Abstract
This study included deidentified antibiotic susceptibility results from outpatient urinary Escherichia coli isolates from Washington state which were tested at a large clinical laboratory during 2013–2019. Isolates were categorized as representing the first, second, third, or fourth-or-greater occurrence of infection in data from individual patients. We used logistic regression with the outcome of resistance, adjusting for year of antimicrobial susceptibility test, patient sex, patient age, and facility type. In cases of subsequent infection, we found a significant risk of resistance to levofloxacin, ciprofloxacin, ceftriaxone, trimethoprim-sulfa, nitrofurantoin, ampicillin, gentamicin, and amoxicillin-clavulanate. Our findings suggest that Escherichia coli isolates from recurrent urinary tract infections have a higher rate of resistance to most tested antibiotics than isolates from the first urinary tract infection in a given year. However, susceptibility frequencies did not differ significantly between antibiograms constructed using only the first occurrence in a patient and those constructed using all subsequent occurrences. These findings suggest that the traditional approach of including only the first occurrence of urinary Escherichia coli in a patient may underestimate levels of antibiotic resistance in a community. Such underestimation could negatively affect empiric therapeutic choices, health outcomes, and treatment costs.
1. Introduction
Clinicians routinely use cumulative antibiograms in selecting empirical antimicrobial therapies. As noted by Abbo and Hooton, treatment for uncomplicated urinary tract infections (UTIs) should be empiric and guided by local antimicrobial resistance patterns [1]. Clinical and Laboratory Standards Institute (CLSI) guidelines recommend that these antibiograms analyze data yearly and include only final, verified results as follows: bacteria species with ≥30 isolates; and the first isolate only for a species/patient/analysis period [2].
UTIs are more common in women than men, and represent one of the most frequent causes of outpatient visits [3]. Recurrence can be a significant problem among patients with UTIs, with reported recurrence rates of 30–50% in women in the first year; in addition, the risk increases with age [1,4].
The most common pathogen causing UTIs in American adults is the Gram-negative bacterium Escherichia coli (EC), which is known to acquire resistance to multiple antibiotics and increase the likelihood of a recurrent UTI [5,6]. An expert consensus statement on recurrent UTIs recommended that “Clinicians should use first-line therapy (i.e., nitrofurantoin, trimethoprim-sulfamethoxazole, fosfomycin) dependent on the local antibiogram for the treatment of symptomatic UTIs in women (Strong Recommendation; Evidence Level: Grade B) [7]”. This statement demonstrates the importance of local antibiograms providing accurate information. There is agreement that antibiograms are a useful tool for improving antibiotic use, appropriateness, and costs [8]. However, there are multiple ways to present data outside of the traditional antibiogram that provide useful information for empirical treatments such as combination antibiograms and syndromic antibiograms [9]. The question of how to address multiple isolates from an individual patient when constructing a cumulative antibiogram has sparked some debate [10]. Although the recommendation from authorities such as CLSI is to remove repeat isolates to prevent the antibiogram presenting a greater percentage of resistance [11], this approach may underestimate the actual prevalence of resistance to pathogens that tend to cause recurrent infections, such as EC. Previous studies have identified common risk factors for multidrug-resistant urinary tract infections, with the most prevalent of these factors being previous antibiotic usage, followed by urinary catheterization, previous hospitalization and being a nursing home resident [12]. To explore the potential impact of excluding results from recurrent UTIs on antibiogram accuracy, we used a large set of outpatient test result data from a commercial reference laboratory to assess differences in resistance patterns between first and subsequent isolates from patients.
2. Materials and Methods
We performed a retrospective cross-sectional analysis of antibiotic susceptibility test result data from a commercial reference clinical laboratory (Quest Diagnostics, Secaucus, NJ, USA). EC isolates were collected from urinary sources in outpatient settings during 2013–2019 in Washington State, i.e., over a seven-year period. Under a previously described academic–corporate research agreement [13], these data were made available to the University of Washington without personal identifiers. We restricted the analysis to only urinary tract infections (based on ICD codes) with EC isolates. Those with missing patient IDs and whose gender or age was inconsistent across the study period were excluded from the analysis; those excluded constituted less than 10% of the data. Unique patient IDs were used to categorize patients into bins of 1st occurrence, 2nd occurrence, 3rd occurrence, and 4th-or-greater occurrence of EC UTI by year. The 10 antibiotics in this analysis were chosen based on the frequency of their use in outpatient settings, to represent resistance within the local community.
