Colocalization Analysis Reveals Shared Genetic Loci Contributing to Gout and Metabolite Levels
Abstract
1. Introduction
2. Materials and Methods
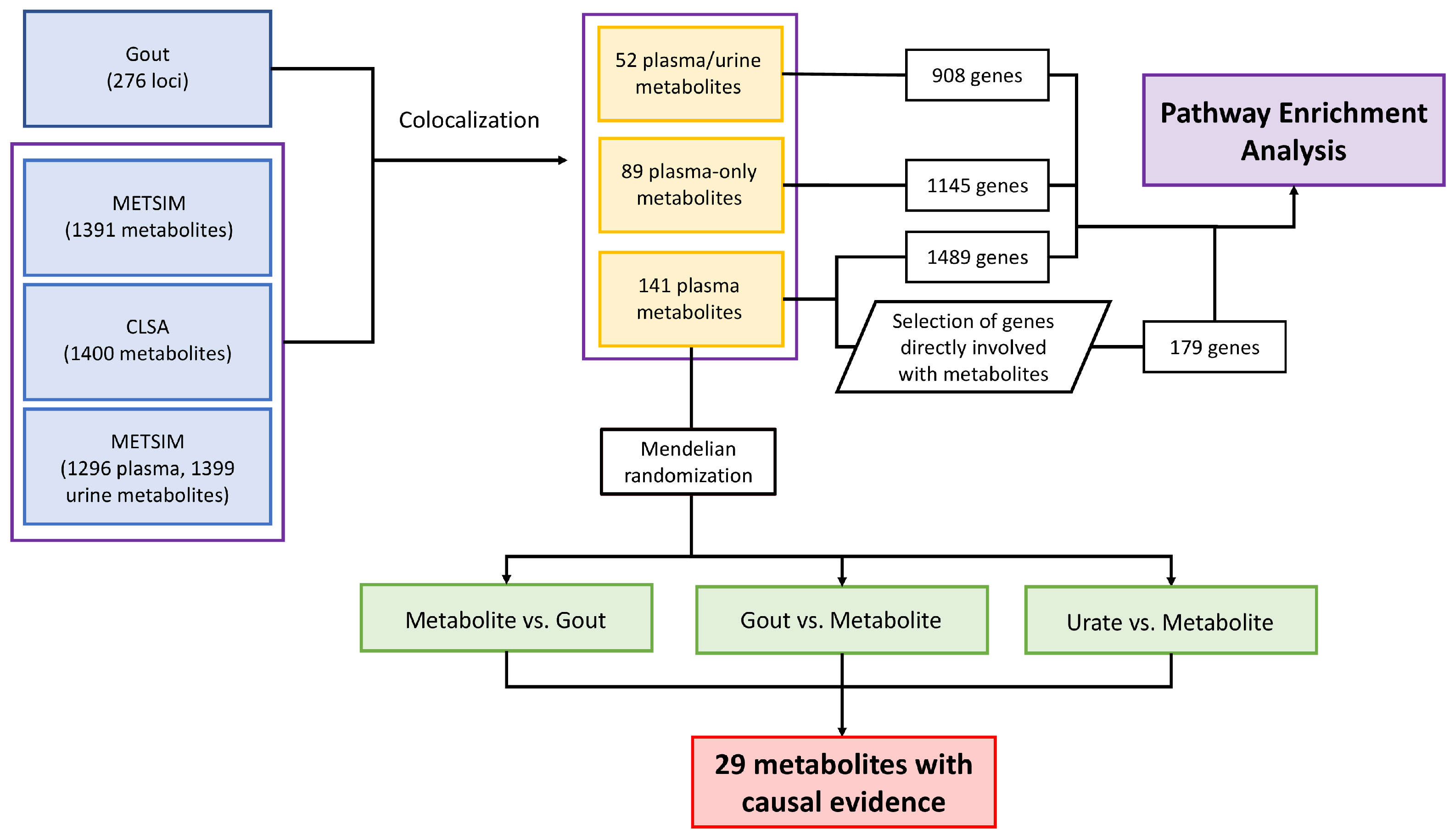
2.1. Summary Data and Meta-Analysis
2.2. Colocalization Analysis
2.3. Mapping of Metabolite Names
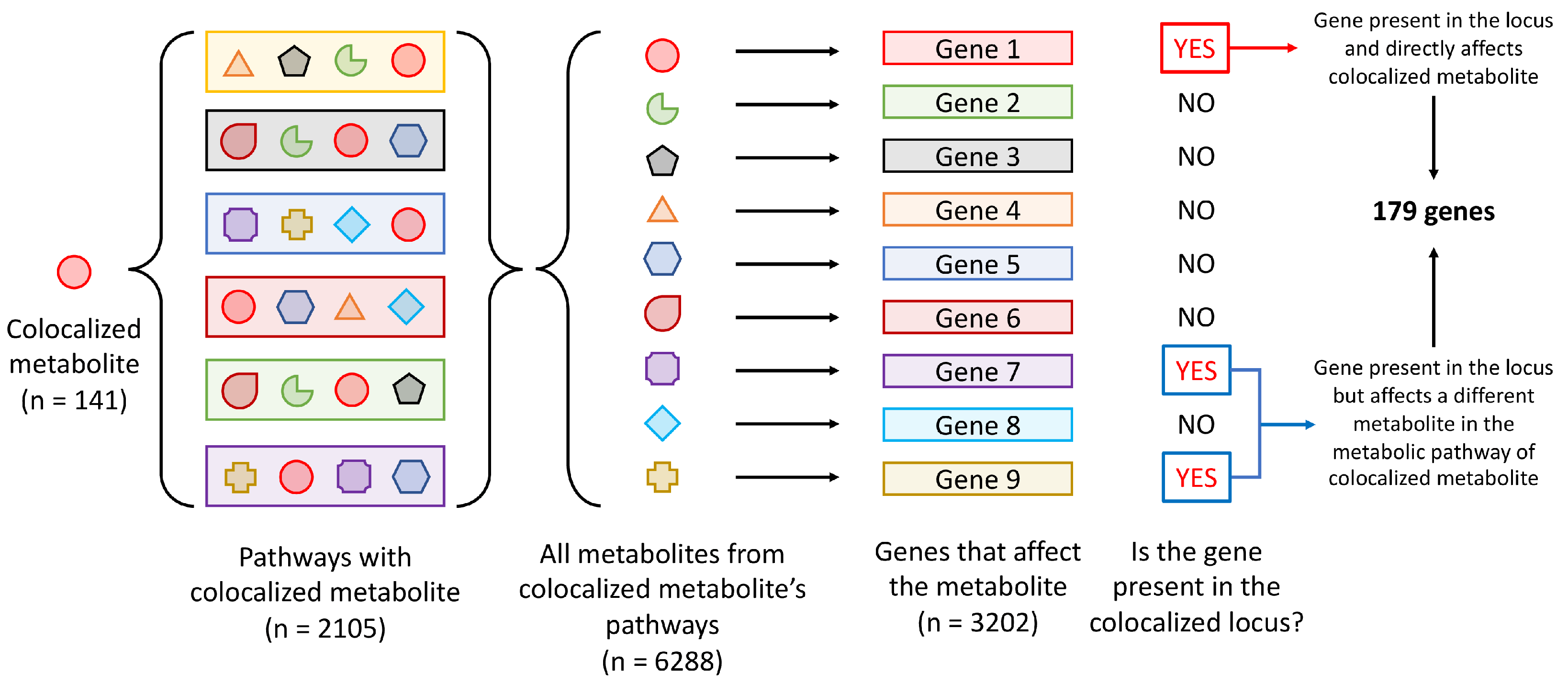
2.4. Pathway Enrichment Analysis
2.5. Mendelian Randomization
3. Results
3.1. Genetic Colocalization of Gout and Metabolite Quantitative Trait Loci
3.2. Pathway Enrichment Analysis of Genes at Loci That Colocalized with Metabolite Levels
3.3. Mendelian Randomization of Colocalized Metabolites with Gout and Urate
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CLSA | Canadian Longitudinal Study on Aging |
| FDR | False-Discovery Rate |
| GCKD | German Chronic Kidney Disease |
| GWAS | Genome-Wide Association Study |
| HMDB | The Human Metabolome Database |
| LD | Linkage Disequilibrium |
| METSIM | Metabolic Syndrome in Men |
| MSU | Monosodium Urate |
| QTL | Quantitative Trait Loci |
References
- Dalbeth, N.; Gosling, A.L.; Gaffo, A.; Abhishek, A. Gout. Lancet 2021, 397, 1843–1855. [Google Scholar] [CrossRef] [PubMed]
- Martinon, F.; Pétrilli, V.; Mayor, A.; Tardivel, A.; Tschopp, J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 2006, 440, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Tin, A.; Marten, J.; Halperin Kuhns, V.L.; Li, Y.; Wuttke, M.; Kirsten, H.; Sieber, K.B.; Qiu, C.; Gorski, M.; Yu, Z.; et al. Target genes, variants, tissues and transcriptional pathways influencing human serum urate levels. Nat. Genet. 2019, 51, 1459–1474. [Google Scholar] [CrossRef] [PubMed]
- Major, T.J.; Takei, R.; Matsuo, H.; Leask, M.P.; Sumpter, N.A.; Topless, R.K.; Shirai, Y.; Wang, W.; Cadzow, M.J.; Phipps-Green, A.J.; et al. A genome-wide association analysis reveals new pathogenic pathways in gout. Nat. Genet. 2024, 56, 2392–2406. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, Z.; Hou, X.; Lu, D.; Yuan, X.; Lu, J.; Wang, C.; Han, L.; Cui, L.; Liu, Z.; et al. Replication of Gout/Urate Concentrations GWAS Susceptibility Loci Associated with Gout in a Han Chinese Population. Sci. Rep. 2017, 7, 4094. [Google Scholar] [CrossRef]
