Serendipitous Discovery of Desert Hairy Scorpion Mitogenomes as Bycatch in Venom Data via Nanopore Sequencing
Abstract
1. Introduction
2. Materials and Methods
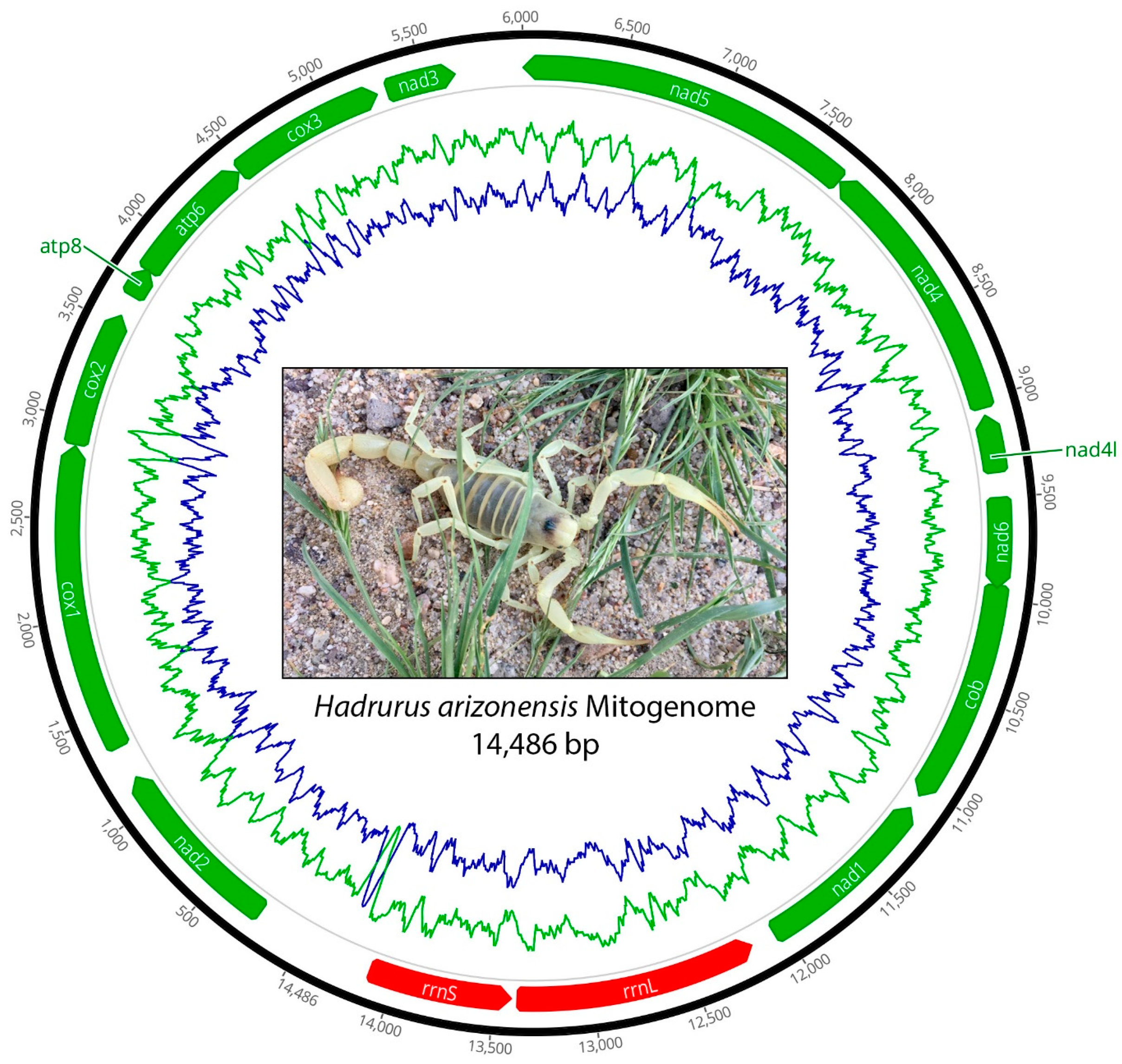
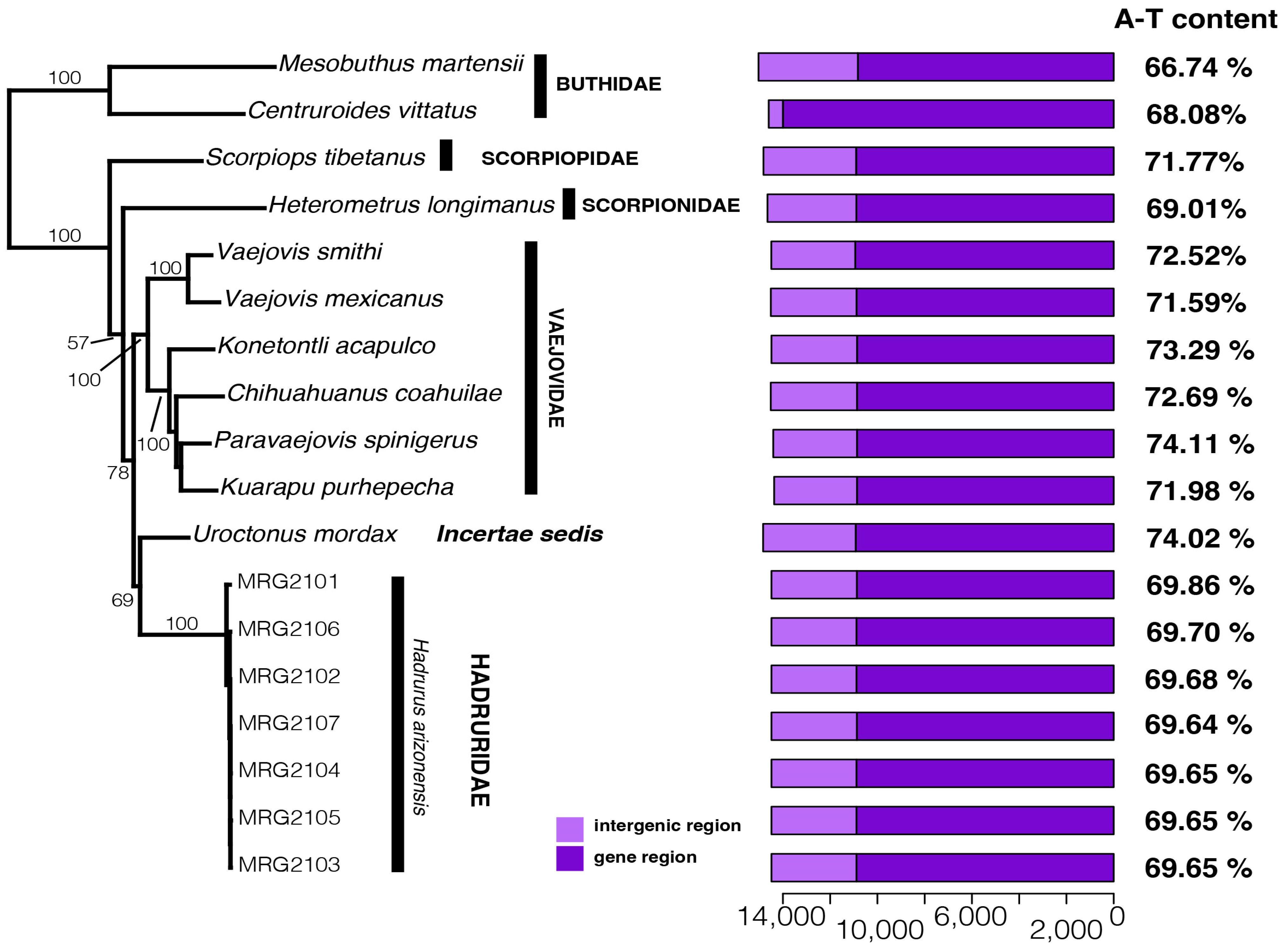
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruiz-Mendoza, P.X.; Jasso-Martínez, J.M.; Gutiérrez-Rodríguez, J.; Samacá-Sáenz, E.; Zaldívar-Riverón, A. Mitochondrial genome characterization and mitogenome phylogenetics in the central Mexican Stenopelmatus talpa complex (Orthoptera: Stenopelmatidae: Stenopelmatini). Rev. Mex. Biodivers. 2023, 94, e945094. [Google Scholar] [CrossRef]
- Kong, D.; Gan, Z.; Li, X. Phylogenetic relationships and adaptation in deep-sea carideans revealed by mitogenomes. Gene 2023, 896, 148054. [Google Scholar] [CrossRef] [PubMed]
- Galen, S.C.; Borner, J.; Perkins, S.L.; Weckstein, J.D. Phylogenomics from transcriptomic “bycatch” clarify the origins and diversity of avian trypanosomes in North America. PLoS ONE 2020, 15, e0240062. [Google Scholar] [CrossRef] [PubMed]
- Ghanavi, H.R.; Twort, V.G.; Duplouy, A. Exploring bycatch diversity of organisms in whole genome sequencing of Erebidae moths (Lepidoptera). Sci. Rep. 2021, 11, 24499. [Google Scholar] [CrossRef] [PubMed]
- Santibáñez-López, C.E.; Graham, M.R.; Sharma, P.P.; Ortiz, E.; Possani, L.D. Hadrurid scorpion toxins: Evolutionary conservation and selective pressures. Toxins 2019, 11, 637. [Google Scholar] [CrossRef] [PubMed]
- Santibáñez-López, C.E.; Ojanguren-Affilastro, A.A.; Sharma, P.P. Another one bites the dust: Taxonomic sampling of a key genus in phylogenomic datasets reveals more non-monophyletic groups in traditional scorpion classification. Invertebr. Syst. 2020, 34, 133–143. [Google Scholar] [CrossRef]
