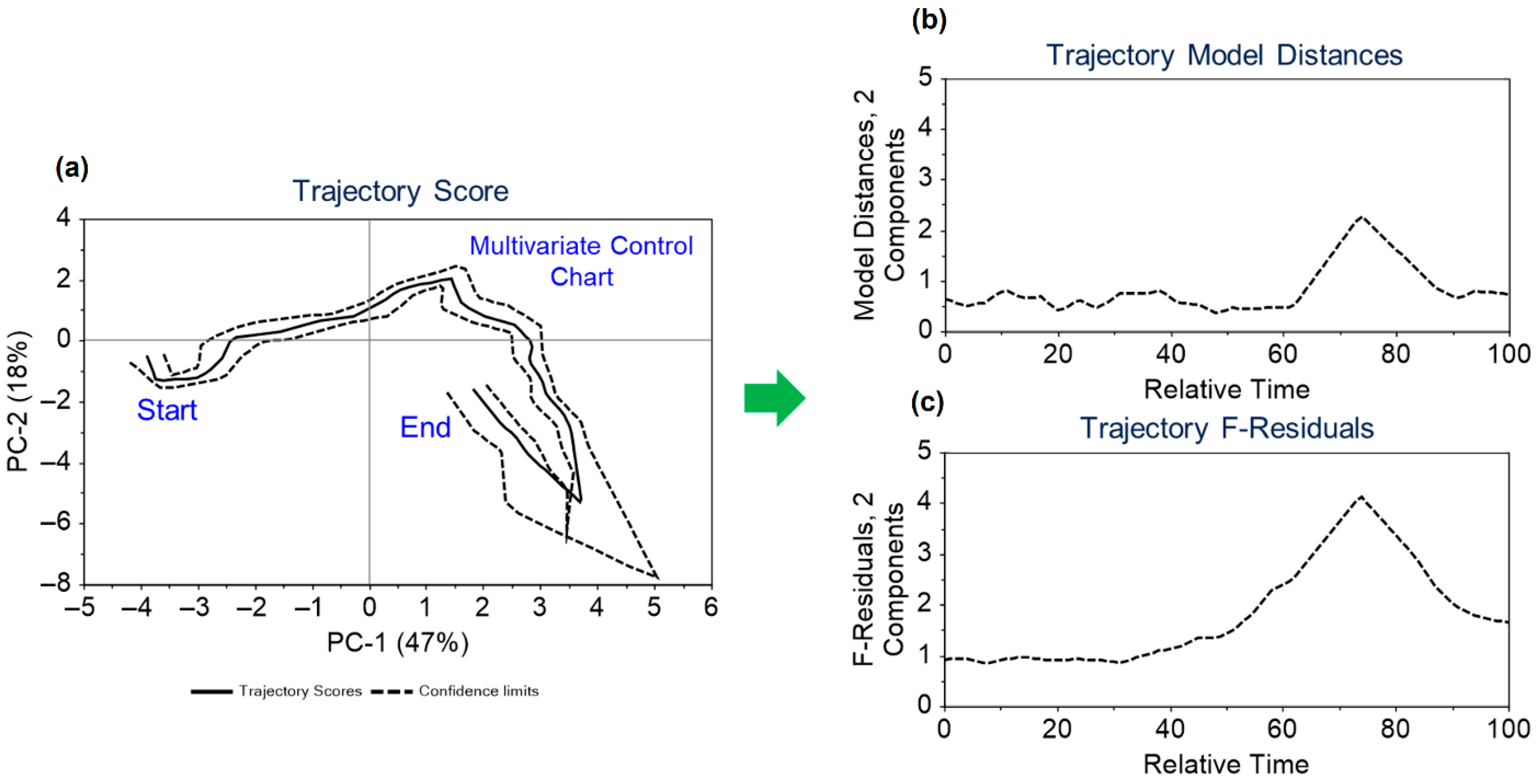
Dynamic variables from 2 real batches and 20 in silico batches were utilized to develop the trajectory model for monitoring these variables. The data were mean-centred, and two principal components were considered in creating the trajectory models. The maximum grid range for dimensions 1 and 2, along with batch coverage, was set at 8, 9, and 80%, respectively. The model’s validity was confirmed through cross-validation. The model results are depicted in
Figure 5.
Figure 5a shows the two-dimensional trajectory score, highlighting a common trajectory for batches over relative time and indicating the start and end points of the trajectory, which correspond to the inoculation and harvesting times. In the
Figure 5a, solid lines represent the average trajectory, while dashed lines indicate the two standard deviation limits. The estimated dynamic model distances and F-residuals control limits are illustrated throughout the trajectory in
Figure 5b,c. Trajectory scores, model distances, and F-residual plots are monitored for any potential deviations. The application of the approach outlined in
Figure 5 is further contextualized in
Section 3.5 within the framework of the discussion on the illustrative case study.