- Nakayama, A.; Nakatochi, M.; Kawamura, Y.; Yamamoto, K.; Nakaoka, H.; Shimizu, S.; Higashino, T.; Koyama, T.; Hishida, A.; Kuriki, K.; et al. Subtype-specific gout susceptibility loci and enrichment of selection pressure on ABCG2 and ALDH2 identified by subtype genome-wide meta-analyses of clinically defined gout patients. Ann. Rheum. Dis. 2020, 79, 657–665. [Google Scholar] [CrossRef]
- Kawamura, Y.; Nakaoka, H.; Nakayama, A.; Okada, Y.; Yamamoto, K.; Higashino, T.; Sakiyama, M.; Shimizu, T.; Ooyama, H.; Ooyama, K.; et al. Genome-wide association study revealed novel loci which aggravate asymptomatic hyperuricaemia into gout. Ann. Rheum. Dis. 2019, 78, 1430–1437. [Google Scholar] [CrossRef]
- Zhou, W.; Kanai, M.; Wu, K.H.H.; Rasheed, H.; Tsuo, K.; Hirbo, J.B.; Wang, Y.; Bhattacharya, A.; Zhao, H.; Namba, S.; et al. Global Biobank Meta-analysis Initiative: Powering genetic discovery across human disease. Cell Genom. 2022, 2, 100192. [Google Scholar] [CrossRef]
- Renaudin, F.; Orliaguet, L.; Castelli, F.; Fenaille, F.; Prignon, A.; Alzaid, F.; Combes, C.; Delvaux, A.; Adimy, Y.; Cohen-Solal, M.; et al. Gout and pseudo-gout-related crystals promote GLUT1-mediated glycolysis that governs NLRP3 and interleukin-1β activation on macrophages. Ann. Rheum. Dis. 2020, 79, 1506–1514. [Google Scholar] [CrossRef]
- Lyu, S.; Rao, Y.; Liu, P.; Yang, S.; Chen, W.; Yang, H.; Ke, S.; OuYang, H.; He, M.; Feng, Y. Metabolomics analysis reveals four biomarkers associated with the gouty arthritis progression in patients with sequential stages. Semin. Arthritis Rheum. 2022, 55, 152022. [Google Scholar] [CrossRef]
- Joshi, A.D.; McCormick, N.; Yokose, C.; Yu, B.; Tin, A.; Terkeltaub, R.; Merriman, T.R.; Eliassen, A.H.; Curhan, G.C.; Raffield, L.M.; et al. Prediagnostic Glycoprotein Acetyl Levels and Incident and Recurrent Flare Risk Accounting for Serum Urate Levels: A Population-Based, Prospective Study and Mendelian Randomization Analysis. Arthritis Rheumatol. 2023, 75, 1648–1657. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liang, N.; Tao, Y.; Yin, H. Metabolomics in Hyperuricemia and Gout. Gout Urate Cryst. Depos. Dis. 2023, 1, 49–61. [Google Scholar] [CrossRef]
- Chen, Y.; Lu, T.; Pettersson-Kymmer, U.; Stewart, I.D.; Butler-Laporte, G.; Nakanishi, T.; Cerani, A.; Liang, K.Y.H.; Yoshiji, S.; Willett, J.D.S.; et al. Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat. Genet. 2023, 55, 44–53. [Google Scholar] [CrossRef]
