Abstract
Skin cancer is one of the most prevalent and potentially lethal cancers worldwide, highlighting the need for accurate and timely diagnosis. Convolutional neural networks (CNNs) have demonstrated strong potential in automating skin lesion classification. In this study, we propose a multi-class classification model using EfficientNet-B0, a lightweight yet powerful CNN architecture, trained on the HAM10000 dermoscopic image dataset. All images were resized to 224 × 224 pixels and normalized using ImageNet statistics to ensure compatibility with the pre-trained network. Data augmentation and preprocessing addressed class imbalance, resulting in a balanced dataset of 7512 images across seven diagnostic categories. The baseline model achieved 77.39% accuracy, which improved to 89.36% with transfer learning by freezing the convolutional base and training only the classification layer. Full network fine-tuning with test-time augmentation increased the accuracy to 96%, and the final model reached 97.15% when combined with Monte Carlo dropout. These results demonstrate EfficientNet-B0’s effectiveness for automated skin lesion classification and its potential as a clinical decision support tool.
1. Introduction
Skin cancer continues to be one of the most widespread and life-threatening types of cancer globally, making early and precise diagnosis essential for effective treatment. Advancements in medical image analysis have increasingly turned to deep learning techniques, offering improved accuracy and efficiency in the detection and classification of various skin cancer types. Ashfaq et al. [1] proposed ‘DermaVision’, a deep learning-based platform for precise skin cancer diagnosis and classification, highlighting the potential of DL models in real-time healthcare applications. Similarly, Kavitha et al. [2] implemented deep learning techniques to detect and classify skin cancer using various convolutional neural networks (CNNs), showing high precision on benchmark datasets. Naeem et al. [3] provided a comprehensive overview of malignant melanoma classification using DL, analyzing datasets, performance metrics, and challenges in real-world deployment. Transfer learning has proven to be an effective strategy for improving the diagnostic performance. For example, Balaha and Hassan [4] enhanced the classification accuracy by combining deep transfer learning with the sparrow search optimization technique. Alotaibi and AlSaeed [5] further enhanced the model performance using deep attention mechanisms combined with transfer learning. In a comparative analysis, Djaroudib et al. [6] emphasized that data quality plays a more critical role than data quantity in training effective transfer learning models. Numerous review articles have been published to assess the existing approaches. Nazari and Garcia [7] presented an in-depth analysis of automated skin cancer detection methods based on clinical imagery, while Naqvi et al. [8] focused on deep learning methods and their limitations. Naseri and Safaei [9] presented a systematic literature review on melanoma diagnosis and prognosis using both ML and DL techniques, underscoring the importance of robust datasets and ensemble learning. Magalhaes et al. [10] synthesized DL techniques for skin cancer detection and outlined key research directions. Model integration and ensemble techniques have gained attention for improving diagnostic reliability. Imran et al. [11] proposed a method combining decisions from multiple deep learners, achieving high classification accuracy. Moturi et al. [12] leveraged CNN techniques for efficient melanoma detection, while Kreouzi et al. [13] developed a deep learning approach to distinguish malignant melanoma from benign nevi using dermoscopic images. To address the multi-class nature of skin lesions, Tahir et al. [14] introduced DSCC_Net, a DL model capable of multi-class classification using dermoscopy images. Similarly, Naeem et al. [15] developed SNC_Net, which integrates handcrafted and DL-based features for enhanced skin cancer detection. Zia Ur Rehman et al. [16] employed explainable DL to classify skin cancer lesions, making AI predictions interpretable for clinicians. Karki et al. [17] combined segmentation, augmentation, and transfer learning techniques to improve early skin cancer detection. Gouda et al. [18] employed CNNs for classifying lesion images and reported promising results on standard datasets. Traditional ML methods also contribute; Natha and Rajeswari [19] used classification models like SVM and Random Forests to detect cancer from extracted image features. Although most studies focus on skin cancer, Das et al. [20] explored brain cancer prediction using CNNs and chatbot integration for smart healthcare, indicating the broader applicability of these techniques. Ashafuddula and Islam [21] proposed intensity value-based estimation combined with CNNs for differentiating melanoma and nevus moles. Rashad et al. [22] demonstrated an automated skin cancer screening system using deep learning techniques, highlighting its potential for scalable screening solutions. De et al. [23] demonstrated high accuracy with their hybrid CNN-DenseNet model, but its limitation lies in its high computational complexity, which may restrict its real-time clinical deployment. Abohashish et al. [24] proposed a hybrid deep learning model for enhanced melanoma and non-melanoma skin cancer classification, achieving improved accuracy on dermoscopic images. García et al. [25] developed a super-resolution-based framework for skin cancer classification, demonstrating that image enhancement prior to classification improves the diagnostic performance. In another study, Zhang et al. [26] introduced a colorectal polyp detection approach that integrates super-resolution techniques with YOLO-based deep learning, effectively improving the detection accuracy in colonoscopy images. Lastly, Ahmed et al. [27] presented a simultaneous super-resolution and classification framework for lung disease analysis, including carcinomas, which enhanced both the image quality and classification reliability.