2.1. Creation of Antibiograms and Comparison of First-Only and All Isolate Antibiograms
Standard antibiograms show the proportion of isolates that are susceptible for a specific bacterium–antibiotic combination. Breakpoints are determined using minimum inhibitory concentration results and CLSI interpretation. For this study, susceptibility was calculated as the total number of susceptible isolates divided by the total number tested for a specific bacterium–antibiotic combination in each year. Antibiograms were created in the standard CLSI format [2], with inclusion of AST results from recurrent UTIs in individual patients. We then compared antibiograms generated using only first-occurrence UTIs in individual patients in a given year with antibiograms that also included second, third, and fourth-or-greater occurrences.
2.2. Logistic Regression Model
We performed a logistic regression analysis using a binary outcome for resistance and looked at the association of resistance and patient occurrence in the data across 10 antibiotics of interest: ampicillin (AMP), amoxicillin-clavulanate (AML), ciprofloxacin (CIP), nitrofurantoin (NIT), trimethoprim-sulfa (SXT), ceftriaxone (CRO), gentamicin (GEN), ertapenem (ERT), imipenem (IMP) and levofloxacin (LVO). All models were adjusted for sex, age in years, the type of facility from where the sample was collected, and the year of testing. We were unable to adjust for additional confounders such as patient comorbidities and prescription patterns due to lack of data.
3. Results
3.1. Multiple Occurrences of Infections in Patients
During the 7-year study period, 40,865 (38,202 women; 93%) patients had EC isolated from urine cultures. Of the 54,584 total urinary EC infections in these patients across all years, 46,920 (86%) were first infections, 5656 (10%) were second infections, 1324 (2%) were third infections, and 684 (1%) were fourth-or-greater infections. The proportion of females varied slightly across bins: 94% for the first-occurrence group, 93% for both the second- and third-occurrence groups, and 88% for the fourth-or-greater-occurrence group. The average female age varied across the occurrence bins (p < 2 × 10−16): 48 years for the first-occurrence group, 53 years for the second-occurrence group, 56 years for the third-occurrence group, and 58 years for the fourth-or-greater-occurrence group. Compared with females, males tended to have a higher average age (p < 2 × 10−16) across occurrence bins: 58 years for first occurrence, 61 years for second, 65 years for third, and 60 years for fourth or greater (Table 1).

Table 1.
Patient demographics from all years (2013–2019). Columns indicate the number of times a patient occurred in the data for 1st, 2nd, 3rd, and 4th+ occurrences, with “All” including all patient occurrences grouped together.
3.2. Antibiogram Comparison
Looking at raw, unadjusted counts, for all antibiotics, the proportion of susceptible isolates tended to decrease from the first occurrence to the fourth-or-greater occurrence (Table 2). However, the proportion of susceptible isolates did not differ significantly between the first occurrence and all occurrences combined.

Table 2.
2013–2019 urinary E. coli antibiogram across patient occurrences (All, 1st, 2nd, 3rd, and 4th or greater).
3.3. Logistic Regression
A logistic regression analysis using a binary outcome for resistance and prediction of patient reoccurrence was performed for the 10 antibiotics of interest, controlling for year of antimicrobial susceptibility test, patient sex, patient age, and facility type. For all antibiotics except ertapenem and imipenem, the odds of resistance increased significantly with subsequent UTI occurrences following the first occurrence (Table 3).

Table 3.
Odds ratios (OR) of antimicrobial resistance for each subsequent urinary Escherichia coli isolate occurrence after the first.