- Schlosser, P.; Scherer, N.; Grundner-Culemann, F.; Monteiro-Martins, S.; Haug, S.; Steinbrenner, I.; Uluvar, B.; Wuttke, M.; Cheng, Y.; Ekici, A.B.; et al. Genetic studies of paired metabolomes reveal enzymatic and transport processes at the interface of plasma and urine. Nat. Genet. 2023, 55, 995–1008. [Google Scholar] [CrossRef]
- Yin, X.; Chan, L.S.; Bose, D.; Jackson, A.U.; VandeHaar, P.; Locke, A.E.; Fuchsberger, C.; Stringham, H.M.; Welch, R.; Yu, K.; et al. Genome-wide association studies of metabolites in Finnish men identify disease-relevant loci. Nat. Commun. 2022, 13, 1644. [Google Scholar] [CrossRef]
- Arts, R.J.W.; Novakovic, B.; ter Horst, R.; Carvalho, A.; Bekkering, S.; Lachmandas, E.; Rodrigues, F.; Silvestre, R.; Cheng, S.C.; Wang, S.Y.; et al. Glutaminolysis and Fumarate Accumulation Integrate Immunometabolic and Epigenetic Programs in Trained Immunity. Cell Metab. 2016, 24, 807–819. [Google Scholar] [CrossRef]
- Willer, C.J.; Li, Y.; Abecasis, G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010, 26, 2190–2191. [Google Scholar] [CrossRef]
- Giambartolomei, C.; Vukcevic, D.; Schadt, E.E.; Franke, L.; Hingorani, A.D.; Wallace, C.; Plagnol, V. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet. 2014, 10, e1004383. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.R-project.org/ (accessed on 22 December 2024).
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Cohen, G.J.; Dice, J.; Friedrichs, M.; Gupta, K.; Hayes, W.; Kitschelt, I.; Lee, S.J.; Marsh, W.B.; Mislang, N.; Shaton, M.; et al. The U.S. syndicated loan market: Matching data. J. Financ. Res. 2021, 44, 695–723. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yavorska, O.O.; Burgess, S. MendelianRandomization: An R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 2017, 46, 1734–1739. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, s13742–015–0047–8. [Google Scholar] [CrossRef]
- Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Abecasis, G.R.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Gloudemans, M.J.; Balliu, B.; Nachun, D.; Schnurr, T.M.; Durrant, M.G.; Ingelsson, E.; Wabitsch, M.; Quertermous, T.; Montgomery, S.B.; Knowles, J.W.; et al. Integration of genetic colocalizations with physiological and pharmacological perturbations identifies cardiometabolic disease genes. Genome Med. 2022, 14, 31. [Google Scholar] [CrossRef]