The motivations for the paper are as follows: Skin cancer is one of the most prevalent and life-threatening cancers worldwide, and its early and accurate diagnosis is essential for improving patient outcomes. Traditional diagnostic methods are time-consuming, require specialized expertise, and are often not scalable for large populations. Although deep learning techniques have recently shown great promise in medical image analysis, challenges such as dataset imbalance, computational efficiency, and prediction reliability still remain. Therefore, there is a pressing need for lightweight and robust deep learning frameworks that can deliver accurate and interpretable multi-class skin cancer classification in real-world settings.
The primary objectives of this study are as follows:
- To develop a lightweight and efficient multi-class skin lesion classification framework using EfficientNet-B0.
- To mitigate dataset imbalance by applying augmentation and downsampling techniques for achieving fair class representation.
- To enhance classification robustness and accuracy by integrating transfer learning, full-network fine-tuning, test-time augmentation (TTA), and Monte Carlo dropout (MC dropout).
- To benchmark the proposed model against state-of-the-art CNN architectures for comprehensive validation.
- To design and deploy a web-based skin cancer detection system capable of providing automated classification, confidence scores, and lesion visualization for practical dermatology applications.
The key contributions of this research are as follows:
- Development of an EfficientNet-B0-based classification model tailored for seven diagnostic categories of skin lesions, ensuring high accuracy and computational efficiency.
- Construction of a balanced dataset from HAM10000, consisting of 7512 dermoscopic images, achieved through augmentation of minority classes and downsampling of majority classes.
- Integration of test-time augmentation (TTA) and Monte Carlo dropout (MC dropout) for improved uncertainty quantification and reliable predictions.
- Achievement of a peak classification accuracy of 97.15%, outperforming several state-of-the-art CNN models, validated through confusion matrix, ROC–AUC, precision–recall, and F1-score analyses.
- Deployment of the proposed model on a web-based clinical decision support platform, demonstrating strong applicability in dermatology practice.
2. Materials and Methods
In this section, the proposed architecture, workflow, and algorithm for skin lesion classification are discussed.
2.1. Proposed Architecture
Skin lesion images are initially preprocessed by resizing them to a fixed resolution and normalizing them using standard ImageNet statistics. The dataset is then divided into training, validation, and test sets to ensure unbiased model evaluation. An EfficientNet-B0 backbone is employed as the feature extractor and fine-tuned on the training data. The model is validated on the validation set after each epoch, and the checkpoint with the best performance is retained for inference. At test time, two complementary inference branches are utilized to improve robustness and reliability. The first branch, MC dropout, keeps the dropout layers active and performs 10 stochastic forward passes to quantify the predictive uncertainty and compute the mean prediction. The second branch, test-time augmentation (TTA), applies deterministic transformations—including horizontal and vertical flips, color jitter, and rotations of ±45°—to the input images, generating multiple predictions per sample. The outputs from both branches are ensembled to calculate final class probabilities and confidence scores. The predicted diagnosis corresponds to the class with the highest confidence. This architecture allows the model to capture both variability in the input data and model uncertainty, enhancing the classification reliability for skin lesion images. Figure 1 illustrates the proposed architecture and workflow, including the preprocessing, model training, and dual inference pipelines. The class abbreviations used throughout this work are akiec, bcc, bkl, df, mel, nv, and vasc. Figure 1 illustrates the overall process and classification approach.
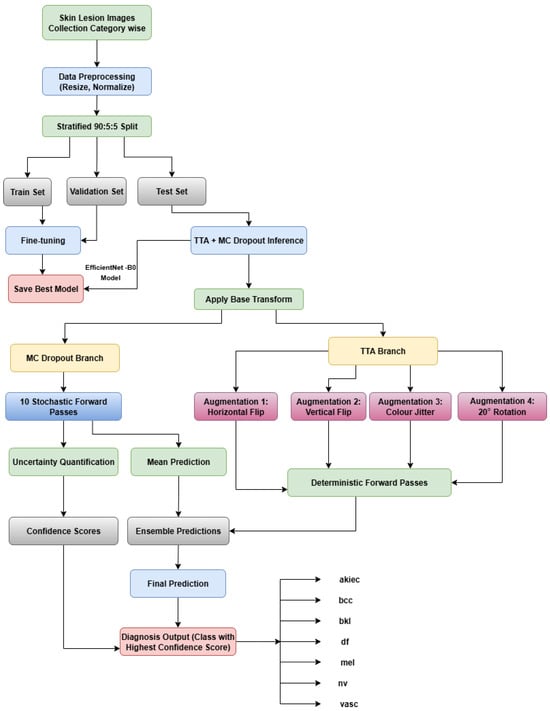
Figure 1.