4. Discussion
Our results suggest that isolates from subsequent UTIs in the same patient had increased odds of resistance to the antibiotics of interest, with effects that varied by antibiotic class. This finding highlights the limitations of standard antibiogram reporting practice. Including only the first urinary EC from a single patient per year may underestimate levels of reservoir resistance in a community among certain patients. While our analysis did not find a significant impact on the antibiogram percent susceptibility by including subsequent infections, there may be advantages to including all isolates in an antibiogram. This approach provides a truer estimate of resistance in a population and avoids the risk of underestimation of resistance, which could impact the effectiveness of empiric therapeutic choices and healthcare outcomes based on the antibiogram being used. Isolates in our study that would be excluded from a standard CLSI format for antibiograms (n = 7664) were from patients who tended to be older (Table 1) and were more resistant (Figure 1) than first-occurrence patients. It may be worthwhile to consider whether these excluded antimicrobial susceptibility test results might provide insights into the level of bacterial resistance in the community. Their inclusion may help clinicians select the most appropriate antibiotics for the treatment of reoccurring UTIs.
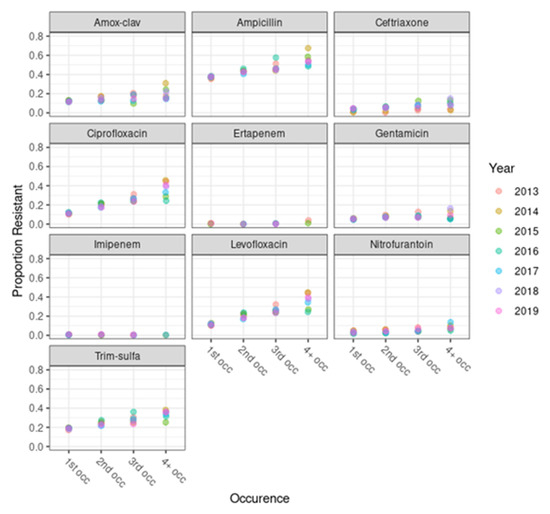
Figure 1.
Proportion of antimicrobial resistant urinary Escherichia coli isolates across patient occurrence categories, by antibiotic. The x-axis indicates the four occurrence bins of patients in the data: 1st, 2nd, 3rd, or 4th+ occurrence. The y-axis indicates the proportion of isolates with resistance in each year, with different years represented by different colors, as shown in the key.
We found significant differences in resistance between the different occurrence groups of EC UTI patients, with isolates from individuals with multiple UTI occurrences having decreased antibiotic susceptibility. An isolate’s odds of resistance increased significantly with each increase in a patient’s number of occurrences in the data for all antibiotics except ertapenem and imipenem. This increase in resistance was more pronounced for quinolones (e.g., ciprofloxacin and levofloxacin) than for nitrofurantoin.
One of the simplest explanations for the increase in resistance with subsequent UTI occurrences is the presumed repeated administration of antibiotics. In a recent systematic review, prior antibiotic use was identified as a major risk factor for resistance for community acquired infections with extended spectrum beta-lactamase (EBSL)-producing EC [14]. It is logical that resistance to other drugs would be similarly affected. The same review found that previous hospitalization and UTI history were also risk factors for ESBL UTIs [14].
The increase, with each subsequent infection, in the likelihood of resistance to multiple agents should perhaps not come as a surprise: changes in the global epidemiology of UTI have been driven by the emergence of multi-drug-resistant (MDR) EC strains such as Sequence Type (ST) 131 [15], which encode resistance to the fluoroquinolones (ciprofloxacin and levofloxacin) on their chromosomes while also carrying accessory plasmids that may encode multiple determinants of resistance, including those against ampicillin, gentamicin, trimethoprim-sulfa, and even ceftriaxone [16]. While emergence of MDR EC strains in general may increase chances of first-line treatment failure in any UTI patient, EC ST131 specifically may be prone to produce recurrent UTI independent of antibiotic resistance traits [17].
Because the standard laboratory antibiogram is issued with an annual cadence, longitudinal trends such as the global spread of EC ST131 cannot be represented beyond their impact on the index isolates among the preceding year’s infectious episodes. Even without reporting on longitudinal trends, creation of an annual UTI syndrome-specific antibiogram that includes stratification by subsequent infection might alert clinicians and antibiotic stewards to the presence in their communities of a reservoir of strains to which multi-drug resistance has been genetically linked.