- Santibáñez-López, C.E.; González-Santillán, E.; Monod, L.; Sharma, P.P. Phylogenomics facilitates stable scorpion systematics: Reassessing the relationships of Vaejovidae and a new higher-level classification of Scorpiones (Arachnida). Mol. Phylogenet. Evol. 2019, 135, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Santibáñez-López, C.E.; Ojanguren-Affilastro, A.A.; Graham, M.R.; Sharma, P.P. Congruence between ultraconserved element-based matrices and phylotranscriptomic datasets in the scorpion Tree of Life. Cladistics 2023, 39, 533–547. [Google Scholar] [CrossRef] [PubMed]
- Shimwell, C.; Atkinson, L.; Graham, M.R.; Murdoch, B. A first molecular characterization of the scorpion telson microbiota of Hadrurus arizonensis and Smeringurus mesaensis. PLoS ONE 2023, 18, e0277303. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, K.; Parkhill, J.; Crook, J.; Horsnell, T.; Rice, P.; Rajandream, M.; Barrell, B. Artemis: Sequence visualization and annotation. Bioinformatics 2000, 474, 944–945. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP v6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Ban, X.C.; Shao, Z.K.; Wu, L.J.; Sun, J.T.; Xue, X.F. Highly diversified mitochondrial genomes provide new evidence for interordinal relationships in the Arachnida. Cladistics 2022, 38, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, W.T.; Zhang, Q.L.; Liu, M.; Xing, C.W.; Cao, Y.; Luo, F.Z.; Yuan, M.L. Mitochondrial phylogenomics provides insights into the phylogeny and evolution of spiders (Arthropoda: Araneae). Zool. Res. 2022, 43, 566–584. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeeseler, A.; Lanfear, R. IQTREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Revell, L. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Fet, V.; Soleglad, M.E.; Barker, M.D. Phylogenetic analysis of the “hirsutus” group of the genus Hadrurus Thorell (Scorpiones: Iuridae) based on morphology and mitochondrial DNA. In Scorpions 2001. In Memoriam Gary A. Polis; Fet, V., Selden, P.A., Eds.; British Arachnological Society: Burnham Beeches, UK, 2001; pp. 139–160. [Google Scholar]
- Gantenbein, B.; Fet, V.; Largiader, C.; Scholl, A. First DNA phylogeny of the genus Euscorpius Thorell 1876 (Scorpiones, Euscorpiidae) and its bearing on the taxonomy and biogeography of this genus. Biogeographica 1999, 75, 59–72. [Google Scholar]