Proposed architecture.
2.2. Sequence Diagram for Proposed Work
The process starts with a user uploading a skin lesion image, which is preprocessed by resizing it to 224 × 224 and normalizing it with ImageNet statistics. The image undergoes two inference pipelines: (i) test-time augmentation (TTA)—five transformations (original, horizontal flip, vertical flip, color jitter, and 20° rotation) are applied, and predictions are generated using the EfficientNet-B0 model; (ii) MC dropout inference—the dropout layers remain active to perform 10 stochastic forward passes for uncertainty estimation. Both pipelines produce softmax probabilities, which are aggregated by averaging the TTA probabilities and calculating the MC dropout variance. The final output is the class label with the highest probability (akiec, bcc, bkl, df, mel, nv, vasc) along with a confidence score. Figure 2 illustrates a strong workflow for classifying skin lesions that combines deep learning and uncertainty quantification, which are depicted in this diagram.
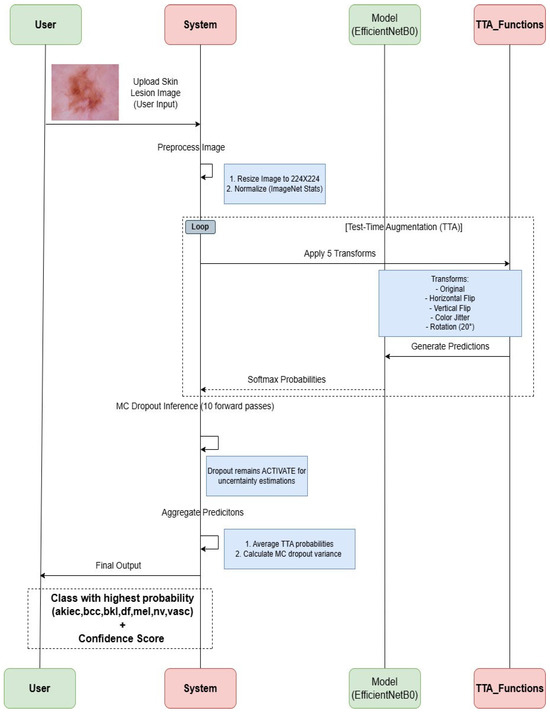
Figure 2.
Proposed sequence diagram for single skin lesion image classification.
Algorithm 1 classifies skin lesion images using EfficientNet-B0 with TTA and MC dropout to produce final class labels and confidence scores.
| Algorithm 1 Skin Lesion Image Classification using EfficientNet-B0 with Enhanced TTA and MC Dropout |
| Input: Set of images X = {x1, x2, …, xn} Trained model weights W Augmentations A = {Original, HFlip, VFlip, ColorJitter, Rotation} Number of MC Dropout passes M (e.g., M = 10) Device device ∈ {cpu, cuda} Output: Predicted class labels = {, , …, } and confidence scores S = {s1, s2, …, sn} // xa = Augmented version of image x using a // pa,m = Softmax output for augmentation a at dropout pass m // P = List of all predictions across TTA and MC Dropout // pavg = Mean prediction vector 1: Model Setup 2: Load EfficientNet-B0 model 3: Replace classifier with final layer for number of classes 4: Load trained weights W into model 5: Move model to device 6: Set model to evaluation mode 7: for all layer m in model do 8: if m is Dropout then 9: Set m to train mode // Enable MC Dropout at inference 10: end if 11: end for 12: Initialize ← [], S ← [] 13: for all image x in X do 14: Preprocess x (resize 224 × 224, normalize, convert to tensor) 15: Initialize P ← [] 16: for all augmentation a in A do 17: Apply augmentation a to x, result is xa 18: for m = 1 to M do 19: pa,m ← softmax(model(xa)) 20: Append pa,m to P 21: end for 22: end for 23: pavg ← mean(P) 24: ← arg max(pavg) 25: s ← max(pavg) 26: Append to , s to S 27: end for 28: return , S |
2.3. Computational Details
The computational analysis employed the EfficientNet-B0 CNN, pre-trained on ImageNet, for the multi-class classification of dermoscopic images from the HAM10000 dataset (10,015 images, seven categories). Experiments were conducted using Python 3.11.0 with Keras 2.11.0 on CPU/GPU platforms. Data were stratified for balanced classes, and training used the Adam optimizer with cross-entropy loss, data augmentation, and batch processing. Test-time augmentation and Monte Carlo dropout were applied during inference to enhance robustness and estimate uncertainty. The best validation model was selected, and all metrics were computed on a designated test set to ensure reproducibility.
