Histone Arginine Methylation in the Kidneys of Rana sylvatica During Freeze–Thaw Cycle
Abstract
1. Introduction
2. Results
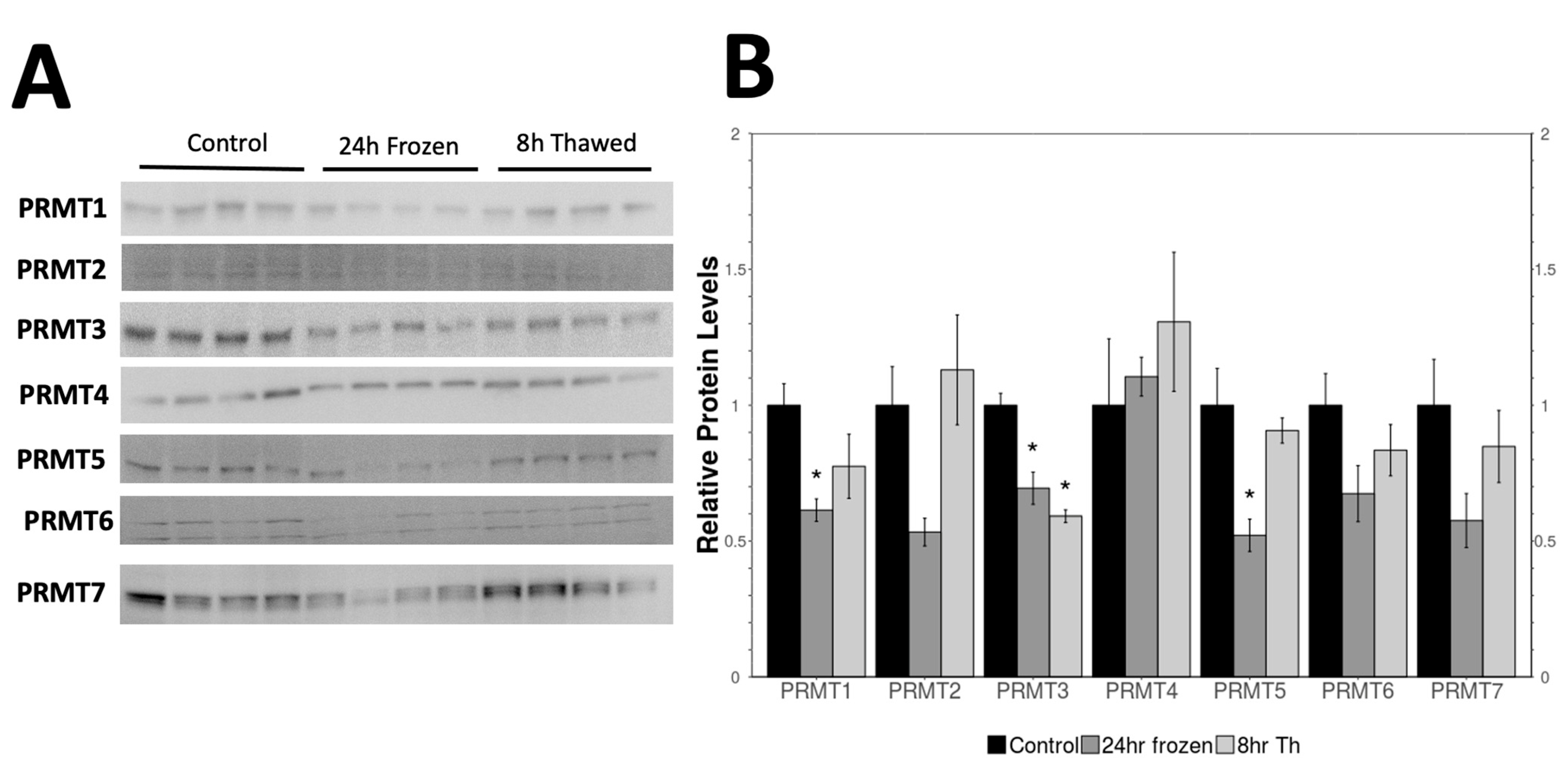
2.1. Expression of PRMTs in the Control, Frozen, and Thawed Samples
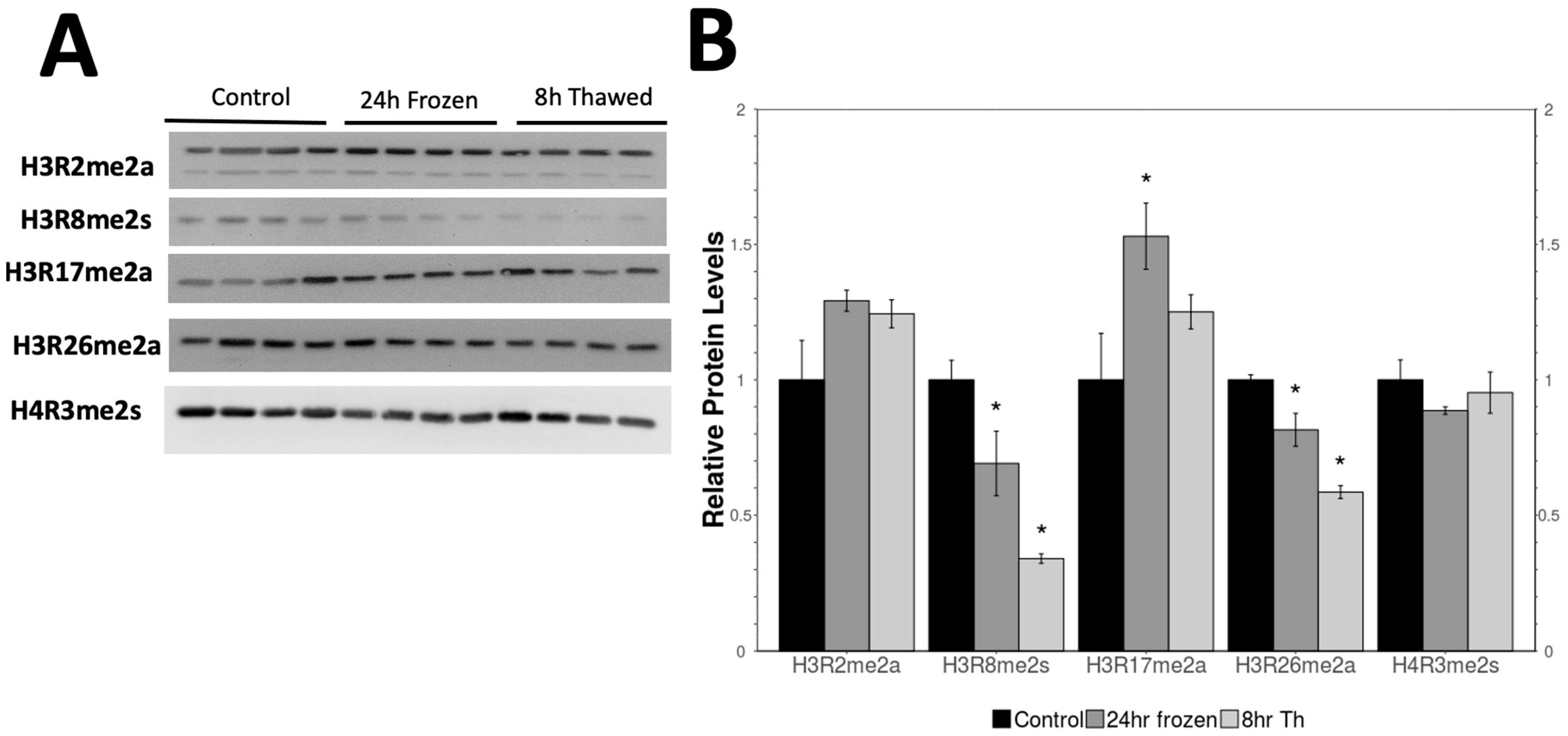
2.2. Analysis of Methylated Histone Levels over the Freeze–Thaw Cycle
2.3. Expression of Readers and Demethylases
3. Discussion
3.1. Role of Protein Arginine Methyltransferases (PRMTs) in Freezing Tolerance
3.2. Histone Arginine Methylation and Gene Regulation During Freezing Stress
4. Materials and Methods
4.1. Ethics Statement
4.2. Animal Collection
4.3. Total Protein Isolation
4.4. Histone Isolation
4.5. Western Immunoblotting
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Gibney, E.R.; Nolan, C.M. Epigenetics and gene expression. Heredity 2010, 105, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002, 12, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002, 12, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Wani, A.A. Histone Modifications: Crucial Elements for Damage Response and Chromatin Restoration. J. Cell. Physiol. 2010, 223, 283. [Google Scholar] [CrossRef]
- Martin, C.; Zhang, Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005, 6, 838–849. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, Z.; Jia, J.; Du, T.; Zhang, N.; Tang, Y.; Fang, Y.; Fang, D. Overview of Histone Modification. Adv. Exp. Med. Biol. 2021, 1283, 1–16. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef]
- Miller, J.L.; Grant, P.A. The Role of DNA Methylation and Histone Modifications in Transcriptional Regulation in Humans. Sub-Cell. Biochem. 2013, 61, 289. [Google Scholar] [CrossRef]
- Zhang, J.; Jing, L.; Li, M.; He, L.; Guo, Z. Regulation of histone arginine methylation/demethylation by methylase and demethylase. Mol. Med. Rep. 2019, 19, 3963. [Google Scholar] [CrossRef]
- Wang, T.S.; Cheng, J.K.; Lei, Q.Y.; Wang, Y.P. A Switch for Transcriptional Activation and Repression: Histone Arginine Methylation. In RNA Technologies; Springer: Cham, Switzerland, 2019; pp. 521–541. [Google Scholar] [CrossRef]
- Lorton, B.M.; Shechter, D. Cellular consequences of arginine methylation. Cell. Mol. Life Sci. 2019, 76, 2933–2956. [Google Scholar] [CrossRef]
