SNP rs3737883 in PPFIA4 Gene Associated with Atrial Fibrillation Risk: A Case–Control Study in a Chinese Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. SNP Genotyping
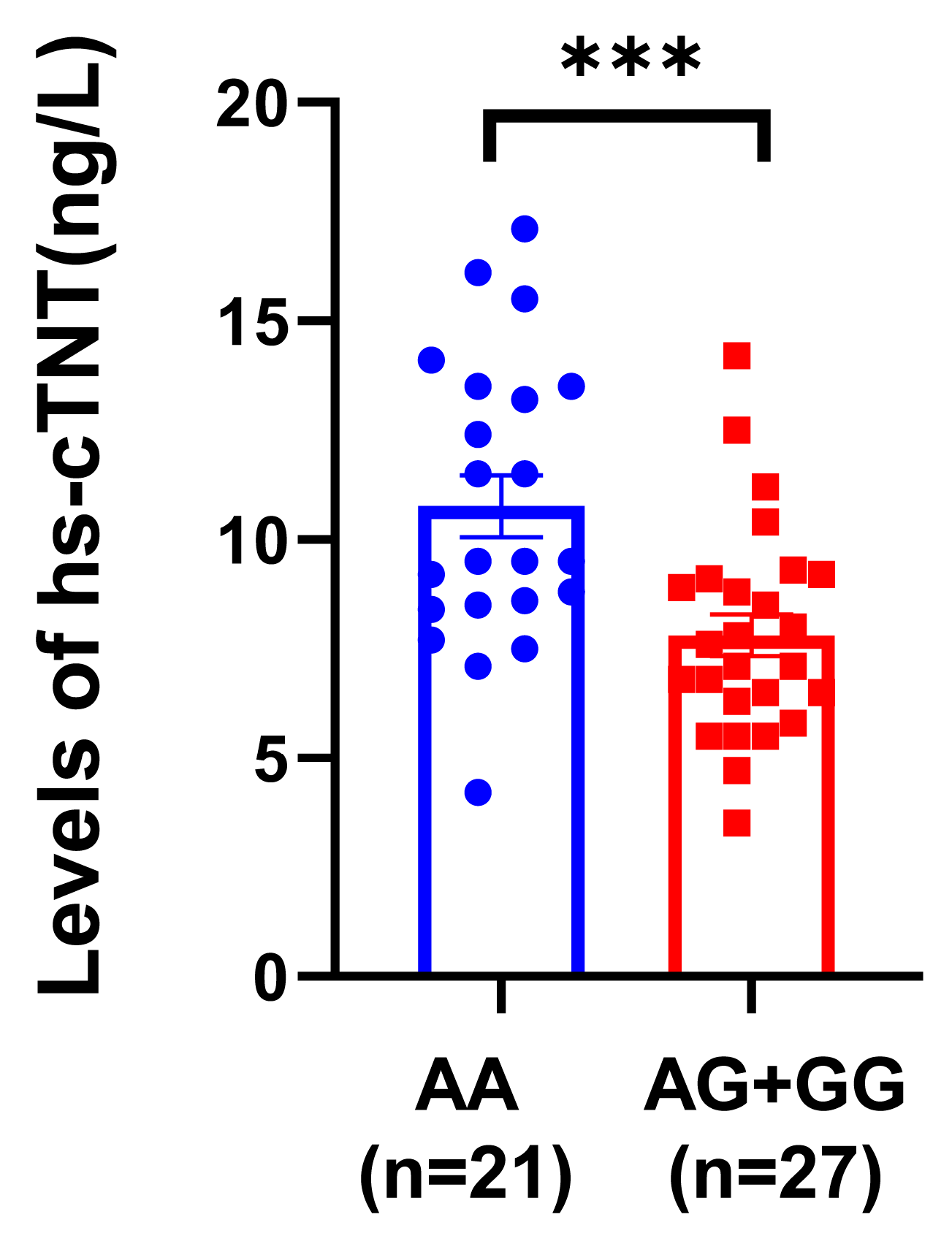
2.3. hs-cTnT Test
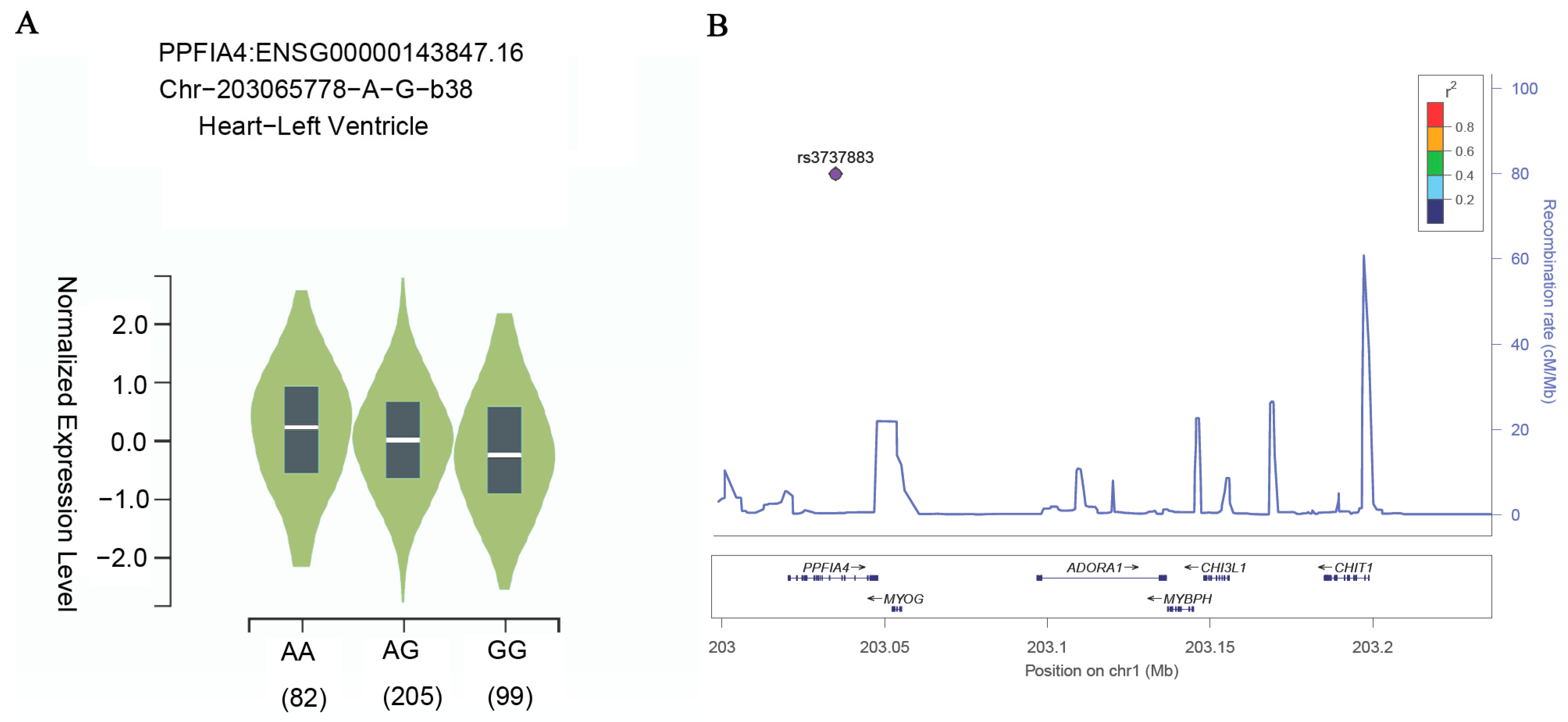
2.4. In Silico eQTL Analysis
2.5. Statistical Analysis
3. Results
3.1. Study Populations
3.2. Allele A of rs3737883 Significantly Associated with Increasing AF Risk in a Chinese Population
3.3. The Effect of rs3737883 on AF Was Larger in Sub-Group with Hypertension than Sub-Group Without Hypertension
3.4. The Risk A Allele of rs3737883 Associated with a Higher Expression Level of PPFIA4 in Left Ventricle Tissue in the Human Heart
3.5. The Risk A Allele of rs3737883 Is Associated with a Higher Serum hs-cTnT Level in AF Patients
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AF | atrial fibrillation |
| hs-cTnT | high-sensitivity cardiac troponin T |
| GWAS | Genome-wide association studies |
| eQTL | expression quantitative trait loci |
| SNP | single nucleotide polymorphism |
| OR | odds ratio |
References
- Go, A.S.; Hylek, E.M.; Phillips, K.A.; Chang, Y.; Henault, L.E.; Selby, J.V.; Singer, D.E. Prevalence of diagnosed atrial fibrillation in adults: National implications for rhythm management and stroke prevention: The AnTicoagulation and Risk Factors in Atrial Fibrillation (ATRIA) Study. JAMA 2001, 285, 2370–2375. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Ling, J.; Xiong, Q.; Chen, W.; Zou, L.; Ling, Z. Global, regional, and national burden of atrial fibrillation/flutter related to metabolic risks over three decades: Estimates from the global burden of disease study 2019. Eur. Heart J. Qual. Care Clin. Outcomes 2024, 10, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Andrade, J.G.; Jackson, L.R.; Diaz, J.C.; Chung, M.; Hurwitz, J.; Morillo, C.A.; Lip, G.Y.H.; Lopez-Cabanillas, N. Atrial fibrillation in the Americas. Lancet Reg. Health Am. 2025, 47, 101110. [Google Scholar] [CrossRef]