Our study has several limitations. Our analysis assumes that all of a patient’s antibiotic susceptibility test results were captured within the Quest data system. However, the “first” occurrence represents only the first occurrence in the data for that associated year and may not be the patients’ actual first EC UTI. By classifying UTIs based on ICD codes, a diagnosis is dependent on the provider and reliance on ICD codes alone may introduce misclassification errors. There is limited external validation, as this analysis assumes that all relevant antibiotic susceptibility tests are captured within the dataset; however, this might not be a true reflection of the broader population. We did not have the clinical data available to know if the antibiotic treatment of patients in this dataset was empirical or based on antibiograms. Another limitation is the lack of data on other potential confounders, such as rural versus urban populations, socioeconomic status, and insurance type. By not controlling for these confounders, the association between patient occurrence and resistance may be biased. Our data set did not include clinical history data; therefore, we were unable to assess the effects of other host factors on resistance [5]. In future research, it would be beneficial to understand the financial burden of underestimating resistance such as higher treatment costs, extended hospital stays, and increased testing. It would also be helpful to evaluate the real-world impact of revised antibiogram methodologies in the context of health and economic outcomes.
While our analysis did not find a significant impact on overall antibiogram susceptibility frequencies, there is potential for underestimation of resistance which could impact accuracy of empiric therapeutic choices, healthcare outcomes, and treatment costs.
The adoption of antibiograms that include recurrent isolates requires several logistical and policy changes to ensure effective implementation, with key considerations including the following: (1) A standardized method for collecting and recording isolate data should be implemented across different departments or facilities. This includes ensuring that the identification of recurrent isolates is consistent and accurately documented in electronic health records (EHRs). Laboratory databases should be integrated with clinical databases to allow for efficient tracking of recurrent infections and their susceptibility patterns. (2) Formal policies should be developed that define what constitutes a recurrent isolate and outline how they should be included in antibiograms. This may involve reviewing definitions from established guidelines and organizations. Policies that encourage laboratories to routinely analyze data on recurrent isolates, including frequency and susceptibility trends, should be implemented. (3) Training for laboratory staff and clinicians on the importance of recurrent isolates and how they can influence treatment strategies should be provided. This will help ensure that staff understand the relevance of including these isolates in antibiograms. Educational materials should be created for healthcare providers that explain how changes to antibiogram reporting will affect patient management. (4) Collaboration should be fostered between microbiologists, infectious disease specialists, pharmacists, and epidemiologists to ensure that all perspectives are considered in the development and utilization of revised antibiograms. Multidisciplinary committees should be established to review and interpret data related to recurrent isolates, providing insights that can guide antimicrobial stewardship efforts. (5) If necessary, laboratory information systems (LISs) should be implemented or updated to handle recurrent isolate data appropriately and generate antibiograms that reflect this information. Analytic tools that can assist in identifying patterns in recurrent infections and their susceptibility should be developed to facilitate better decision-making. (6) Feedback loops should be created where clinicians receive regular updates on antibiogram data, emphasizing the importance of recurrent isolates. Healthcare providers should be encouraged to use the revised antibiograms in their empirical treatment recommendations and provide evidence of its impact on patient outcomes. (7) Lastly, quality assurance protocols should be developed to regularly assess the accuracy and completeness of data included in the antibiograms, ensuring that recurrent isolates are not overlooked. The impact of the new antibiogram approach on clinical outcomes and antibiotic resistance patterns should be monitored, making adjustments based on clinical feedback and new evidence.
Any increased complexity in developing and interpreting antibiograms can hinder their usefulness in clinical settings and, in turn, make it difficult for clinicians to make informed decisions regarding antibiotic therapy. To combat this barrier, healthcare professionals may need additional education on how to utilize these antibiograms effectively, and how to interpret the data and integrate it into their clinical practice. Even with this training, variability in local resistance patterns means that there is also a need for continuous updating of antibiograms that can complicate the tools’ relevance and application in various clinical scenarios. Resource limitations, such as time constraints and availability of laboratory support, can impact the implementation and use of antibiograms. In addition to resource limitations, a lack of standardized protocols for constructing and applying antibiograms across different institutions may lead to inconsistencies and errors in prescribing.
