Advances in Conventional and Extended Fluorescence Correlation Spectroscopy for the Analysis of Biological Clusters and Aggregates
Abstract
1. Introduction
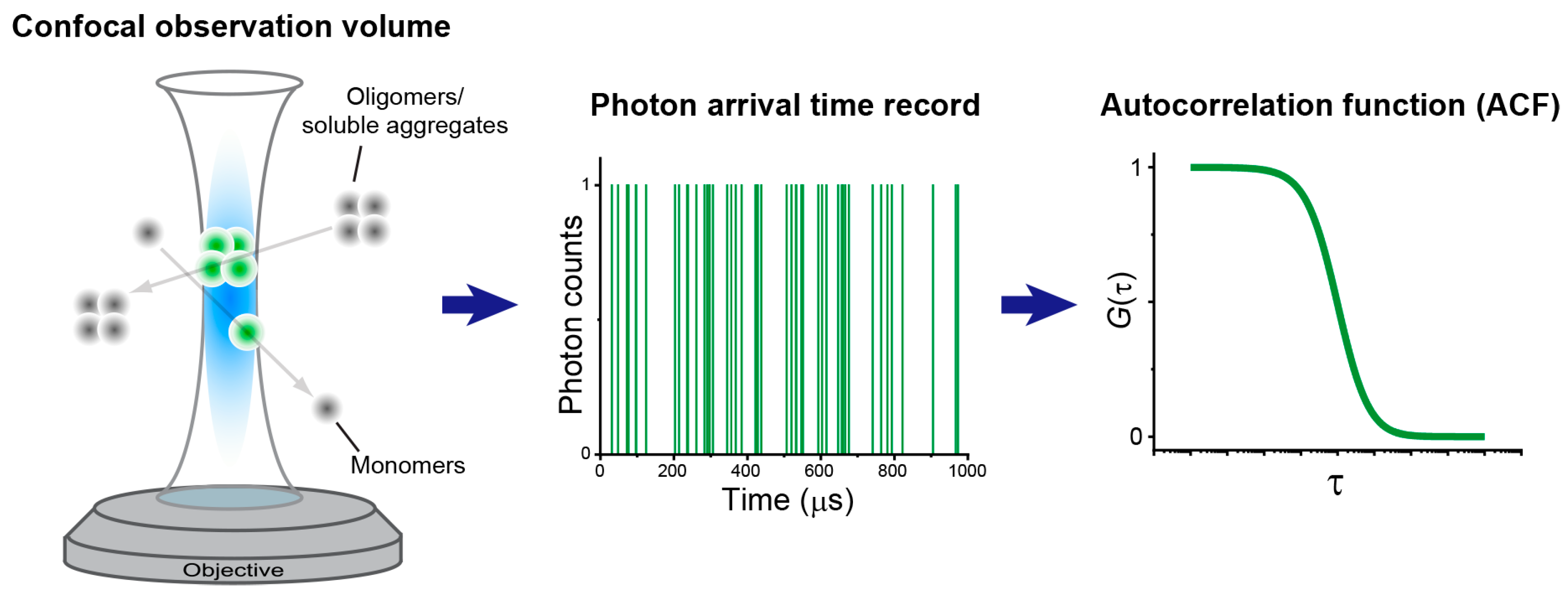
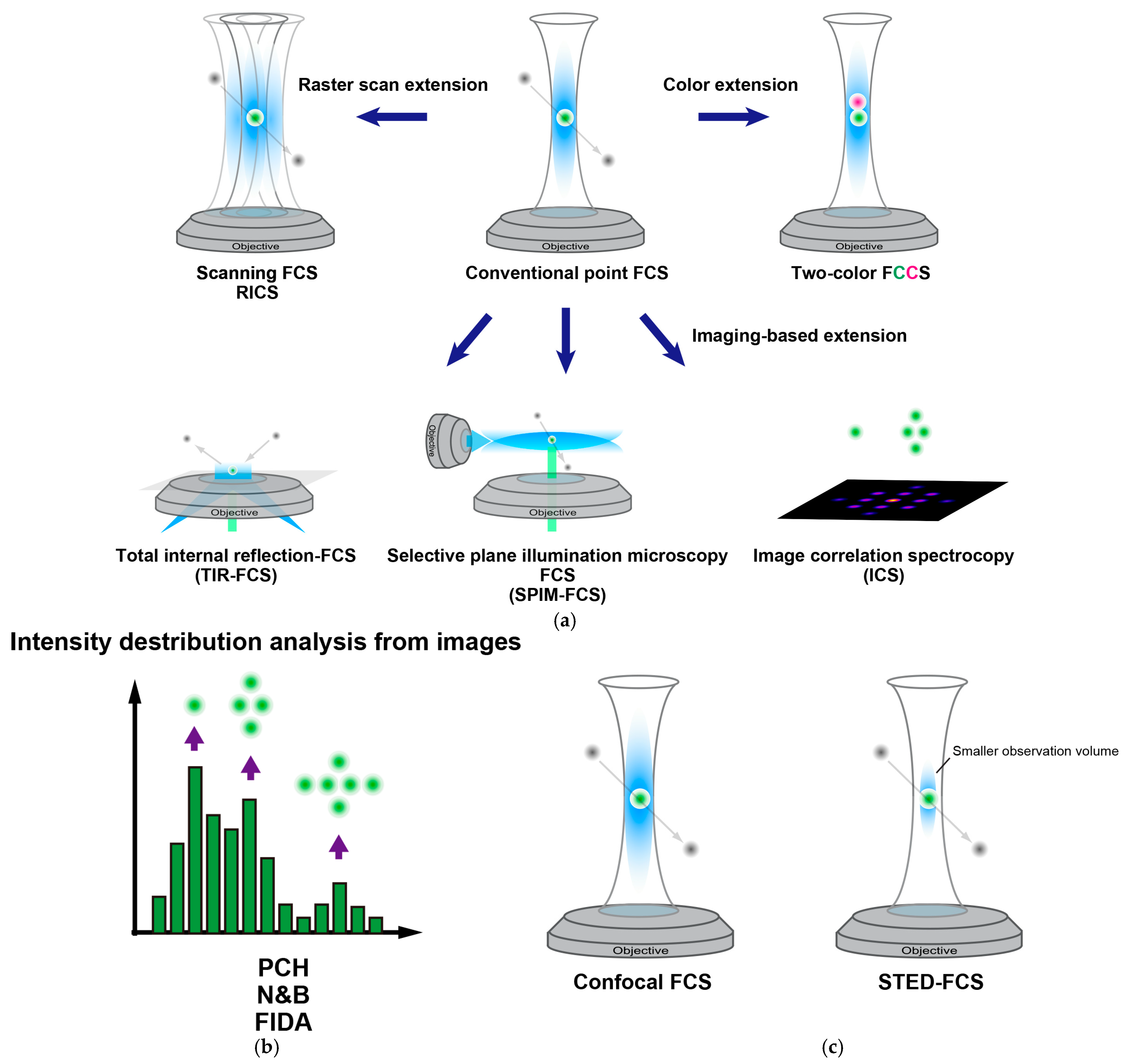
2. Principles and Extensions of FCS
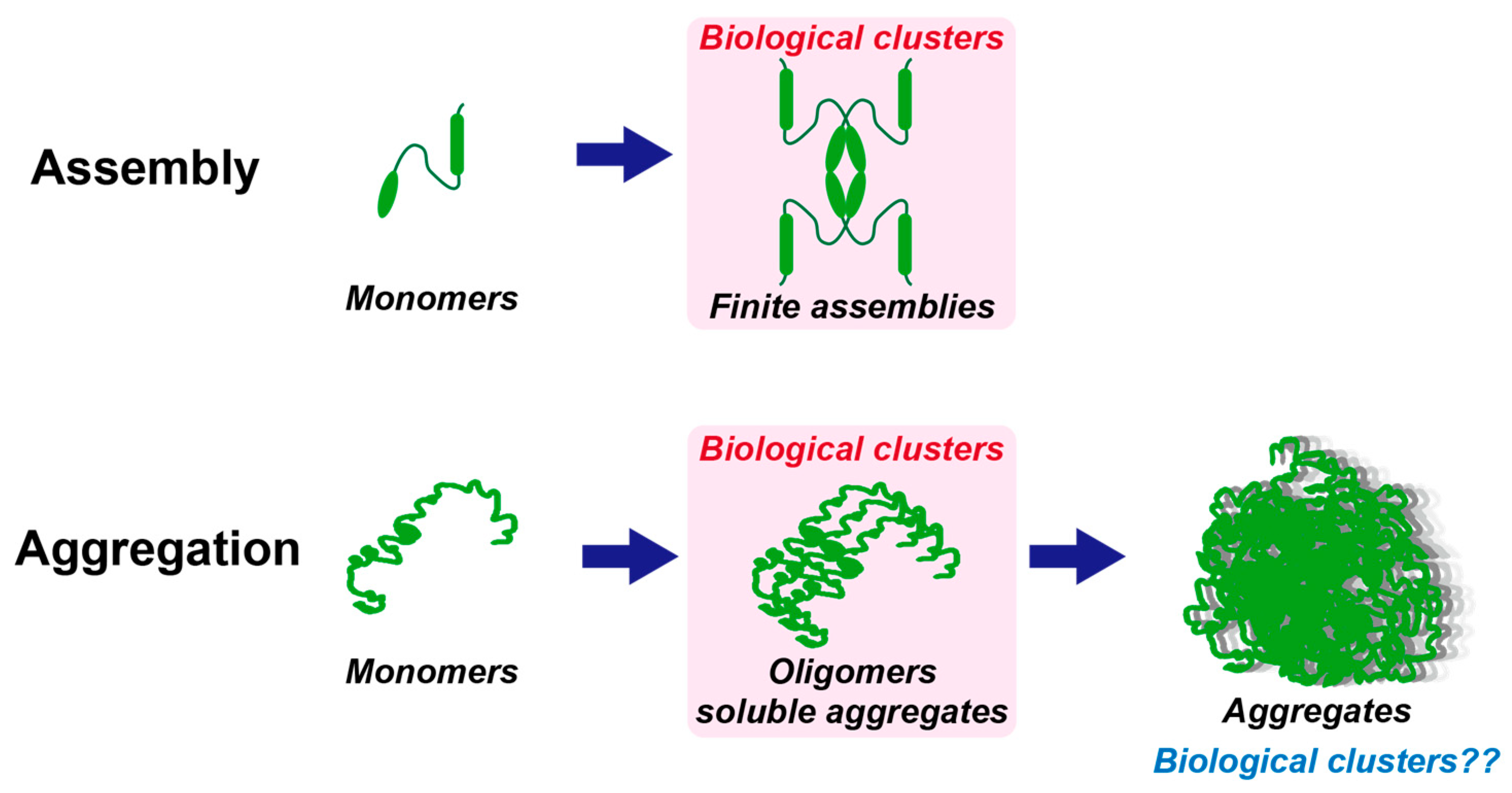
3. Definition and Properties of Biological Clusters
4. Applications of FCS to Biological Clusters and Aggregates
4.1. Protein Clusters
4.2. Protein Aggregates
4.3. Nucleic Acid Clusters
4.4. Lipid Clusters
5. Experimental Considerations and Artifacts
6. Future Perspectives
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| DNA | Deoxyribonucleic acid |
| FRAP | Fluorescence recovery after photobleaching |
| RNA | Ribonucleic acid |
References
- Toledo, P.L.; Gianotti, A.R.; Vazquez, D.S.; Ermacora, M.R. Protein nanocondensates: The next frontier. Biophys. Rev. 2023, 15, 515–530. [Google Scholar] [CrossRef]
- Alberti, S.; Hyman, A.A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 196–213. [Google Scholar] [CrossRef]
- Ripin, N.; Parker, R. Formation, function, and pathology of RNP granules. Cell 2023, 186, 4737–4756. [Google Scholar] [CrossRef]
- Levental, I.; Levental, K.R.; Heberle, F.A. Lipid Rafts: Controversies Resolved, Mysteries Remain. Trends Cell Biol. 2020, 30, 341–353. [Google Scholar] [CrossRef]
- Morris, R.; Black, K.A.; Stollar, E.J. Uncovering protein function: From classification to complexes. Essays Biochem. 2022, 66, 255–285. [Google Scholar] [CrossRef]
- Haustein, E.; Schwille, P. Fluorescence correlation spectroscopy: Novel variations of an established technique. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Magde, D.; Elson, E.; Webb, W.W. Thermodynamic Fluctuations in a Reacting System—Measurement by Fluorescence Correlation Spectroscopy. Phys. Rev. Lett. 1972, 29, 705–708. [Google Scholar] [CrossRef]
- Elson, E.L.; Magde, D. Fluorescence correlation spectroscopy. I. Conceptual basis and theory. Biopolymers 1974, 13, 1–27. [Google Scholar] [CrossRef]
- Magde, D.; Elson, E.L.; Webb, W.W. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers 1974, 13, 29–61. [Google Scholar] [CrossRef] [PubMed]
- Ehrenberg, M.; Rigler, R. Rotational brownian motion and fluorescence intensify fluctuations. Chem. Phys. 1974, 4, 390–401. [Google Scholar] [CrossRef]
- Borejdo, J. Motion of myosin fragments during actin-activated ATPase: Fluorescence correlation spectroscopy study. Biopolymers 1979, 18, 2807–2820. [Google Scholar] [CrossRef]