- Olesen, M.S.; Jespersen, T.; Nielsen, J.B.; Liang, B.; Moller, D.V.; Hedley, P.; Christiansen, M.; Varro, A.; Olesen, S.; Haunso, S.; et al. Mutations in sodium channel beta-subunit SCN3B are associated with early-onset lone atrial fibrillation. Cardiovasc. Res. 2011, 89, 786–793. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, S.; Bendahhou, S.; Wang, X.; Wang, Y.; Xu, W.; Jin, H.; Sun, H.; Su, X.; Zhuang, Q.; et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science 2003, 299, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, S.; Yoo, S.; Chakrabarti, S.; Zhang, T.; Ke, T.; Oberti, C.; Yong, S.L.; Fang, F.; Li, L.; et al. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell 2008, 135, 1017–1027. [Google Scholar] [CrossRef]
- Gudbjartsson, D.F.; Arnar, D.O.; Helgadottir, A.; Gretarsdottir, S.; Holm, H.; Sigurdsson, A.; Jonasdottir, A.; Baker, A.; Thorleifsson, G.; Kristjansson, K.; et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 2007, 448, 353–357. [Google Scholar] [CrossRef]
- Zhang, R.; Tian, X.; Gao, L.; Li, H.; Yin, X.; Dong, Y.; Yang, Y.; Xia, Y. Common Variants in the TBX5 Gene Associated with Atrial Fibrillation in a Chinese Han Population. PLoS ONE 2016, 11, e0160467. [Google Scholar] [CrossRef]
- Yuan, S.; Chen, J.; Ruan, X.; Li, Y.; Abramowitz, S.A.; Wang, L.; Jiang, F.; Xiong, Y.; Levin, M.G.; Voight, B.F.; et al. Cross-population GWAS and proteomics improve risk prediction and reveal mechanisms in atrial fibrillation. Nat. Commun. 2025, 16, 6426. [Google Scholar] [CrossRef]
- Roselli, C.; Chaffin, M.D.; Weng, L.; Aeschbacher, S.; Ahlberg, G.; Albert, C.M.; Almgren, P.; Alonso, A.; Anderson, C.D.; Aragam, K.G.; et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat. Genet. 2018, 50, 1225–1233. [Google Scholar] [CrossRef]
- Feghaly, J.; Zakka, P.; London, B.; Macrae, C.A.; Refaat, M.M. Genetics of Atrial Fibrillation. J. Am. Heart Assoc. 2018, 7, e009884. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.B.; Thorolfsdottir, R.B.; Fritsche, L.G.; Zhou, W.; Skov, M.W.; Graham, S.E.; Herron, T.J.; Mccarthy, S.; Schmidt, E.M.; Sveinbjornsson, G.; et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 2018, 50, 1234–1239. [Google Scholar] [CrossRef]