Including recurrent isolates in discussions about antibiotic guidelines can help improve treatment strategies but may also highlight problems with current practices. On the one hand, considering these recurrent isolates fits well with antibiotic stewardship goals. It encourages doctors to tailor treatments specifically for these recurrent infections. By focusing on effective management, it can reduce the use of broad-spectrum antibiotics and help preserve healthy bacteria. On the other hand, the incidence of recurrent isolates may challenge existing guidelines and show that standard treatments are not sufficient for certain patients. If these infections arise from resistant strains, or if initial treatments are inadequate, revisions in guidelines should be considered. This could mean using better diagnostic methods, applying targeted treatments, or exploring alternative options. Dealing with recurrent isolates requires collaboration among healthcare experts, including infectious-disease specialists and pharmacists. This teamwork can ensure that stewardship programs adapt to new resistance patterns and meet patient needs effectively. In summary, considering recurrent isolates in antibiotic stewardship highlights the need for personalized care and suggests that current guidelines be reviewed so that complex infections are better addressed.
Author Contributions
Conceptualization, L.F., A.E.S., S.J.W., H.K. and P.R.; methodology, L.F., A.E.S., S.J.W., H.K. and P.R.; formal analysis, L.F.; resources, P.R.; data curation, L.F., A.E.S., H.K. and P.R.; writing—original draft preparation, L.F., A.E.S., J.R., S.J.W., H.K. and P.R.; writing—review and editing, L.F., A.E.S., J.R., S.J.W., H.K. and P.R.; visualization, L.F.; supervision, A.E.S., H.K. and P.R.; funding acquisition, H.K and P.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
All study protocols were reviewed and approved by the University of Washington’s Institutional Review Board (STUDY00000042: GAZER, approved date: 4 August 2016).
Informed Consent Statement
Informed consent was not required as patient-level data were deidentified.
Data Availability Statement
The data that support the findings are not publicly available.
Acknowledgments
This work was supported by a Memorandum of Understanding/Data Use agreement between Quest Diagnostics Clinical Laboratories Inc. and the University of Washington.
Conflicts of Interest
S.J.W. reports consulting for, and having equity in MicrobiomX, as well as a patent pending for a point-of-care fecal transplant device licensed to MicrobiomX. A.S. and J.R. are, respectively, current and previous employees of Quest Diagnostics, and both own stock in the company. All other authors report no potential conflicts.
Abbreviations
The following abbreviations are used in this manuscript:
| UTI | Urinary tract infection |
| EC | Escherichia coli |
| ESBL | Extended spectrum beta-lactamase |
| AMP | Ampicillin |
| AML | Amoxicillin-clavulanate |
| CIP | Ciprofloxacin |
| NIT | Nitrofurantoin |
| SXT | Trimethoprim-sulfa |
| CRO | Ceftriaxone |
| GEN | Gentamicin |
| ERT | Ertapenem |
| IMP | Imipenem |
| LVO | Levofloxacin |
References
- Abbo, L.M.; Hooton, T.M. Antimicrobial stewardship and urinary tract infections. Antibiotics 2014, 3, 174–192. [Google Scholar] [CrossRef] [PubMed]
- Clinical & Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2018; p. 296. [Google Scholar]
- Foxman, B. Epidemiology of urinary tract infections: Incidence, morbidity, and economic costs. Dis. Mon. 2003, 49, 53–70. [Google Scholar] [CrossRef] [PubMed]