- Thompson, N.L.; Axelrod, D. Immunoglobulin surface-binding kinetics studied by total internal reflection with fluorescence correlation spectroscopy. Biophys. J. 1983, 43, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Elson, E.L. Analysis of confocal laser-microscope optics for 3-D fluorescence correlation spectroscopy. Appl. Opt. 1991, 30, 1185–1195. [Google Scholar] [CrossRef]
- Rigler, R.; Mets, U.; Widengren, J.; Kask, P. Fluorescence Correlation Spectroscopy with High Count Rate and Low-Background—Analysis of Translational Diffusion. Eur. Biophys. J. Biophy 1993, 22, 169–175. [Google Scholar] [CrossRef]
- Elson, E.L. Fluorescence correlation spectroscopy: Past, present, future. Biophys. J. 2011, 101, 2855–2870. [Google Scholar] [CrossRef]
- Kim, S.A.; Heinze, K.G.; Schwille, P. Fluorescence correlation spectroscopy in living cells. Nat. Methods 2007, 4, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, A.; Kinjo, M. State-of-the-Art Fluorescence Fluctuation-Based Spectroscopic Techniques for the Study of Protein Aggregation. Int. J. Mol. Sci. 2018, 19, 964. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Meuvis, J.; Hendrix, J.; Carl, S.A.; Engelborghs, Y. Early aggregation steps in alpha-synuclein as measured by FCS and FRET: Evidence for a contagious conformational change. Biophys. J. 2010, 98, 1302–1311. [Google Scholar] [CrossRef]
- Chakraborty, M.; Kuriata, A.M.; Nathan Henderson, J.; Salvucci, M.E.; Wachter, R.M.; Levitus, M. Protein oligomerization monitored by fluorescence fluctuation spectroscopy: Self-assembly of rubisco activase. Biophys. J. 2012, 103, 949–958. [Google Scholar] [CrossRef]
- Tsutsumi, M.; Muto, H.; Myoba, S.; Kimoto, M.; Kitamura, A.; Kamiya, M.; Kikukawa, T.; Takiya, S.; Demura, M.; Kawano, K.; et al. In vivo fluorescence correlation spectroscopy analyses of FMBP-1, a silkworm transcription factor. Febs Open Bio 2016, 6, 106–125. [Google Scholar] [CrossRef]
- Kohler, J.; Hur, K.H.; Mueller, J.D. Statistical analysis of the autocorrelation function in fluorescence correlation spectroscopy. Biophys. J. 2024, 123, 667–680. [Google Scholar] [CrossRef]
- Zhao, M.; Jin, L.; Chen, B.; Ding, Y.; Ma, H.; Chen, D. Afterpulsing and its correction in fluorescence correlation spectroscopy experiments. Appl. Opt. 2003, 42, 4031–4036. [Google Scholar] [CrossRef]
- Sun, G.; Guo, S.M.; Teh, C.; Korzh, V.; Bathe, M.; Wohland, T. Bayesian model selection applied to the analysis of fluorescence correlation spectroscopy data of fluorescent proteins in vitro and in vivo. Anal. Chem. 2015, 87, 4326–4333. [Google Scholar] [CrossRef]