- Litt, M.; Qiu, Y.; Huang, S. Histone arginine methylations: Their roles in chromatin dynamics and transcriptional regulation. Biosci. Rep. 2009, 29, 131. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.W.; Cho, Y.; Bae, G.U.; Kim, S.N.; Kim, Y.K. Protein arginine methyltransferases: Promising targets for cancer therapy. Exp. Mol. Med. 2021, 53, 788–808. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Al-attar, R.; Storey, K.B. Lessons from nature: Leveraging the freeze-tolerant wood frog as a model to improve organ cryopreservation and biobanking. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2022, 261, 110747. [Google Scholar] [CrossRef]
- Luu, B.E.; Storey, K.B. Solving Donor Organ Shortage with Insights from Freeze Tolerance in Nature: Activating endogenous antioxidant systems with non-coding RNA to precondition donor organs. BioEssays News Rev. Mol. Cell. Dev. Biol. 2018, 40, 1800092. [Google Scholar] [CrossRef]
- Bedford, M.T.; Richard, S. Arginine methylation an emerging regulator of protein function. Mol. Cell 2005, 18, 263–272. [Google Scholar] [CrossRef]
- Di Lorenzo, A.; Bedford, M.T. Histone arginine methylation. FEBS Lett. 2011, 585, 2024–2031. [Google Scholar] [CrossRef]
- Chang, B.; Chen, Y.; Zhao, Y.; Bruick, R.K. JMJD6 is a histone arginine demethylase. Science 2007, 318, 444–447. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y. Peptidylarginine deiminases in citrullination, gene regulation, health and pathogenesis. Biochim. Biophys. Acta 2013, 1829, 1126–1135. [Google Scholar] [CrossRef]
- Walport, L.J.; Hopkinson, R.J.; Chowdhury, R.; Schiller, R.; Ge, W.; Kawamura, A.; Schofield, C.J. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 2016, 7, 11974. [Google Scholar] [CrossRef]
- Hawkins, L.J.; Storey, K.B. Histone methylation in the freeze-tolerant wood frog (Rana sylvatica). J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2018, 188, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Taiwo, O.O.; Breedon, S.A.; Storey, K.B. Epigenetic Regulation by Histone Methylation and Demethylation in Freeze-Tolerant Frog Kidney. Cell Biochem. Funct. 2024, 42, e70036. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, J.P.; Lee, R.E. Cryoprotection by urea in a terrestrially hibernating frog. J. Exp. Biol. 2005, 208 Pt 21, 4079–4089. [Google Scholar] [CrossRef] [PubMed]