- Reus, L.M.; Boltz, T.; Francia, M.; Bot, M.; Ramesh, N.; Koromina, M.; Pijnenburg, Y.A.L.; den Braber, A.; van der Flier, W.M.; Visser, P.J.; et al. Quantitative trait loci mapping of circulating metabolites in cerebrospinal fluid to uncover biological mechanisms involved in brain-related phenotypes. Mol. Psychiatry 2025. [Google Scholar] [CrossRef]
- Austin Doyle, L.; Ross, D.D. Multidrug resistance mediated by the breast cancer resistance protein BCRP (ABCG2). Oncogene 2003, 22, 7340–7358. [Google Scholar] [CrossRef]
- Chen-Xu, M.; Yokose, C.; Rai, S.K.; Pillinger, M.H.; Choi, H.K. Contemporary Prevalence of Gout and Hyperuricemia in the United States and Decadal Trends: The National Health and Nutrition Examination Survey, 2007–2016. Arthritis Rheumatol. 2019, 71, 991–999. [Google Scholar] [CrossRef]
- Netea, M.G.; Quintin, J.; van der Meer, J.W.M. Trained Immunity: A Memory for Innate Host Defense. Cell Host Microbe 2011, 9, 355–361. [Google Scholar] [CrossRef]
- van der Heijden, C.D.; Noz, M.P.; Joosten, L.A.; Netea, M.G.; Riksen, N.P.; Keating, S.T. Epigenetics and Trained Immunity. Antioxidants Redox Signal. 2018, 29, 1023–1040. [Google Scholar] [CrossRef] [PubMed]
- Arts, R.J.W.; Carvalho, A.; La Rocca, C.; Palma, C.; Rodrigues, F.; Silvestre, R.; Kleinnijenhuis, J.; Lachmandas, E.; Gonçalves, L.G.; Belinha, A.; et al. Immunometabolic Pathways in BCG-Induced Trained Immunity. Cell Rep. 2016, 17, 2562–2571. [Google Scholar] [CrossRef]
- Xiao, M.; Yang, H.; Xu, W.; Ma, S.; Lin, H.; Zhu, H.; Liu, L.; Liu, Y.; Yang, C.; Xu, Y.; et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012, 26, 1326–1338. [Google Scholar] [CrossRef] [PubMed]
- Straton, A.R.; Kischkel, B.; Crișan, T.O.; Joosten, L.A.B. Epigenomic Reprogramming in Gout. Gout Urate Cryst. Depos. Dis. 2024, 2, 325–338. [Google Scholar] [CrossRef]
- Li, G.; Li, D.; Wang, T.; He, S. Pyrimidine Biosynthetic Enzyme CAD: Its Function, Regulation, and Diagnostic Potential. Int. J. Mol. Sci. 2021, 22, 10253. [Google Scholar] [CrossRef]