- Enderlein, J. Machine learning and advanced statistical analysis for fluorescence correlation spectroscopy. Biophys. J. 2024, 123, 651–652. [Google Scholar] [CrossRef]
- Bacia, K.; Kim, S.A.; Schwille, P. Fluorescence cross-correlation spectroscopy in living cells. Nat. Methods 2006, 3, 83–89. [Google Scholar] [CrossRef]
- Mikuni, S.; Kodama, K.; Sasaki, A.; Kohira, N.; Maki, H.; Munetomo, M.; Maenaka, K.; Kinjo, M. Screening for FtsZ Dimerization Inhibitors Using Fluorescence Cross-Correlation Spectroscopy and Surface Resonance Plasmon Analysis. PLoS ONE 2015, 10, e0130933. [Google Scholar] [CrossRef]
- Hwang, L.C.; Gosch, M.; Lasser, T.; Wohland, T. Simultaneous multicolor fluorescence cross-correlation spectroscopy to detect higher order molecular interactions using single wavelength laser excitation. Biophys. J. 2006, 91, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Previte, M.J.; Pelet, S.; Kim, K.H.; Buehler, C.; So, P.T. Spectrally resolved fluorescence correlation spectroscopy based on global analysis. Anal. Chem. 2008, 80, 3277–3284. [Google Scholar] [CrossRef] [PubMed]
- Dunsing, V.; Petrich, A.; Chiantia, S. Multicolor fluorescence fluctuation spectroscopy in living cells via spectral detection. eLife 2021, 10, e69687. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Muller, J.D.; So, P.T.; Gratton, E. The photon counting histogram in fluorescence fluctuation spectroscopy. Biophys. J. 1999, 77, 553–567. [Google Scholar] [CrossRef] [PubMed]
- Digman, M.A.; Dalal, R.; Horwitz, A.F.; Gratton, E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys. J. 2008, 94, 2320–2332. [Google Scholar] [CrossRef]
- Godin, A.G.; Rappaz, B.; Potvin-Trottier, L.; Kennedy, T.E.; De Koninck, Y.; Wiseman, P.W. Spatial Intensity Distribution Analysis Reveals Abnormal Oligomerization of Proteins in Single Cells. Biophys. J. 2015, 109, 710–721. [Google Scholar] [CrossRef]
- Vetri, V.; Ossato, G.; Militello, V.; Digman, M.A.; Leone, M.; Gratton, E. Fluctuation methods to study protein aggregation in live cells: Concanavalin A oligomers formation. Biophys. J. 2011, 100, 774–783. [Google Scholar] [CrossRef]
- Scales, N.; Swain, P.S. Resolving fluorescent species by their brightness and diffusion using correlated photon-counting histograms. PLoS ONE 2019, 14, e0226063. [Google Scholar] [CrossRef]
- Yu, L.; Lei, Y.; Ma, Y.; Liu, M.; Zheng, J.; Dan, D.; Gao, P. A Comprehensive Review of Fluorescence Correlation Spectroscopy. Front. Phys. 2021, 9, 644450. [Google Scholar] [CrossRef]