- Gantenbein, B.; Largiadèr, C.R. The phylogeographic importance of the Strait of Gibraltar as a gene flow barrier in terrestrial arthropods: A case study with the scorpion Buthus occitanus as model organism. Mol. Phylogenet. Evol. 2003, 28, 119–130. [Google Scholar] [CrossRef]
- Graham, M.R.; Jaeger, J.R.; Prendini, L.; Riddle, B.R. Phylogeography of the Arizona hairy scorpion (Hadrurus arizonensis) supports a model of biotic assembly in the Mojave Desert and adds a new Pleistocene refugium. J. Biogeogr. 2013, 40, 1298–1312. [Google Scholar] [CrossRef]
- Santibáñez-López, C.E.; Aharon, S.; Ballesteros, J.A.; Gainett, G.; Baker, C.M.; González-Santillán, E.; Harvey, M.S.; Hassan, M.K.; Almaaty, A.H.A.; Monod, L.; et al. Phylogenomics of scorpions reveal contemporaneous diversification of scorpion mammalian predators and mammal-active sodium channel toxins. Syst. Biol. 2022, 71, 1281–1289. [Google Scholar] [CrossRef] [PubMed]
- Ghanavi, H.R.; Twort, V.; Hartman, T.J.; Zahiri, R.; Wahlberg, N. The (non) accuracy of mitochondrial genomes for family-level phylogenetics in Erebidae (Lepidoptera). Zool. Scr. 2022, 51, 695–707. [Google Scholar] [CrossRef]
- Martin, S.; Heavens, D.; Lan, Y.; Horsfield, S.; Clark, M.D.; Leggett, R.M. Nanopore adaptive sampling: A tool for enrichment of low abundance species in metagenomic samples. Genome Biol. 2022, 23, 11. [Google Scholar] [CrossRef]
- Abramson, N.I.; Bodrov, S.Y.; Bondareva, O.V.; Genelt-Yanovskiy, E.A.; Petrova, T.V. A mitochondrial genome phylogeny of voles and lemmings (Rodentia: Arvicolinae): Evolutionary and taxonomic implications. PLoS ONE 2021, 16, e0248198. [Google Scholar] [CrossRef]
- Pan, D.; Shi, B.; Du, S.; Gu, T.; Wang, R.; Xing, Y.; Zhang, Z.; Chen, J.; Cumberlidge, N.; Sun, H. Mitogenome phylogeny reveals Indochina Peninsula origin and spatiotemporal diversification of freshwater crabs (Potamidae: Potamiscinae) in China. Cladistics 2022, 38, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Dietrich, C.H.; Yu, X.; Jiao, M.; Dai, R.; Yang, M. Mitogenomic phylogeny of Typhlocybinae (Hemiptera: Cicadellidae) reveals homoplasy in tribal diagnostic morphological traits. Ecol. Evol. 2022, 12, e8982. [Google Scholar] [CrossRef]
