Discovery and Characterization of 7,8-Dihydropyrido[4,3-d]pyrimidines as SARS-CoV-2 Entry Inhibitors
Abstract
1. Introduction
2. Results and Discussion
2.1. High-Throughput Screening (HTS) Assay Development
2.2. Screening of a Small Molecule Library
2.3. HTS Hit Validation
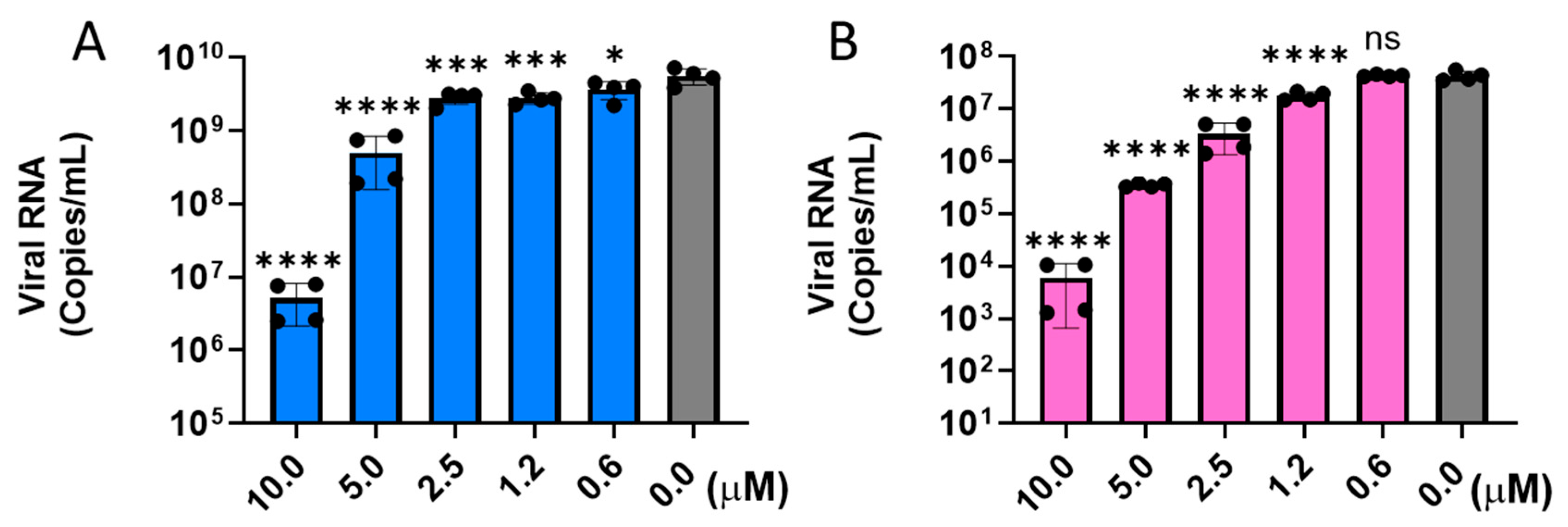
2.4. Mechanism of Action
2.5. In Silico Predicted Parameters
2.6. ADME/Tox Profile
3. Materials and Methods
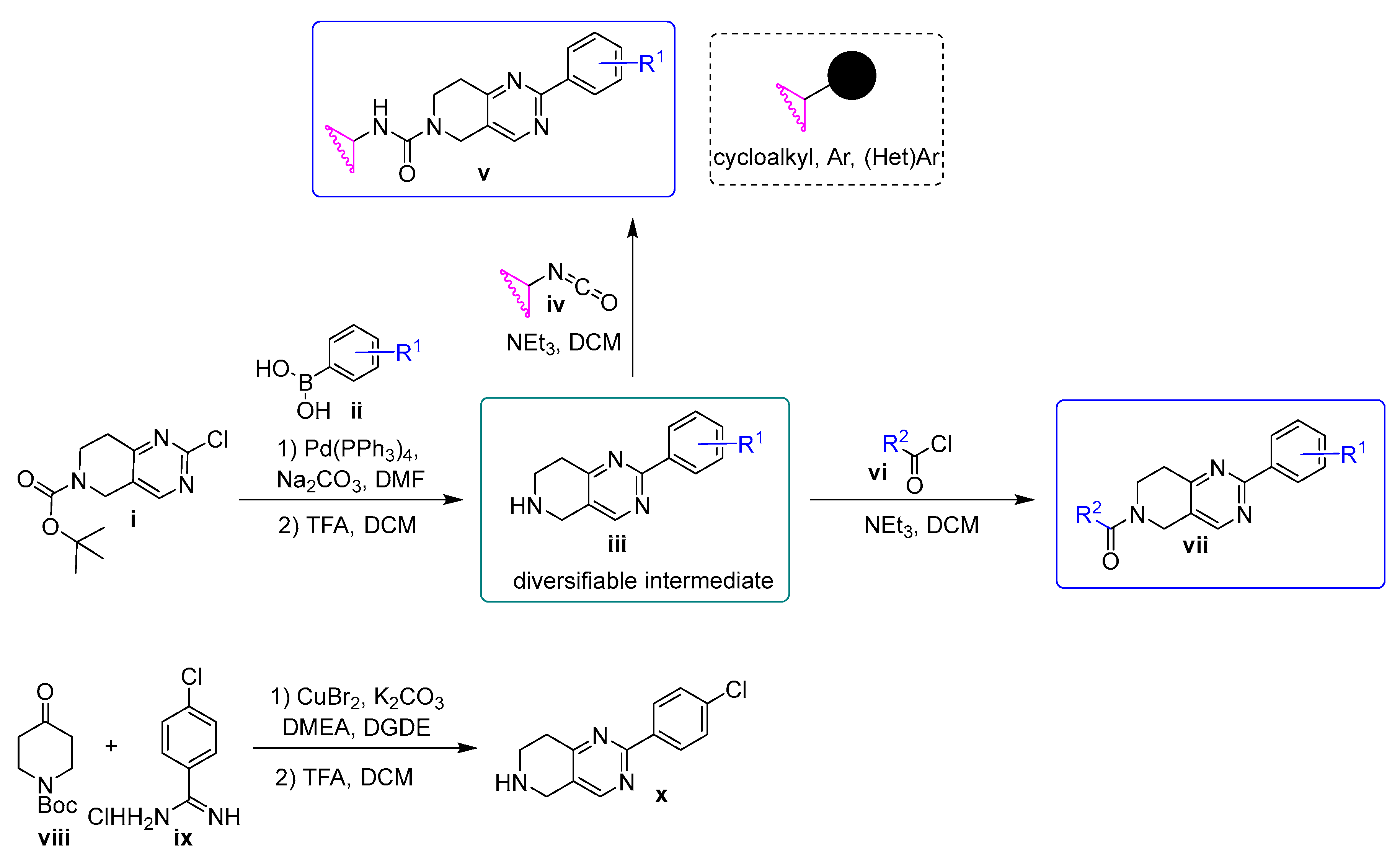
3.1. Chemistry
3.2. General Method A: Suzuki Coupling
3.3. General Method B: Boc Deprotection
3.4. General Method C: Synthesis of Ureas
3.5. General Method D: Synthesis of Amides
- 2-(4-Chlorophenyl)-N-cyclohexyl-7,8-dihydropyrido[4,3-d]pyrimidine-6(5H)-carboxamide (1).
- N-Cyclohexyl-2-(4-(trifluoromethyl)phenyl)-7,8-dihydropyrido[4,3-d]pyrimidine-6(5H)-carboxamide (2).
- N-Cyclohexyl-2-(3,4-difluorophenyl)-7,8-dihydropyrido[4,3-d]pyrimidine-6(5H)-carboxamide (3).
- N-Cyclohexyl-2-phenyl-7,8-dihydropyrido[4,3-d]pyrimidine-6(5H)-carboxamide (4).