- Digman, M.A.; Wiseman, P.W.; Horwitz, A.R.; Gratton, E. Detecting protein complexes in living cells from laser scanning confocal image sequences by the cross correlation raster image spectroscopy method. Biophys. J. 2009, 96, 707–716. [Google Scholar] [CrossRef]
- Digman, M.A.; Brown, C.M.; Sengupta, P.; Wiseman, P.W.; Horwitz, A.R.; Gratton, E. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys. J. 2005, 89, 1317–1327. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P.; Wohland, T. Applications of imaging fluorescence correlation spectroscopy. Curr. Opin. Chem. Biol. 2014, 20, 29–35. [Google Scholar] [CrossRef]
- Petrasek, Z.; Schwille, P. Precise measurement of diffusion coefficients using scanning fluorescence correlation spectroscopy. Biophys. J. 2008, 94, 1437–1448. [Google Scholar] [CrossRef]
- Fujioka, Y.; Alam, J.M.; Noshiro, D.; Mouri, K.; Ando, T.; Okada, Y.; May, A.I.; Knorr, R.L.; Suzuki, K.; Ohsumi, Y.; et al. Phase separation organizes the site of autophagosome formation. Nature 2020, 578, 301–305. [Google Scholar] [CrossRef]
- Widengren, J.; Rigler, R.; Mets, Ü. Triplet-state monitoring by fluorescence correlation spectroscopy. J. Fluoresc. 1994, 4, 255–258. [Google Scholar] [CrossRef]
- Widengren, J.; Mets, U.; Rigler, R. Fluorescence Correlation Spectroscopy of Triplet-States in Solution—A Theoretical and Experimental-Study. J. Phys. Chem. 1995, 99, 13368–13379. [Google Scholar] [CrossRef]
- Hinde, E.; Pandzic, E.; Yang, Z.; Ng, I.H.; Jans, D.A.; Bogoyevitch, M.A.; Gratton, E.; Gaus, K. Quantifying the dynamics of the oligomeric transcription factor STAT3 by pair correlation of molecular brightness. Nat. Commun. 2016, 7, 11047. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, J.; Mikuni, S.; Kinjo, M. Multipoint fluorescence correlation spectroscopy using spatial light modulator. Biomed. Opt. Express 2018, 9, 5881–5890. [Google Scholar] [CrossRef]
- Ma, Y.; Pandzic, E.; Nicovich, P.R.; Yamamoto, Y.; Kwiatek, J.; Pageon, S.V.; Benda, A.; Rossy, J.; Gaus, K. An intermolecular FRET sensor detects the dynamics of T cell receptor clustering. Nat. Commun. 2017, 8, 15100. [Google Scholar] [CrossRef] [PubMed]
- Kastrup, L.; Blom, H.; Eggeling, C.; Hell, S.W. Fluorescence Fluctuation Spectroscopy in Subdiffraction Focal Volumes. Phys. Rev. Lett. 2005, 94, 178104. [Google Scholar] [CrossRef] [PubMed]