- Ding, L.; Zhou, Q.; Sun, Y.; Feoktistova, N.Y.; Liao, J. Two novel cricetine mitogenomes: Insight into the mitogenomic characteristics and phylogeny in Cricetinae (Rodentia: Cricetidae). Genomics 2020, 112, 1716–1725. [Google Scholar] [CrossRef] [PubMed]
- Irisarri, I.; Uribe, J.E.; Eernisse, D.J.; Zardoya, R. A mitogenomic phylogeny of chitons (Mollusca: Polyplacophora). BMC Evol. Biol. 2020, 20, 22. [Google Scholar] [CrossRef] [PubMed]
- Catanese, G.; Morey, G.; Verger, F.; Grau, A.M. The Nursehound Scyliorhinus stellaris mitochondrial genome—Phylogeny, relationships among Scyliorhinidae and variability in waters of the Balearic Islands. Int. J. Mol. Sci. 2022, 23, 10355. [Google Scholar] [CrossRef] [PubMed]


| Identifier | Passed Reads | Trimmed Reads | Trimmed Read Length | Mapped Reads | Coverage |
|---|---|---|---|---|---|
| MRG2101 | 444,000 | 435,340 | 200–36,686 | 1411 | 23–89 |
| MRG2102 | 1,272,000 | 515,045 | 200–30,400 | 1220 | 29–93 |
| MRG2103 | 372,000 | 362,438 | 200–40,124 | 1327 | 38–83 |
| MRG2104 | 236,000 | 235,969 | 50–25,996 | 709 | 14–64 |
| MRG2105 | 972,000 | 892,446 | 200–48,837 | 7838 | 39–145 |
| MRG2106 | 844,000 | 841,533 | 105–34,665 | 2693 | 81–170 |
| MRG2107 | 1,760,000 | 892,394 | 500–82,653 | 2563 | 84–227 |
| Gene | Full Gene Name | Length (bp) | Variable Sites (S) | Percent Variable | Haplotypes (h) | Haplotype Diversity (Hd) | Nucleotide Diversity (π) |
|---|---|---|---|---|---|---|---|
| 12S | 12S ribosomal RNA | 721 | 8 | 1.11% | 4 | 0.714 | 0.00317 |
| 16S | 16S ribosomal RNA | 1197 | 23 | 1.92% | 6 | 0.952 | 0.00597 |
| ATP6 | ATP synthase membrane subunit 6 | 666 | 22 | 3.30% | 7 | 1.000 | 0.01044 |
| ATP8 | ATP synthase membrane subunit 8 | 156 | 4 | 2.56% | 4 | 0.714 | 0.00733 |