3.3. Real-Time Monitoring and Diagnostics Criteria
The developed models were deployed on the online platform ‘Aspen Process Pulse version 12.2’ developed by Aspen Technology, Inc. The univariate monitoring of the process describes the characteristics in just one dimension and only the upper and lower limit or confidence bands around the mean value. For the multivariate process monitoring, the following parameters were used for fault detection: Hotelling’s T2 and F-residual limits.
Hotelling’s
T2, Equation (1), describes the behaviour of the process in the state space and identifies the correlation structure of the variables in the model using the covariance matrix used to build the model [
26,
27,
28].
where
x is observation vector with
p variables, vector
is the estimated mean for each variable, and
t refers to the transpose operation. S is the estimated covariance of the matrix. The
T2 can be calculated and plotted for each new data point [
29]. The model distances in trajectory modelling are estimated similarly to the calculation of Hotelling’s
T2 statistic [
20].
F-residuals represent the prediction ability of the model. When the model is robust, the residuals are smaller. Also, when the projected data point from the process has variable correlation, as seen by the model from the training dataset, the residuals for the projection are small. Large residuals can indicate unusual process behaviour and a potential fault in the process [
29].
For the PCA models, we used Hotelling’s T2 and F-residual limits at a 95% confidence interval as fault detection thresholds for identifying outliers. In the trajectory models, fault detection limits were set using F-residuals and model distances with a 95% confidence interval. Due to the dynamic nature of trajectory models, the limits are inherently dynamic. However, we established a static limit for outlier detection by considering only the highest value for F-residuals and Hotelling’s T2/model distances within the model. Errors or process changes were detected through signs of drift, excessive noise, or by tracking residuals/distance values over time. The models were set up to acquire and project new data every 5 min on score plots, as well as residual and Hotelling’s T2/model distances plots. Recognizing that biomanufacturing processes are comparatively slower than chemical ones, we set fault detection thresholds at 12 and 24 consecutive points outside the residual and Hotelling’s T2/model distance limits for warning and alarm alerts. These corresponded to process durations of 1 and 2 h, respectively. This approach is specifically designed to be effective, drawing on insights from monitoring real-time batches.
3.4. Model Lifecycle
Figure 6 illustrates the lifecycle of the MSPM model for biopharmaceutical manufacturing in the low-N scenario, which begins with limited historical data and progresses toward the development of the final version. Prototype MSPM models are iteratively refined by expanding the volume of real data and generating new in silico data through retraining the GPSS model. This iterative process incorporates additional data from new batches, progressively enhancing the model’s data. Once a prototype demonstrates robust performance and meets predefined sensitivity criteria, it is designated as the final version. Additionally, as sufficient batches of real data become available, subsequent models can be developed solely using real data, thereby eliminating the reliance on in silico data generated by the GPSS model. Throughout the process, regular engagement with manufacturing subject matter experts is critical to identify false positive and false negative detections, with the aim of enhancing the model’s performance and validating its accuracy as part of the continuous improvement framework.
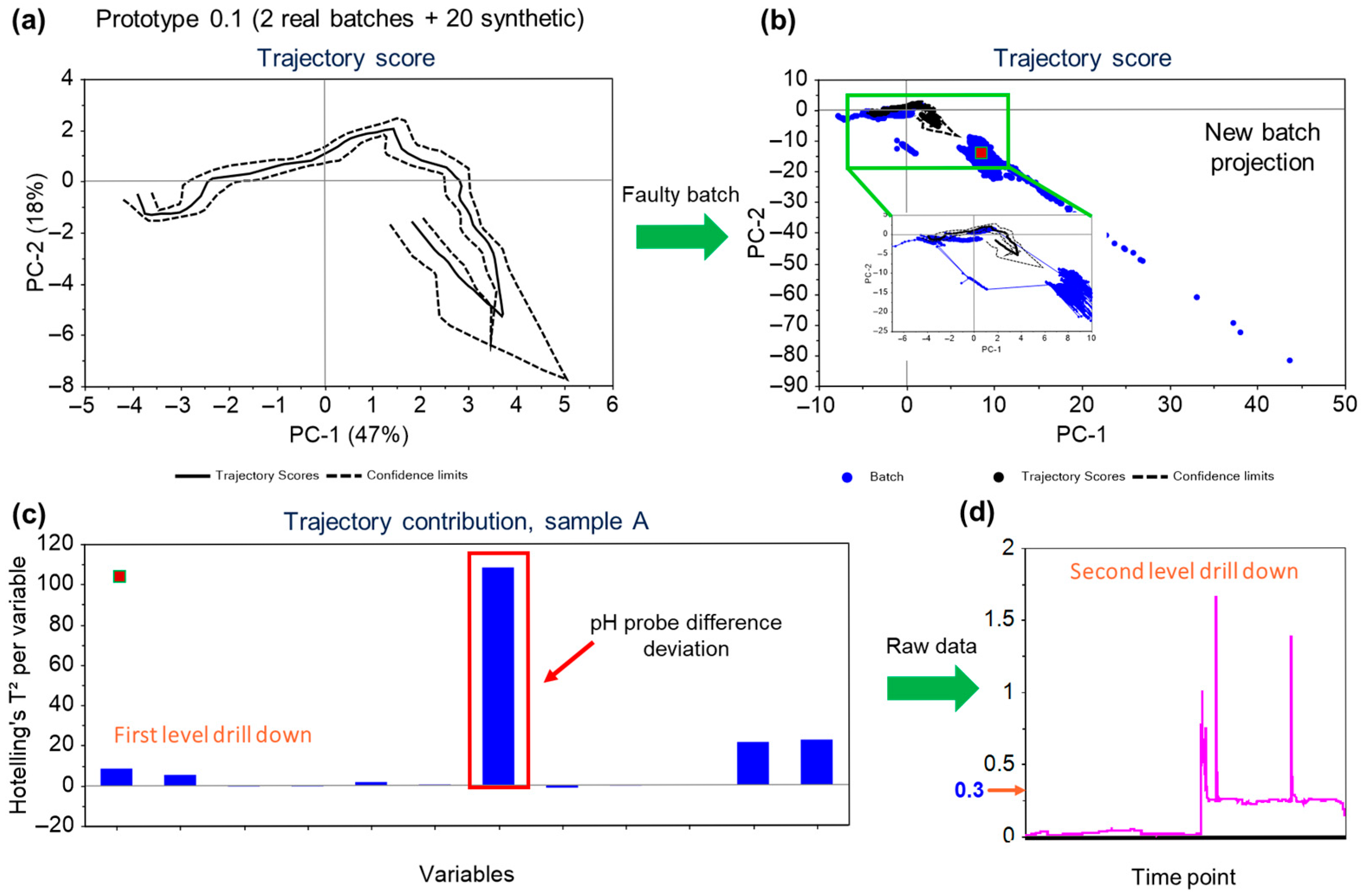
3.5. Case Study of the Application of the MSPM Model—Detection of the Drifting pH and Failure of a pH Probe