- Carneiro, A.B.; Iaciura, B.M.F.; Nohara, L.L.; Lopes, C.D.; Veas, E.M.C.; Mariano, V.S.; Bozza, P.T.; Lopes, U.G.; Atella, G.C.; Almeida, I.C.; et al. Lysophosphatidylcholine Triggers TLR2- and TLR4-Mediated Signaling Pathways but Counteracts LPS-Induced NO Synthesis in Peritoneal Macrophages by Inhibiting NF-κB Translocation and MAPK/ERK Phosphorylation. PLoS ONE 2013, 8, e76233. [Google Scholar] [CrossRef]
- Koh, J.H.; Yoon, S.J.; Kim, M.; Cho, S.; Lim, J.; Park, Y.; Kim, H.S.; Kwon, S.W.; Kim, W.U. Lipidome profile predictive of disease evolution and activity in rheumatoid arthritis. Exp. Mol. Med. 2022, 54, 143–155. [Google Scholar] [CrossRef]
- Fuchs, B.; Schiller, J.; Wagner, U.; Häntzschel, H.; Arnold, K. The phosphatidylcholine/lysophosphatidylcholine ratio in human plasma is an indicator of the severity of rheumatoid arthritis: Investigations by 31P NMR and MALDI-TOF MS. Clin. Biochem. 2005, 38, 925–933. [Google Scholar] [CrossRef]
- Knuplez, E.; Marsche, G. An Updated Review of Pro- and Anti-Inflammatory Properties of Plasma Lysophosphatidylcholines in the Vascular System. Int. J. Mol. Sci. 2020, 21, 4501. [Google Scholar] [CrossRef]
- Major, T.J.; Dalbeth, N.; Stahl, E.A.; Merriman, T.R. An update on the genetics of hyperuricaemia and gout. Nat. Rev. Rheumatol. 2018, 14, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Kolz, M.; Johnson, T.; Sanna, S.; Teumer, A.; Vitart, V.; Perola, M.; Mangino, M.; Albrecht, E.; Wallace, C.; Farrall, M.; et al. Meta-Analysis of 28,141 Individuals Identifies Common Variants within Five New Loci That Influence Uric Acid Concentrations. PLoS Genet. 2009, 5, e1000504. [Google Scholar] [CrossRef]
- Saeed, S.; Quintin, J.; Kerstens, H.H.D.; Rao, N.A.; Aghajanirefah, A.; Matarese, F.; Cheng, S.C.; Ratter, J.; Berentsen, K.; van der Ent, M.A.; et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 2014, 345, 1251086. [Google Scholar] [CrossRef] [PubMed]
- Bekkering, S.; Arts, R.J.W.; Novakovic, B.; Kourtzelis, I.; van der Heijden, C.D.C.C.; Li, Y.; Popa, C.D.; ter Horst, R.; van Tuijl, J.; Netea-Maier, R.T.; et al. Metabolic Induction of Trained Immunity through the Mevalonate Pathway. Cell 2018, 172, 135–146.e9. [Google Scholar] [CrossRef]