- Sankaran, J.; Wohland, T. Current capabilities and future perspectives of FCS: Super-resolution microscopy, machine learning, and in vivo applications. Commun. Biol. 2023, 6, 699. [Google Scholar] [CrossRef]
- Koukos, P.I.; Bonvin, A.M.J.J. Integrative Modelling of Biomolecular Complexes. J. Mol. Biol. 2020, 432, 2861–2881. [Google Scholar] [CrossRef]
- Ariyoshi, M.; Fukagawa, T. An updated view of the kinetochore architecture. Trends Genet. 2023, 39, 941–953. [Google Scholar] [CrossRef]
- Liu, W.; Wang, H.; Xu, C. Antigen Receptor Nanoclusters: Small Units with Big Functions. Trends Immunol. 2016, 37, 680–689. [Google Scholar] [CrossRef]
- Housmans, J.A.J.; Wu, G.; Schymkowitz, J.; Rousseau, F. A guide to studying protein aggregation. FEBS J. 2023, 290, 554–583. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Rinauro, D.J.; Chiti, F.; Vendruscolo, M.; Limbocker, R. Misfolded protein oligomers: Mechanisms of formation, cytotoxic effects, and pharmacological approaches against protein misfolding diseases. Mol. Neurodegener. 2024, 19, 20. [Google Scholar] [CrossRef]
- Mahen, R. cNap1 bridges centriole contact sites to maintain centrosome cohesion. PLoS Biol. 2022, 20, e3001854. [Google Scholar] [CrossRef]
- Kilpatrick, L.E.; Hill, S.J. The use of fluorescence correlation spectroscopy to characterise the molecular mobility of G protein-coupled receptors in membrane microdomains: An update. Biochem. Soc. Trans. 2021, 49, 1547–1554. [Google Scholar] [CrossRef]
- Kaliszewski, M.J.; Shi, X.; Hou, Y.; Lingerak, R.; Kim, S.; Mallory, P.; Smith, A.W. Quantifying membrane protein oligomerization with fluorescence cross-correlation spectroscopy. Methods 2018, 140–141, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Bag, N.; Holowka, D.A.; Baird, B.A. Imaging FCS delineates subtle heterogeneity in plasma membranes of resting mast cells. Mol. Biol. Cell 2020, 31, 709–723. [Google Scholar] [CrossRef] [PubMed]
- Garai, K.; Sureka, R.; Maiti, S. Detecting amyloid-beta aggregation with fiber-based fluorescence correlation spectroscopy. Biophys. J. 2007, 92, L55–L57. [Google Scholar] [CrossRef]
- Lazaro-Alfaro, A.; Nicholas, S.L.N.; Sanabria, H. FRET-FCS: Advancing comprehensive insights into complex biological systems. Biophys. J. 2025, 124, 3329–3341. [Google Scholar] [CrossRef]
- Wennmalm, S.; Chmyrov, V.; Widengren, J.; Tjernberg, L. Highly Sensitive FRET-FCS Detects Amyloid beta-Peptide Oligomers in Solution at Physiological Concentrations. Anal. Chem. 2015, 87, 11700–11705. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.; Gracia, P.; Navarro, S.; Pena-Diaz, S.; Pujols, J.; Cremades, N.; Pallares, I.; Ventura, S. alpha-Helical peptidic scaffolds to target alpha-synuclein toxic species with nanomolar affinity. Nat. Commun. 2021, 12, 3752. [Google Scholar] [CrossRef]