| CYTB | Cytochrome b | 1119 | 42 | 3.75% | 6 | 0.952 | 0.01170 |
| COI | Cytochrome c oxidase I | 1539 | 39 | 2.53% | 7 | 1.000 | 0.00792 |
| COII | Cytochrome c oxidase II | 670 | 15 | 2.24% | 7 | 1.000 | 0.00739 |
| COIII | Cytochrome c oxidase III | 781 | 20 | 2.56% | 4 | 0.714 | 0.00756 |
| NAD1 | NADH dehydrogenase subunit 1 | 924 | 26 | 2.81% | 7 | 1.000 | 0.00948 |
| NAD2 | NADH dehydrogenase subunit 2 | 962 * | 25 | 2.60% | 7 | 1.000 | 0.00832 |
| NAD3 | NADH dehydrogenase subunit 3 | 348 | 14 | 4.02% | 5 | 0.905 | 0.01286 |
| NAD4 | NADH dehydrogenase subunit 4 | 1275 | 39 | 3.06% | 6 | 0.952 | 0.01083 |
| NAD4L | NADH dehydrogenase subunit 4L | 288 | 10 | 3.47% | 4 | 0.810 | 0.01157 |
| NAD5 | NADH dehydrogenase subunit 5 | 1698 | 53 | 3.12% | 7 | 1.000 | 0.00937 |
| NAD6 | NADH dehydrogenase subunit 6 | 453 | 14 | 3.09% | 6 | 0.952 | 0.01092 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graham, M.R.; Santibáñez-López, C.E.; Zehnpfennig, J.R.; Tillman, D.S.; Murdoch, B. Serendipitous Discovery of Desert Hairy Scorpion Mitogenomes as Bycatch in Venom Data via Nanopore Sequencing. Arthropoda 2024, 2, 119-129. https://doi.org/10.3390/arthropoda2020009
Graham MR, Santibáñez-López CE, Zehnpfennig JR, Tillman DS, Murdoch B. Serendipitous Discovery of Desert Hairy Scorpion Mitogenomes as Bycatch in Venom Data via Nanopore Sequencing. Arthropoda. 2024; 2(2):119-129. https://doi.org/10.3390/arthropoda2020009
Chicago/Turabian StyleGraham, Matthew R., Carlos E. Santibáñez-López, Jessica R. Zehnpfennig, Dylan S. Tillman, and Barbara Murdoch. 2024. "Serendipitous Discovery of Desert Hairy Scorpion Mitogenomes as Bycatch in Venom Data via Nanopore Sequencing" Arthropoda 2, no. 2: 119-129. https://doi.org/10.3390/arthropoda2020009
APA StyleGraham, M. R., Santibáñez-López, C. E., Zehnpfennig, J. R., Tillman, D. S., & Murdoch, B. (2024). Serendipitous Discovery of Desert Hairy Scorpion Mitogenomes as Bycatch in Venom Data via Nanopore Sequencing. Arthropoda, 2(2), 119-129. https://doi.org/10.3390/arthropoda2020009