- Lee, J.; Kim, T.; Yang, P.; Lim, H.E.; Choi, E.; Shim, J.; Shin, E.; Uhm, J.; Kim, J.; Joung, B.; et al. Korean atrial fibrillation network genome-wide association study for early-onset atrial fibrillation identifies novel susceptibility loci. Eur. Heart J. 2017, 38, 2586–2594. [Google Scholar] [CrossRef]
- Shi, L.; Li, C.; Wang, C.; Xia, Y.; Wu, G.; Wang, F.; Xu, C.; Wang, P.; Li, X.; Wang, D.; et al. Assessment of association of rs2200733 on chromosome 4q25 with atrial fibrillation and ischemic stroke in a Chinese Han population. Hum. Genet. 2009, 126, 843–849. [Google Scholar] [CrossRef]
- Wang, P.; Qin, W.; Wang, P.; Huang, Y.; Liu, Y.; Zhang, R.; Li, S.; Yang, Q.; Wang, X.; Chen, F.; et al. Genomic Variants in NEURL, GJA1 and CUX2 Significantly Increase Genetic Susceptibility to Atrial Fibrillation. Sci. Rep. 2018, 8, 3297. [Google Scholar] [CrossRef]
- Li, C.; Wang, F.; Yang, Y.; Fu, F.; Xu, C.; Shi, L.; Li, S.; Xia, Y.; Wu, G.; Cheng, X.; et al. Significant association of SNP rs2106261 in the ZFHX3 gene with atrial fibrillation in a Chinese Han GeneID population. Hum. Genet. 2011, 129, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, C.; Wang, X.; Xu, C.; Wu, M.; Wang, P.; Tu, X.; Wang, Q.K. Significant Association Between CAV1 Variant rs3807989 on 7p31 and Atrial Fibrillation in a Chinese Han Population. J. Am. Heart Assoc. 2015, 4, e001980. [Google Scholar] [CrossRef] [PubMed]
- Little, J.; Higgins, J.P.T.; Ioannidis, J.P.A.; Moher, D.; Gagnon, F.; Von Elm, E.; Khoury, M.J.; Cohen, B.; Davey-Smith, G.; Grimshaw, J.; et al. STrengthening the REporting of genetic association studies (STREGA)—An extension of the STROBE statement. Genet. Epidemiol. 2009, 33, 581–598. [Google Scholar] [CrossRef]
- January, C.T.; Wann, L.S.; Calkins, H.; Chen, L.Y.; Cigarroa, J.E.; Cleveland, J.C.J.; Ellinor, P.T.; Ezekowitz, M.D.; Field, M.E.; Furie, K.L.; et al. 2019 AHA/ACC/HRS Focused Update of the 2014 AHA/ACC/HRS Guideline for the Management of Patients With Atrial Fibrillation: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. J. Am. Coll. Cardiol. 2019, 74, 104–132. [Google Scholar] [CrossRef]