- Storey, K.B. Glycolysis and the regulation of cryoprotectant synthesis in liver of the freeze tolerant wood frog. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 1987, 157, 373–380. [Google Scholar] [CrossRef]
- Do Amaral, M.C.F.; Lee, R.E.; Costanzo, J.P. Enzymatic Regulation of Glycogenolysis in a Subarctic Population of the Wood Frog: Implications for Extreme Freeze Tolerance. PLoS ONE 2013, 8, e79169. [Google Scholar] [CrossRef]
- Sinclair, B.J.; Stinziano, J.R.; Williams, C.M.; MacMillan, H.A.; Marshall, K.E.; Storey, K.B. Real-time measurement of metabolic rate during freezing and thawing of the wood frog, Rana sylvatica: Implications for overwinter energy use. J. Exp. Biol. 2013, 216, 292–302. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Molecular Physiology of Freeze Tolerance in Vertebrates. Physiol. Rev. 2017, 97, 623–665. [Google Scholar] [CrossRef]
- Sims, R.J.; Nishioka, K.; Reinberg, D. Histone lysine methylation: A signature for chromatin function. Trends Genet. 2003, 19, 629–639. [Google Scholar] [CrossRef]
- Gorr, T.A. Hypometabolism as the ultimate defence in stress response: How the comparative approach helps understanding of medically relevant questions. Acta Physiol. 2017, 219, 409–440. [Google Scholar] [CrossRef]
- Gui, S.; Gathiaka, S.; Li, J.; Qu, J.; Acevedo, O.; Hevel, J.M. A Remodeled Protein Arginine Methyltransferase 1 (PRMT1) Generates Symmetric Dimethylarginine. J. Biol. Chem. 2014, 289, 9320. [Google Scholar] [CrossRef] [PubMed]
- Bedford, M.T.; Clarke, S.G. Protein Arginine Methylation in Mammals: Who, What, and Why. Mol. Cell 2009, 33, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tewary, S.K.; Zheng, Y.G.; Ho, M.C. Protein arginine methyltransferases: Insights into the enzyme structure and mechanism at the atomic level. Cell. Mol. Life Sci. 2019, 76, 2917–2932. [Google Scholar] [CrossRef]