- Beam, M.; Silva, M.C.; Morimoto, R.I. Dynamic imaging by fluorescence correlation spectroscopy identifies diverse populations of polyglutamine oligomers formed in vivo. J. Biol. Chem. 2012, 287, 26136–26145. [Google Scholar] [CrossRef]
- Kitamura, A.; Fujimoto, A.; Kawashima, R.; Lyu, Y.; Sasaki, K.; Hamada, Y.; Moriya, K.; Kurata, A.; Takahashi, K.; Brielmann, R.; et al. Hetero-oligomerization of TDP-43 carboxy-terminal fragments with cellular proteins contributes to proteotoxicity. Commun. Biol. 2024, 7, 743. [Google Scholar] [CrossRef]
- Das, B.; Roychowdhury, S.; Mohanty, P.; Rizuan, A.; Chakraborty, J.; Mittal, J.; Chattopadhyay, K. A Zn-dependent structural transition of SOD1 modulates its ability to undergo phase separation. EMBO J. 2023, 42, e111185. [Google Scholar] [CrossRef] [PubMed]
- Abu-Arish, A.; Pandzic, E.; Luo, Y.; Sato, Y.; Turner, M.J.; Wiseman, P.W.; Hanrahan, J.W. Lipid-driven CFTR clustering is impaired in cystic fibrosis and restored by corrector drugs. J. Cell Sci. 2022, 135, jcs259002. [Google Scholar] [CrossRef] [PubMed]
- Kolin, D.L.; Wiseman, P.W. Advances in image correlation spectroscopy: Measuring number densities, aggregation states, and dynamics of fluorescently labeled macromolecules in cells. Cell Biochem. Biophys. 2007, 49, 141–164. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamamoto, J.; Kinjo, M.; Noda, N. Absolute Quantification of RNA Molecules Using Fluorescence Correlation Spectroscopy with Certified Reference Materials. Anal. Chem. 2018, 90, 10865–10871. [Google Scholar] [CrossRef]
- Chabot, N.M.; Purkanti, R.; Del Panta Ridolfi, A.; Nogare, D.D.; Oda, H.; Kimura, H.; Jug, F.; Dal Co, A.; Vastenhouw, N.L. Local DNA compaction creates TF-DNA clusters that enable transcription. bioRxiv 2024. [Google Scholar] [CrossRef]
- Kitamura, A.; Tornmalm, J.; Demirbay, B.; Piguet, J.; Kinjo, M.; Widengren, J. Trans-cis isomerization kinetics of cyanine dyes reports on the folding states of exogeneous RNA G-quadruplexes in live cells. Nucleic Acids Res. 2023, 51, e27. [Google Scholar] [CrossRef]
- Eggeling, C.; Ringemann, C.; Medda, R.; Schwarzmann, G.; Sandhoff, K.; Polyakova, S.; Belov, V.N.; Hein, B.; von Middendorff, C.; Schonle, A.; et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 2009, 457, 1159–1162. [Google Scholar] [CrossRef] [PubMed]