This case study covers the detection of the failure of a pH probe through the batch trajectory model. The relative functioning of the primary and secondary pH probes in the bioreactors is monitored using a variable called pH probe difference. This value for the variable is calculated by subtracting the values measured by primary and secondary pH probes fitted within the bioreactor vessel. In an ideal condition, the value for this variable should be zero, but for normal functioning, the tolerance for the accepted value for the pH probe difference variable is up to 0.07.
In the given instance, the secondary pH probe had failed and the MSPM models were able to pick up the trend in the variable, with an excursion reported on the trajectory scores (as well as on the F-residuals for the model).
Figure 7a,b illustrates the developed score plot and the score plot with the projected faulty batch, respectively. The projected batch points (blue solid points in
Figure 7b) are shown to deviate from the normal trajectory limits.
The variables contributing to the projection of the points can be confirmed from the correlations plot
Figure 7c. As seen in
Figure 7d, value for the pH probe difference variable had jumped to more than 1.5. An immediate action was taken to replace the probe, and the trend returned to the normal functioning limits. The pH of the bioreactor was not significantly affected as it is controlled using the primary probe with the secondary probe as a back-up. In this case, although the process itself was not impacted due to the secondary pH probe failure, the equipment defect was promptly addressed leveraging the MSPM model-based observations. This scenario exemplifies the crucial role of process monitoring during manufacturing operations to ensure the consistency and the robustness of the process. It not only enables swift responses to deviations in process parameters but also facilitates the early detection and resolution of equipment failures.
4. Conclusions
In this article, we demonstrate the application of the Multivariate Statistical Process Monitoring (MSPM) models in the detection and correction of atypicality within biopharmaceutical manufacturing processes during production, compared to the historical multivariate dataspace. By integrating both real and in silico data in low-N scenarios utilizing the GPSS model, we developed robust models that can monitor both static and dynamic variables effectively using the online platform. This is particularly significant as low-N scenarios are common in biopharmaceutical processes, especially during early CMC phases of development, and our successful implementation of the GPSS model emphasizes the applicability and efficacy of this approach. To effectively monitor all process parameters, we developed two models: the PCA model, which includes static variables that traditionally serve as process set points; and the trajectory model, which captures the relationships between dynamic variables that evolve throughout the batch process. The detection parameters Hotelling T2/model distances and F-residuals were used to identify the excursions. When multiple consecutive excursions were flagged going beyond the confidence limits, an alarm was triggered indicating process deviation to alert our production engineers. This enabled them to take timely corrective action and investigate the root cause, preventing potential failures and ensuring that the process remained in optimal production condition. Utilizing multivariate statistical process modelling to identify process atypicality, as opposed to relying solely on univariate trends, offers significant industrial advantages. This approach can significantly reduce costs by preventing batch failures in a preventative manner, as well as confirming equipment health on an ongoing basis, when appropriately implemented within the manufacturing setting. Furthermore, it enhances the efficiency and the productivity of the biopharmaceutical manufacturing processes. Thus, deploying this method is highly recommended during the routine production of biopharmaceuticals.