- Miyazawa, K.; Ito, K.; Ito, M.; Zou, Z.; Kubota, M.; Nomura, S.; Matsunaga, H.; Koyama, S.; Ieki, H.; Akiyama, M.; et al. Cross-ancestry genome-wide analysis of atrial fibrillation unveils disease biology and enables cardioembolic risk prediction. Nat. Genet. 2023, 55, 187–197. [Google Scholar] [CrossRef]
- Okamura, S.; Ochi, H.; Onohara, Y.; Nakashima, M.; Akiyama, R.; Tokuyama, T.; Okubo, Y.; Ikeuchi, Y.; Miyauchi, S.; Miyamoto, S.; et al. GJA1 gene polymorphism is a genetic predictor of recurrence after pulmonary vein isolation in patients with paroxysmal atrial fibrillation. Heart Rhythm. 2022, 19, 2044–2050. [Google Scholar] [CrossRef]
- Yang, Y.; Bartz, T.M.; Brown, M.R.; Guo, X.; Zilhao, N.R.; Trompet, S.; Weiss, S.; Yao, J.; Brody, J.A.; Defilippi, C.R.; et al. Identification of Functional Genetic Determinants of Cardiac Troponin T and I in a Multiethnic Population and Causal Associations With Atrial Fibrillation. Circ.-Genom. Precis. Med. 2021, 14, e003460. [Google Scholar] [CrossRef]
- Ahlberg, G.; Andreasen, L.; Ghouse, J.; Bertelsen, L.; Bundgaard, H.; Haunso, S.; Svendsen, J.H.; Olesen, M.S. Genome-wide association study identifies 18 novel loci associated with left atrial volume and function. Eur. Heart J. 2021, 42, 4523–4534. [Google Scholar] [CrossRef]
- Machado, R.A.; Nogueira, E.N.; Martelli-Junior, H.; Reis, S.R.; Persuhn, D.C.; Coletta, R.D. 2p24.2 (rs7552) is a susceptibility locus for nonsyndromic cleft lip with or without cleft palate in the Brazilian population. Clin. Genet. 2018, 93, 1199–1204. [Google Scholar] [CrossRef]
- Zimmerman, R.M.; Hernandez, E.J.; Tristani-Firouzi, M.; Yandell, M.; Steinberg, B.A. Explainable artificial intelligence for stroke risk stratification in atrial fibrillation. Eur. Heart J. Digit. Health 2025, 6, 317–325. [Google Scholar] [CrossRef]