2.4. Data Preparation and Initialization
High-resolution dermoscopic images were gathered from publicly accessible medical datasets in order to classify skin lesion images. Resizing and normalization were performed after the data had been meticulously preprocessed to guarantee consistency in the image size and quality. To facilitate robust model training, stratified splitting and data augmentation were used to achieve a balanced class distribution.
The HAM10000 dataset was sourced from Kaggle, comprising a total of 10,015 dermoscopic images categorized into seven diagnostic classes, as shown in Figure 3:
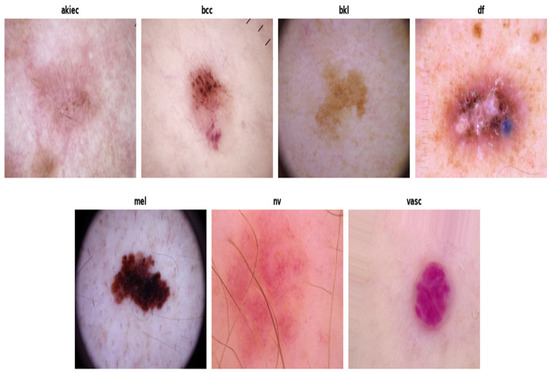
Figure 3.
Sample of data collection. Source: https://www.kaggle.com/datasets/surajghuwalewala/ham1000-segmentation-and-classification (accessed on 10 December 2022).
Given the class imbalance, image augmentation was applied to minority classes (akiec, bcc, df, vasc) to increase their samples to 1000 images each. This was achieved using Keras’ ImageDataGenerator, applying random transformations such as rotation, zoom, shifts, and flips. The majority class ‘nv’ was downsampled to 1300 images through random selection to avoid bias. Classes bkl and mel were retained at their original counts. The final dataset consisted of 7512 images with improved class balance. The balanced dataset was split into training (70%), validation (15%), and testing (15%) subsets via stratified sampling to maintain proportional class distribution across sets. This resulted in 5258 training, 1127 validation, and 1127 test images. All images were resized to a resolution of 224 × 224 pixels, normalized using ImageNet mean and standard deviation values, and converted into tensors for model processing. The dataset was organized in a hierarchical folder structure to ensure compatibility with standard deep learning frameworks, thereby enabling efficient loading and preprocessing during training and evaluation.
2.5. Model Setup and Implementation
The EfficientNet-B0 architecture, which was selected for its ideal balance between accuracy and efficiency, was used to classify the skin lesion images. The final classification layers of the model were adjusted, and early stopping, data augmentation, and hyperparameter tuning were used to improve training and guarantee reliable results.
2.5.1. Model Architecture and Transfer Learning
The EfficientNet-B0 model, pre-trained on the ImageNet dataset, was utilized in this study. To take advantage of its pre-learned features, the convolutional layers were initially kept frozen. The original classification head was replaced with a new fully connected layer configured to predict the seven skin cancer categories.
2.5.2. Model Training
Using the Adam optimizer and CrossEntropyLoss criterion, the model was trained for 15 epochs. During this phase, only the classifier layer’s weights were updated. Training and validation metrics were monitored to assess the learning progress and mitigate overfitting.
2.5.3. Fine-Tuning
To further improve the accuracy, all layers of EfficientNet-B0 were unfrozen for full-model fine-tuning with a reduced learning rate (1 × 10−4). This allowed the entire network to adapt to the specific dataset over another 15 epochs of training.
2.6. Model Evaluation
Final evaluation on the test set showed substantial accuracy improvement after fine-tuning. Additional enhancements included test-time augmentation (TTA), such as horizontal flip, vertical flip, color jitter, and a 20° rotation, as seen in Figure 4, where predictions on multiple augmented versions of test images were averaged to reduce the prediction variance. Furthermore, Monte Carlo dropout was integrated at inference to capture uncertainty, combined with TTA for a robust performance.
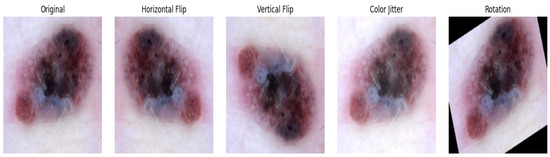
Figure 4.
Skin lesion image preprocessing by test-time augmentation (TTA).