- N-(4-Chlorophenyl)-2-phenyl-7,8-dihydropyrido[4,3-d]pyrimidine-6(5H)-carboxamide (5).
- N-(4-Fluorophenyl)-2-phenyl-7,8-dihydropyrido[4,3-d]pyrimidine-6(5H)-carboxamide (6).
- N-(Benzo[d][1,3]dioxol-5-yl)-2-phenyl-7,8-dihydropyrido[4,3-d]pyrimidine-6(5H)-carboxamide (7).
- N-(3-Fluoro-4-methylphenyl)-2-(4-methoxyphenyl)-7,8-dihydropyrido[4,3-d]pyrimidine-6(5H)-carboxamide (8) was purchased from ChemDiv (Cat# M568-0042).
- 2-(2-Chlorophenyl)-1-(2-(4-chlorophenyl)-7,8-dihydropyrido[4,3-d]pyrimidin-6(5H)-yl)ethan-1-one (9) was purchased from ChemDiv (Cat# M567-0228).
- 2-(4-Chlorophenyl)-1-(2-(4-chlorophenyl)-7,8-dihydropyrido[4,3-d]pyrimidin-6(5H)-yl)ethan-1-one (10) was purchased from ChemDiv (Cat# M567-0223).
- 1-(2-(4-Chlorophenyl)-7,8-dihydropyrido[4,3-d]pyrimidin-6(5H)-yl)-2-(3-methoxyphenyl)ethan-1-one (11) was purchased from ChemDiv (Cat# M567-0210).
- 2-(2-Chlorophenyl)-1-(2-(4-(trifluoromethyl)phenyl)-7,8-dihydropyrido[4,3-d]pyrimidin-6(5H)-yl)ethan-1-one (12).