- van Ouwerkerk, A.F.; Bosada, F.M.; van Duijvenboden, K.; Hill, M.C.; Montefiori, L.E.; Scholman, K.T.; Liu, J.; de Vries, A.A.F.; Boukens, B.J.; Ellinor, P.T.; et al. Identification of atrial fibrillation associated genes and functional non-coding variants. Nat. Commun. 2019, 10, 4755. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Jurgens, S.J.; Xiao, L.; Hill, M.C.; Haggerty, C.M.; Sveinbjornsson, G.; Morrill, V.N.; Marston, N.A.; Weng, L.; Pirruccello, J.P.; et al. Sequencing in over 50,000 cases identifies coding and structural variation underlying atrial fibrillation risk. Nat. Genet. 2025, 57, 548–562. [Google Scholar] [CrossRef] [PubMed]
- Christophersen, I.E.; Rienstra, M.; Roselli, C.; Yin, X.; Geelhoed, B.; Barnard, J.; Lin, H.; Arking, D.E.; Smith, A.V.; Albert, C.M.; et al. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat. Genet. 2017, 49, 946–952. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.; Gore-Panter, S.; Tchou, G.; Castel, L.; Lovano, B.; Moravec, C.S.; Pettersson, G.B.; Roselli, E.E.; Gillinov, A.M.; Mccurry, K.R.; et al. Genetic Control of Left Atrial Gene Expression Yields Insights into the Genetic Susceptibility for Atrial Fibrillation. Circ. Genom. Precis. Med. 2018, 11, e002107. [Google Scholar] [CrossRef]
- Parwani, A.S.; Boldt, L. Atrial fibrillation-induced cardiac troponin I release. Int. J. Cardiol. 2014, 172, 220. [Google Scholar] [CrossRef]
- Costabel, J.P.; Urdapilleta, M.; Lambardi, F.; Campos, R.; Vergara, J.M.; Ariznavarreta, P.; Trivi, M. High-Sensitivity Cardiac Troponin Levels in Supraventricular Tachyarrhythmias. PACE Pacing Clin. Electrophysiol. 2016, 39, 588–591. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Zhao, X.; Li, X.; Li, N.; Hu, X. Cardiac troponin and adverse outcomes in atrial fibrillation: A meta-analysis. Clin. Chim. Acta. 2018, 477, 48–52. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | AF Cases (n = 724) | AF Controls (n = 1475) | p |
|---|---|---|---|
| Age (Years, Mean ± SD) a | 57.41 ± 9.64 | 59.63 ± 10.45 | 1.65 × 10−6 |
| Gender, Female, n (%) | 307 (42.4%) | 685 (46.4%) | 0.08 |
| Diabetes, n (%) | 147 | 326 | 0.33 |