3. Results and Discussion
3.1. Training and Validation Performance
The EfficientNet-B0 model initially trained with frozen convolutional layers showed progressive improvement over 15 epochs. The training accuracy increased from 57.74% to 75.53%, while the validation accuracy improved from 64.10% to 73.40%. The corresponding loss values steadily decreased, indicating effective learning without significant overfitting. Table 1 summarizes the training and validation loss and accuracy across 15 epochs for the EfficientNet-B0 model with frozen base layers.

Table 1.
Training and validation performance metrics across epochs using the EfficientNet-B0 model with frozen base layers.
Based on the results presented in Table 2, EfficientNet-B0 was identified as the most suitable model for final deployment in this study. Prior to this selection, an extensive comparative analysis was performed across multiple deep learning architectures. The performances of these models were assessed using standard training and testing procedures under different train–validation–test splits, as summarized in the comparison table. Among all the evaluated models, EfficientNet-B0 achieved the highest classification accuracy of 89.36% before fine-tuning, particularly with a 70:15:15 train–validation–test split. Owing to this superior baseline performance, EfficientNet-B0 was chosen for further optimization. Fine-tuning of the model resulted in a substantial improvement in accuracy, reaching 95%. To further enhance the generalization and robustness, test-time augmentation (TTA) was applied, which increased the accuracy to 96%. Finally, the incorporation of Monte Carlo dropout during inference led to a peak accuracy of 97.15%, establishing EfficientNet-B0 as the most effective model within the experimental pipeline.

Table 2.
Model accuracy comparison table (before fine-tuning).
3.2. Fine-Tuning Performance
Full fine-tuning of the model with all layers trainable over 15 epochs yielded significant improvements. The training accuracy reached 99.08%, while the validation accuracy peaked at 89.36%. The loss values consistently decreased, demonstrating an enhanced generalization capability. The detailed training and validation performances across epochs are presented in Table 3.

Table 3.
Training and validation performance metrics across epochs during full fine-tuning of the model.
3.3. Test Set Evaluation
The final test accuracy of the fine-tuned EfficientNet-B0 model was 95%. Applying test-time augmentation (TTA) improved the accuracy to 96%, while the combination of TTA with Monte Carlo dropout further increased the test accuracy to 97.15%. A detailed classification report is presented in Table 4.

Table 4.
Classification report summarizing the precision, recall, F1-score, and support for each class in the test set (70/15/15 split: 5258 training, 1127 validation, 1127 testing).
The classification performance metrics for each class are summarized in Table 4. The model achieved perfect precision and recall for the classes df and vasc, indicating flawless classification on these categories. Classes such as akiec and bcc also demonstrate very high precision and recall values (above 0.96), reflecting strong model reliability. The bkl and mel classes show slightly lower recall compared to the others but still maintain F1-scores around 0.95, which indicates a robust performance even for challenging lesion categories. The nv class, having the highest number of samples, achieved stable results with an F1-score of 0.96. Overall, the model attained a final accuracy of 97.15% across all 1127 test samples. The macro-average and weighted average metrics are both approximately 0.97, demonstrating a balanced performance across both majority and minority classes.
3.4. Confusion Matrix
The performance of the classification model was evaluated using multiple metrics to ensure robustness across all lesion types. The confusion matrix, shown in Figure 5, illustrates the model’s ability to correctly classify each class. Classes such as akiec, df, and vasc achieved near-perfect precision and recall, reflecting the model’s strong capability of identifying critical and rare lesion types. Classes like bcc and nv also demonstrate high performances across the precision, recall, and F1-score metrics, indicating reliable classification for more common lesion types. However, classes bkl and mel exhibit comparatively lower F1-scores, likely due to class overlap, visual similarity, or limited representation in the dataset. Overall, the confusion matrix confirms that the model maintains a balanced performance across both majority and minority classes.
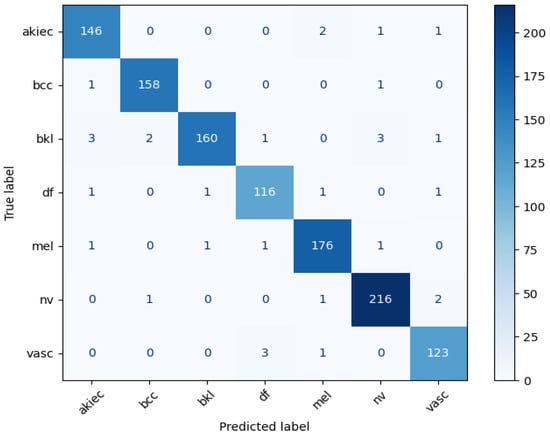
Figure 5.