- Stopa, N.; Krebs, J.E.; Shechter, D. The PRMT5 arginine methyltransferase: Many roles in development, cancer and beyond. Cell. Mol. Life Sci. 2015, 72, 2041–2059. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Zeng, J.; Liu, D.-F.; Zhao, Y.; Yang, H.-L.; Li, Y.; Qiu, P.; Tang, M.-W. Abnormalities of the PRMT1-ADMA-DDAH1 metabolism axis and probucol treatment in diabetic patients and diabetic rats. Ann. Palliat. Med. 2021, 10, 3343–3353. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, H.; Li, J.; Shi, Y.; Zhuang, S.; Liu, N. The Role and Mechanism of Lysine Methyltransferase and Arginine Methyltransferase in Kidney Diseases. Front. Pharmacol. 2022, 13, 1488. [Google Scholar] [CrossRef]
- Qin, J.; Xu, J. Arginine methylation in the epithelial-to-mesenchymal transition. FEBS J. 2021, 289, 7292. [Google Scholar] [CrossRef]
- Xu, J.; Richard, S. Cellular Pathways Influenced by Protein Arginine Methylation: Implications for Cancer. Mol Cell 2021, 81, 4357. [Google Scholar] [CrossRef]
- Liu, S.; Hao, X.; Miao, D.; Zhang, Y. A Study on the Binding Mechanism and the Impact of Key Residue Mutations between SND1 and MTDH Peptide through Molecular Dynamics Simulations. J. Phys. Chem. B 2024, 128, 9074–9085. [Google Scholar] [CrossRef]
- Bloskie, T.; Taiwo, O.O.; Storey, K.B. Reversible Histone Modifications Contribute to the Frozen and Thawed Recovery States of Wood Frog Brains. Biomolecules 2024, 14, 839. [Google Scholar] [CrossRef]
- Zhang, J.; Storey, K.B. RBioplot: An easy-to-use R pipeline for automated statistical analysis and data visualization in molecular biology and biochemistry. PeerJ 2016, 4, e2436. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taiwo, O.O.; Storey, K.B. Histone Arginine Methylation in the Kidneys of Rana sylvatica During Freeze–Thaw Cycle. Kinases Phosphatases 2025, 3, 1. https://doi.org/10.3390/kinasesphosphatases3010001
Taiwo OO, Storey KB. Histone Arginine Methylation in the Kidneys of Rana sylvatica During Freeze–Thaw Cycle. Kinases and Phosphatases. 2025; 3(1):1. https://doi.org/10.3390/kinasesphosphatases3010001
Chicago/Turabian StyleTaiwo, Olawale O., and Kenneth B. Storey. 2025. "Histone Arginine Methylation in the Kidneys of Rana sylvatica During Freeze–Thaw Cycle" Kinases and Phosphatases 3, no. 1: 1. https://doi.org/10.3390/kinasesphosphatases3010001
APA StyleTaiwo, O. O., & Storey, K. B. (2025). Histone Arginine Methylation in the Kidneys of Rana sylvatica During Freeze–Thaw Cycle. Kinases and Phosphatases, 3(1), 1. https://doi.org/10.3390/kinasesphosphatases3010001