- Badii, M.; Gaal, O.I.; Cleophas, M.C.; Klück, V.; Davar, R.; Habibi, E.; Keating, S.T.; Novakovic, B.; Helsen, M.M.; Dalbeth, N.; et al. Urate-induced epigenetic modifications in myeloid cells. Arthritis Res. Ther. 2021, 23, 202. [Google Scholar] [CrossRef]
- Cabău, G.; Crișan, T.O.; Klück, V.; Popp, R.A.; Joosten, L.A.B. Urate-induced immune programming: Consequences for gouty arthritis and hyperuricemia. Immunol. Rev. 2020, 294, 92–105. [Google Scholar] [CrossRef]
- Cobo, I.; Murillo-Saich, J.; Alishala, M.; Guma, M. Epigenetic and Metabolic Regulation of Macrophages during Gout. Gout Urate Cryst. Depos. Dis. 2023, 1, 137–151. [Google Scholar] [CrossRef]


| Study | Total Number of Metabolites | Number of Unique Metabolites Colocalized with Gout Loci | Number of Gout Loci Colocalized with Metabolites | Average Number of Metabolites That Colocalized with Gout Locus * | Average Number of Loci That Colocalized with a Metabolite |
|---|---|---|---|---|---|
| METSIM | 1391 | 633 | 135 | 7.12 | 1.56 |
| CLSA | 1400 | 719 | 137 | 7.96 | 1.55 |
| GCKD (plasma) | 1296 | 482 | 136 | 4.74 | 1.36 |
| GCKD (urine) | 1399 | 430 | 138 | 4.17 | 1.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Gout, Hyperuricemia and Crystal Associated Disease Network. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takei, R.; Sumpter, N.A.; Leask, M.P.; Merriman, T.R. Colocalization Analysis Reveals Shared Genetic Loci Contributing to Gout and Metabolite Levels. Gout Urate Cryst. Depos. Dis. 2025, 3, 6. https://doi.org/10.3390/gucdd3020006
Takei R, Sumpter NA, Leask MP, Merriman TR. Colocalization Analysis Reveals Shared Genetic Loci Contributing to Gout and Metabolite Levels. Gout, Urate, and Crystal Deposition Disease. 2025; 3(2):6. https://doi.org/10.3390/gucdd3020006
Chicago/Turabian StyleTakei, Riku, Nicholas A. Sumpter, Megan P. Leask, and Tony R. Merriman. 2025. "Colocalization Analysis Reveals Shared Genetic Loci Contributing to Gout and Metabolite Levels" Gout, Urate, and Crystal Deposition Disease 3, no. 2: 6. https://doi.org/10.3390/gucdd3020006
APA StyleTakei, R., Sumpter, N. A., Leask, M. P., & Merriman, T. R. (2025). Colocalization Analysis Reveals Shared Genetic Loci Contributing to Gout and Metabolite Levels. Gout, Urate, and Crystal Deposition Disease, 3(2), 6. https://doi.org/10.3390/gucdd3020006