Confusion matrix analysis for proposed model.
3.5. AUC and ROC Curves
To further evaluate the discriminative ability of the proposed EfficientNet-B0 model, Receiver Operating Characteristic (ROC) curve analysis was performed for each lesion class. As illustrated in Figure 6, the model demonstrates an outstanding classification performance, with all classes achieving an AUC score of 1.00. Both the micro-average and macro-average ROC curves also reach an AUC of 1.00, confirming that the model is capable of reliably separating malignant and benign categories across diverse lesion types. These results strongly support the robustness of the model and align with the final test accuracy of 97.15%.
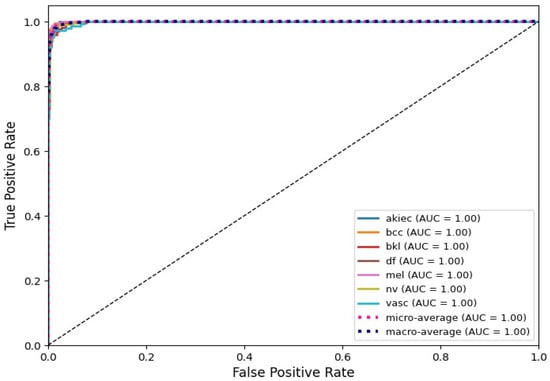
Figure 6.
AUC-ROC analysis showing strong class separation and balanced performance.
3.6. Web-Based Skin Cancer Detection System Results
To evaluate the real-world applicability of the proposed framework, the EfficientNet-B0 model was deployed within a web-based skin cancer detection system. This platform allows for automated lesion classification with confidence scoring and the visual highlighting of clinically relevant regions, providing a practical interface for decision support in dermatology. Representative outputs are shown in Figure 7 and Figure 8. In Figure 7, the system correctly identifies a lesion as benign keratosis, a non-cancerous skin condition. The output includes the predicted class, associated confidence score, and highlighted lesion regions to aid interpretation. Such an automated preliminary evaluation can reduce diagnostic errors and assist clinicians in making efficient and accurate decisions. Figure 8 presents a case of melanoma, a malignant and potentially life-threatening form of skin cancer. The system provides a clear classification along with a confidence score and lesion region visualization, facilitating improved clinical interpretation. By reliably distinguishing malignant from benign lesions, the framework enables prioritization of high-risk cases and supports timely clinical decision making. Overall, the web-based platform demonstrates strong potential as a reliable decision support tool for dermatology practice and early detection initiatives.
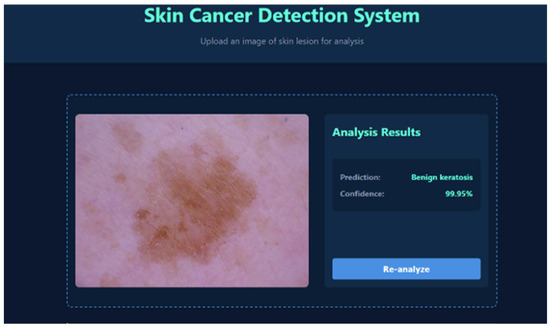
Figure 7.
System output showing predicted benign keratosis.
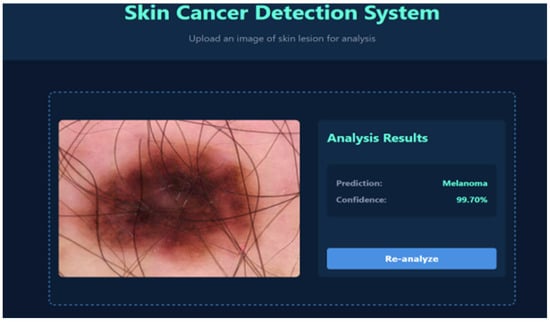
Figure 8.
System output showing predicted melanoma.
4. Conclusions
Skin cancer remains one of the most common and deadly cancers worldwide, underscoring the critical importance of early and accurate detection for successful treatment. Traditional diagnostic methods are often time-consuming and highly dependent on specialized expertise, which has fueled the growing interest in automated diagnostic solutions. Deep learning, and particularly convolutional neural networks (CNNs), has shown remarkable promise in medical image analysis. Among modern architectures, EfficientNet offers an optimal balance between accuracy and computational efficiency. In this study, the EfficientNet-B0 framework was applied for the reliable multi-class classification of skin cancer using dermoscopic images from the HAM10000 dataset. Transfer learning and data preprocessing strategies were employed, followed by the fine-tuning of the entire network to maximize the performance. The proposed model achieved a final test accuracy of 97.15%, representing a substantial improvement over the baseline training. The integration of Monte Carlo dropout and test-time augmentation further enhanced the model’s robustness and generalization. Given its lightweight design and strong performance, EfficientNet-B0 is a suitable candidate for deployment in real-time clinical environments with limited computational resources. Overall, the findings demonstrate that deep CNN-based approaches can significantly improve the accuracy and efficiency of skin lesion diagnosis, ultimately supporting earlier intervention and better patient outcomes.