- Nosseir, S.B.; Lind, L.R.; Winkler, H.A. Recurrent uncomplicated urinary tract infections in women: A review. J. Women’s Health 2012, 21, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Ku, J.H.; Bruxvoort, K.J.; Salas, S.B.; Varley, C.D.; Casey, J.A.; Raphael, E.; Robinson, S.C.; Nachman, K.E.; Lewin, B.J.; Contreras, R.; et al. Multidrug Resistance of Escherichia coli From Outpatient Uncomplicated Urinary Tract Infections in a Large United States Integrated Healthcare Organization. Open Forum Infect. Dis. 2023, 10, ofad287. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B. Urinary Tract Infection Syndromes. Infect. Dis. Clin. North Am. 2014, 28, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Anger, J.; Lee, U.; Ackerman, A.L.; Chou, R.; Chughtai, B.; Clemens, J.Q.; Hickling, D.; Kapoor, A.; Kenton, K.S.; Kaufman, M.R.; et al. Recurrent uncomplicated urinary tract infections in women: AUA/CUA/SUFU guideline. J. Urol. 2019, 202, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Khatri, D.; Freeman, C.; Falconer, N.; de Camargo Catapan, S.; Gray, L.C.; Paterson, D.L. Clinical impact of antibiograms as an intervention to optimize antimicrobial prescribing and patient outcomes—A systematic review. Am. J. Infect. Control 2024, 52, 107–122. [Google Scholar] [CrossRef] [PubMed]
- Klinker, K.P.; Hidayat, L.K.; DeRyke, C.A.; DePestel, D.D.; Motyl, M.; Bauer, K.A. Antimicrobial stewardship and antibiograms: Importance of moving beyond traditional antibiograms. Ther. Adv. Infect. Dis. 2021, 8, 20499361211011373. [Google Scholar] [CrossRef] [PubMed]
- Hindler, J.F.; Stelling, J. Analysis and Presentation of Cumulative Antibiograms: A New Consensus Guideline from the Clinical and Laboratory Standards Institute. Clin. Infect. Dis. 2007, 44, 867–873. [Google Scholar] [CrossRef] [PubMed]
- Simner, P.J.; Hindler, J.A.; Bhowmick, T.; Das, S.; Johnson, J.K.; Lubers, B.V.; Redell, M.A.; Stelling, J.; Erdman, S.M. What’s New in Antibiograms? Updating CLSI M39 Guidance with Current Trends. J. Clin. Microbiol. 2022, 60, e0221021. [Google Scholar] [CrossRef] [PubMed]
- Tenney, J.; Hudson, N.; Alnifaidy, H.; Li, J.T.C.; Fung, K.H. Risk factors for acquiring multidrug-resistant organisms in urinary tract infections: A systematic literature review. Saudi Pharm. J. 2018, 26, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Frisbie, L.; Weissman, S.J.; Kapoor, H.; D’Angeli, M.; Salm, A.; Radcliff, J.; Rabinowitz, P. Antimicrobial Resistance Patterns of Urinary Escherichia coli Among Outpatients in Washington State, 2013–2017: Associations With Age and Sex. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2021, 73, 1066–1074. [Google Scholar] [CrossRef] [PubMed]
- Larramendy, S.; Deglaire, V.; Dusollier, P.; Fournier, J.-P.; Caillon, J.; Beaudeau, F.; Moret, L. Risk Factors of Extended-Spectrum Beta-Lactamases-Producing Escherichia coli Community Acquired Urinary Tract Infections: A Systematic Review. Infect. Drug Resist. 2020, 13, 3945–3955. [Google Scholar] [CrossRef] [PubMed]
- Nicolas-Chanoine, M.H.; Blanco, J.; Leflon-Guibout, V.; Demarty, R.; Alonso, M.P.; Canica, M.M.; Park, Y.J.; Lavigne, J.P.; Pitout, J.; Johnson, J.R. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2008, 61, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D.; Chen, L. The significance of epidemic plasmids on the success of multidrug-resistant drug pandemic extraintestinal pathogenic Escherichia coli. Infect. Dis. Ther. 2023, 12, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Lindblom, A.; Kiszakiewicz, C.; Kristiansson, E.; Yazdanshenas, S.; Kamenska, N.; Karami, N.; Ahren, C. The impact of the ST131 clone on recurrent ESBL-producing E. coli urinary tract infection: A prospective comparative study. Sci. Rep. 2022, 12, 10048. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).