- Schwille, P.; Korlach, J.; Webb, W.W. Fluorescence correlation spectroscopy with single-molecule sensitivity on cell and model membranes. Cytometry 1999, 36, 176–182. [Google Scholar] [CrossRef]
- Rose, M.; Hirmiz, N.; Moran-Mirabal, J.M.; Fradin, C. Lipid Diffusion in Supported Lipid Bilayers: A Comparison between Line-Scanning Fluorescence Correlation Spectroscopy and Single-Particle Tracking. Membranes 2015, 5, 702–721. [Google Scholar] [CrossRef]
- Di Rienzo, C.; Gratton, E.; Beltram, F.; Cardarelli, F. Fast spatiotemporal correlation spectroscopy to determine protein lateral diffusion laws in live cell membranes. Proc. Natl. Acad. Sci. USA 2013, 110, 12307–12312. [Google Scholar] [CrossRef]
- Rubio, V.; McInchak, N.; Fernandez, G.; Benavides, D.; Herrera, D.; Jimenez, C.; Mesa, H.; Meade, J.; Zhang, Q.; Stawikowski, M.J. Development and characterization of fluorescent cholesteryl probes with enhanced solvatochromic and pH-sensitive properties for live-cell imaging. Sci. Rep. 2024, 14, 30777. [Google Scholar] [CrossRef]
- Davis, L.M.; Shen, G. Accounting for triplet and saturation effects in FCS measurements. Curr. Pharm. Biotechnol. 2006, 7, 287–301. [Google Scholar] [CrossRef]
- Hodges, C.; Kafle, R.P.; Hoff, J.D.; Meiners, J.-C. Fluorescence Correlation Spectroscopy with Photobleaching Correction in Slowly Diffusing Systems. J. Fluoresc. 2018, 28, 505–511. [Google Scholar] [CrossRef]
- Pinto-Cámara, R.; Linares, A.; Moreno-Gutiérrez, D.S.; Hernández, H.O.; Martínez-Reyes, J.D.; Rendón-Mancha, J.M.; Wood, C.D.; Guerrero, A. FCSlib: An open-source tool for fluorescence fluctuation spectroscopy analysis for mobility, number and molecular brightness in R. Bioinformatics 2020, 37, 1930–1931. [Google Scholar] [CrossRef] [PubMed]
- Ziarkash, A.W.; Joshi, S.K.; Stipcevic, M.; Ursin, R. Comparative study of afterpulsing behavior and models in single photon counting avalanche photo diode detectors. Sci. Rep. 2018, 8, 5076. [Google Scholar] [CrossRef] [PubMed]
- Nagy, A.; Wu, J.; Berland, K.M. Characterizing observation volumes and the role of excitation saturation in one-photon fluorescence fluctuation spectroscopy. J. Biomed. Opt. 2005, 10, 44015. [Google Scholar] [CrossRef] [PubMed]
- Seltmann, A.; Carravilla, P.; Reglinski, K.; Eggeling, C.; Waithe, D. Neural network informed photon filtering reduces fluorescence correlation spectroscopy artifacts. Biophys. J. 2024, 123, 745–755. [Google Scholar] [CrossRef]