Future work will explore lightweight super-resolution methods and improved uncertainty quantification for enhanced robustness and clinical applicability.
Author Contributions
Conceptualization, supervision, validation, writing—review and editing, methodology, software, visualization, S.D.; data curation, software, formal analysis, editing, investigation, and visualization, R.K.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The original data presented in the study are openly available in [Skin Cancer MNIST: HAM10000] at [https://www.kaggle.com/datasets/kmader/skin-cancer-mnist-ham10000].
Conflicts of Interest
The authors declare no conflicts of interest.
Correction Statement
This article has been republished with a minor correction to the Data Availability Statement. This change does not affect the scientific content of the article.
Abbreviations
This manuscript uses the following abbreviations:
| DL | Deep Learning |
| ML | Machine Learning |
| CNN | Convolutional Neural Network |
| HAM | Human Against Machine |
| TTA | Test-Time Augmentation |
| AKIEC | Actinic Keratoses and Intraepithelial Carcinoma |
| BCC | Basal Cell Carcinoma, |
| DF | Dermatofibroma |
| VASC | Vascular Lesion |
| BCAT | Brain Computer Aptitude Test |
| BKL | Benign Keratosis |
| MEL | Melanoma |
References
- Ashfaq, N.; Suhail, Z.; Khalid, A.; Sarwar, N.; Irshad, A.; Yaman, O.; Alubaidi, A.; Ahmed, F.M.; Almalki, F.A. SkinSight: Advancing deep learning for skin cancer diagnosis and classification. Discov. Comput. 2025, 28, 63. [Google Scholar] [CrossRef]
- Kavitha, C.; Priyanka, S.; Kumar, M.P.; Kusuma, V. Skin Cancer Detection and Classification Using Deep Learning Techniques. Procedia Comput. Sci. 2024, 235, 2793–2802. [Google Scholar] [CrossRef]
- Naeem, A.; Farooq, M.S.; Khelifi, A.; Abid, A. Malignant Melanoma Classification Using Deep Learning: Datasets, Performance Measurements, Challenges and Opportunities. IEEE Access 2020, 8, 110575–110597. [Google Scholar] [CrossRef]
- Balaha, H.M.; Hassan, A.E.-S. Skin cancer diagnosis based on deep transfer learning and sparrow search algorithm. Neural Comput. Appl. 2023, 35, 815–853. [Google Scholar] [CrossRef]
- Alotaibi, A.; AlSaeed, D. Skin Cancer Detection Using Transfer Learning and Deep Attention Mechanisms. Diagnostics 2025, 15, 99. [Google Scholar] [CrossRef]
- Djaroudib, K.; Lorenz, P.; Bouzida, R.B.; Merzougui, H. Skin Cancer Diagnosis Using VGG16 and Transfer Learning: Analyzing the Effects of Data Quality over Quantity on Model Efficiency. Appl. Sci. 2024, 14, 7447. [Google Scholar] [CrossRef]
- Nazari, S.; Garcia, R. Automatic Skin Cancer Detection Using Clinical Images: A Comprehensive Review. Life 2023, 13, 2123. [Google Scholar] [CrossRef]
- Naqvi, M.; Gilani, S.Q.; Syed, T.; Marques, O.; Kim, H.-C. Skin Cancer Detection Using Deep Learning—A Review. Diagnostics 2023, 13, 1911. [Google Scholar] [CrossRef]
- Naseri, H.; Safaei, A.A. Diagnosis and prognosis of melanoma from dermoscopy images using machine learning and deep learning: A systematic literature review. BMC Cancer 2025, 25, 75. [Google Scholar] [CrossRef]
- Magalhaes, C.; Mendes, J.; Vardasca, R. Systematic Review of Deep Learning Techniques in Skin Cancer Detection. BioMedInformatics 2024, 4, 2251–2270. [Google Scholar] [CrossRef]
- Imran, A.; Nasir, A.; Bilal, M.; Sun, G.; Alzahrani, A.; Almuhaimeed, A. Skin Cancer Detection Using Combined Decision of Deep Learners. IEEE Access 2022, 10, 118198–118212. [Google Scholar] [CrossRef]