| Hypertension, n (%) | 273 | 612 | 0.09 |
| Lone AF n (%) | 310 | 0 | n.a. |
| Cohort (n, Case/Control) | Risk Allele | Risk Allele Frequency | Without Adjustment a | With Adjustment b | ||
|---|---|---|---|---|---|---|
| p-obs | OR (95% CI) | p-adj | OR (95% CI) | |||
| Total Cohort (724/1475) | A | 0.70/0.60 | 5.61 × 10−11 | 1.34 (1.22–1.47) | 2.83 × 10−11 | 1.33 (1.22–1.48) |
| Male (417/790) | A | 0.69/0.59 | 9.58 × 10−8 | 1.38 (1.21–1.54) | 1.24 × 10−7 | 1.37 (1.22–1.55) |
| Female (307/685) | A | 0.71/0.61 | 8.73 × 10−5 | 1.31 (1.14–1.51) | 3.44 × 10−5 | 1.30 (1.13–1.50) |
| Hypertension (273/612) | A | 0.71/0.57 | 1.24 × 10−8 | 1.49 (1.29–1.73) | 5.57 × 10−8 | 1.48 (1.27–1.71) |
| Without Hypertension 451/863 | A | 0.70/0.62 | 2.05 × 10−4 | 1.24 (1.10–1.39) | 3.21 × 10−4 | 1.23 (1.10–1.38) |
| Diabetes (147/326) | A | 0.74/0.60 | 1.57 × 10−5 | 1.29 (1.17–1.43) | 6.63 × 10−5 | 1.30 (1.17–1.42) |
| Without Diabetes (577/1149) | A | 0.70/0.60 | 2.27 × 10−7 | 1.57 (1.26–1.95) | 5.14 × 10−7 | 1.55 (1.21–1.92) |
| Lone AF (310/1475) | A | 0.70/0.60 | 3.45 × 10−6 | 1.33 (1.17–1.51) | 6.77 × 10−6 | 1.32 (1.16–1.50) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuo, J.; Wang, P.; Xu, C. SNP rs3737883 in PPFIA4 Gene Associated with Atrial Fibrillation Risk: A Case–Control Study in a Chinese Population. LabMed 2025, 2, 18. https://doi.org/10.3390/labmed2040018
Zhuo J, Wang P, Xu C. SNP rs3737883 in PPFIA4 Gene Associated with Atrial Fibrillation Risk: A Case–Control Study in a Chinese Population. LabMed. 2025; 2(4):18. https://doi.org/10.3390/labmed2040018
Chicago/Turabian StyleZhuo, Jiahui, Pengyun Wang, and Chengqi Xu. 2025. "SNP rs3737883 in PPFIA4 Gene Associated with Atrial Fibrillation Risk: A Case–Control Study in a Chinese Population" LabMed 2, no. 4: 18. https://doi.org/10.3390/labmed2040018
APA StyleZhuo, J., Wang, P., & Xu, C. (2025). SNP rs3737883 in PPFIA4 Gene Associated with Atrial Fibrillation Risk: A Case–Control Study in a Chinese Population. LabMed, 2(4), 18. https://doi.org/10.3390/labmed2040018