- Eilers, Y.; Ta, H.; Gwosch, K.C.; Balzarotti, F.; Hell, S.W. MINFLUX monitors rapid molecular jumps with superior spatiotemporal resolution. Proc. Natl. Acad. Sci. USA 2018, 115, 6117–6122. [Google Scholar] [CrossRef]
- Slenders, E.; Castello, M.; Buttafava, M.; Villa, F.; Tosi, A.; Lanzano, L.; Koho, S.V.; Vicidomini, G. Confocal-based fluorescence fluctuation spectroscopy with a SPAD array detector. Light. Sci. Appl. 2021, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Sanden, T.; Persson, G.; Thyberg, P.; Blom, H.; Widengren, J. Monitoring kinetics of highly environment sensitive states of fluorescent molecules by modulated excitation and time-averaged fluorescence intensity recording. Anal. Chem. 2007, 79, 3330–3341. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Piguet, J.; Baryshnikov, G.; Tornmalm, J.; Demirbay, B.; Ågren, H.; Widengren, J. Imaging Fluorescence Blinking of a Mitochondrial Localization Probe: Cellular Localization Probes Turned into Multifunctional Sensors. J. Phys. Chem. B 2022, 126, 3048–3058. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, E.; Demirbay, B.; Kulkarni, A.; Liu, H.; Piguet, J.; Widengren, J. Fluorescence Bar-Coding and Flowmetry Based on Dark State Transitions in Fluorescence Emitters. J. Phys. Chem. B 2024, 128, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.; Sim, S.R.; Aik, D.Y.K.; Nelanuthala, A.V.S.; Athilingam, T.; Rollin, A.; Wohland, T. Deep learning reduces data requirements and allows real-time measurements in imaging FCS. Biophys. J. 2024, 123, 655–666. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kitamura, A. Advances in Conventional and Extended Fluorescence Correlation Spectroscopy for the Analysis of Biological Clusters and Aggregates. Spectrosc. J. 2025, 3, 31. https://doi.org/10.3390/spectroscj3040031
Kitamura A. Advances in Conventional and Extended Fluorescence Correlation Spectroscopy for the Analysis of Biological Clusters and Aggregates. Spectroscopy Journal. 2025; 3(4):31. https://doi.org/10.3390/spectroscj3040031
Chicago/Turabian StyleKitamura, Akira. 2025. "Advances in Conventional and Extended Fluorescence Correlation Spectroscopy for the Analysis of Biological Clusters and Aggregates" Spectroscopy Journal 3, no. 4: 31. https://doi.org/10.3390/spectroscj3040031
APA StyleKitamura, A. (2025). Advances in Conventional and Extended Fluorescence Correlation Spectroscopy for the Analysis of Biological Clusters and Aggregates. Spectroscopy Journal, 3(4), 31. https://doi.org/10.3390/spectroscj3040031