- Moturi, D.; Surapaneni, R.K.; Avanigadda, V.S.G. Developing an efficient method for melanoma detection using CNN techniques. J. Egypt. Natl. Cancer Inst. 2024, 36, 6. [Google Scholar] [CrossRef]
- Kreouzi, M.; Theodorakis, N.; Feretzakis, G.; Paxinou, E.; Sakagianni, A.; Kalles, D.; Anastasiou, A.; Verykios, V.S.; Nikolaou, M. Deep Learning for Melanoma Detection: A Deep Learning Approach to Differentiating Malignant Melanoma from Benign Melanocytic Nevi. Cancers 2025, 17, 28. [Google Scholar] [CrossRef]
- Tahir, M.; Naeem, A.; Malik, H.; Tanveer, J.; Naqvi, R.A.; Lee, S.-W. DSCC_Net: Multi-Classification Deep Learning Models for Diagnosing of Skin Cancer Using Dermoscopic Images. Cancers 2023, 15, 2179. [Google Scholar] [CrossRef]
- Naeem, A.; Anees, T.; Khalil, M.; Zahra, K.; Naqvi, R.A.; Lee, S.-W. SNC_Net: Skin Cancer Detection by Integrating Handcrafted and Deep Learning-Based Features Using Dermoscopy Images. Mathematics 2024, 12, 1030. [Google Scholar] [CrossRef]
- Zia Ur Rehman, M.; Ahmed, F.; Alsuhibany, S.A.; Jamal, S.S.; Zulfiqar Ali, M.; Ahmad, J. Classification of Skin Cancer Lesions Using Explainable Deep Learning. Sensors 2022, 22, 6915. [Google Scholar] [CrossRef]
- Karki, R.; G C, S.; Rezazadeh, J.; Khan, A. Deep Learning for Early Skin Cancer Detection: Combining Segmentation, Augmentation, and Transfer Learning. Big Data Cogn. Comput. 2025, 9, 97. [Google Scholar] [CrossRef]
- Gouda, W.; Sama, N.U.; Al-Waakid, G.; Humayun, M.; Jhanjhi, N.Z. Detection of Skin Cancer Based on Skin Lesion Images Using Deep Learning. Healthcare 2022, 10, 1183. [Google Scholar] [CrossRef]
- Natha, P.; Rajeswari, P.R. Skin Cancer Detection Using Machine Learning Classification Models. Int. J. Intell. Syst. Appl. Eng. 2023, 12, 139–145. [Google Scholar]
- Das, S.; Kumar, V.; Cicceri, G. Chatbot Enable Brain Cancer Prediction Using Convolutional Neural Network for Smart Healthcare. In Healthcare-Driven Intelligent Computing Paradigms to Secure Futuristic Smart Cities; Chapman and Hall/CRC: Boca Raton, FL, USA, 2024; pp. 268–279. [Google Scholar]
- Ashafuddula, N.I.M.; Islam, R. Melanoma skin cancer and nevus mole classification using intensity value estimation with convolutional neural network. Comput. Sci. 2023, 24, 277–296. [Google Scholar] [CrossRef]
- Rashad, N.M.; Abdelnapi, N.M.; Seddik, A.F.; Sayedelahl, M.A. Automating skin cancer screening: A deep learning. J. Eng. Appl. Sci. 2025, 72, 6. [Google Scholar] [CrossRef]
- De, A.; Mishra, N.; Chang, H.-T. An approach to the dermatological classification of histopathological skin images using a hybridized CNN-DenseNet model. PeerJ Comput. Sci. 2024, 10, e1884. [Google Scholar] [CrossRef]
- Abohashish, S.M.M.; Amin, H.H.; Elsedimy, E.I. Enhanced Melanoma and Non-Melanoma Skin Cancer Classification Using Hybrid Deep Learning Models. Sci. Rep. 2025, 15, 4598. [Google Scholar] [CrossRef]
- Mukadam, S.B.; Patil, H.Y. Skin Cancer Classification Framework Using Enhanced Super Resolution Generative Adversarial Network and Custom Convolutional Neural Network. Appl. Sci. 2023, 13, 1210. [Google Scholar] [CrossRef]
- Wang, S.; Xie, J.; Cui, Y.; Chen, Z. Colorectal Polyp Detection Model by Using Super-Resolution Reconstruction and YOLO. Electronics 2024, 13, 2298. [Google Scholar] [CrossRef]
- Emara, H.M.; Shoaib, M.R.; El-Shafai, W.; Elwekeil, M.; Hemdan, E.E.-D.; Fouda, M.M.; Taha, T.E.; El-Fishawy, A.S.; El-Rabaie, E.-S.M.; El-Samie, F.E.A. Simultaneous Super-Resolution and Classification of Lung Disease Scans. Diagnostics 2023, 13, 1319. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