- 2-(2-Chlorophenyl)-1-(2-(3,4-difluorophenyl)-7,8-dihydropyrido[4,3-d]pyrimidin-6(5H)-yl)ethan-1-one (13).
- 2-(4-Chlorophenyl)-1-(2-(3,4-difluorophenyl)-7,8-dihydropyrido[4,3-d]pyrimidin-6(5H)-yl)ethan-1-one (14).
3.6. Cell Lines
3.7. Virus Stocks
3.8. Generation of Pseudoviruses
3.9. Pseudovirus Neutralization Assay—HTS
3.10. Generation of SARS-CoV-2 Variant S Protein Plasmids
3.11. Pseudovirus Inhibition and Cytotoxicity
3.12. Live Virus Neutralization Assay
3.13. Statistical Analysis
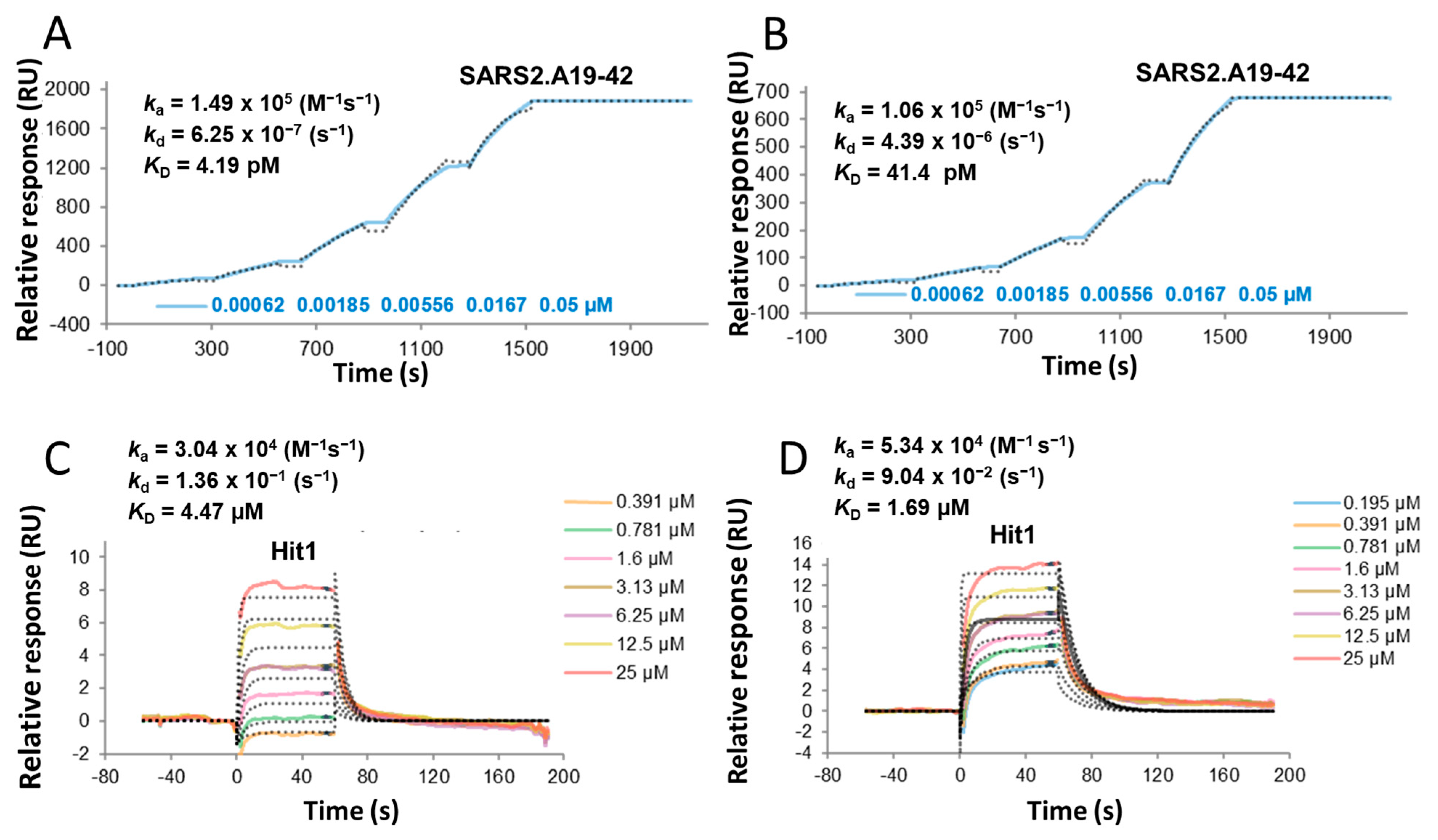
3.14. Assessment of the Binding Properties by SPR
3.15. Liver Microsome Stability Assay
3.16. Plasma Stability Assay
3.17. CYP Inhibition Assay
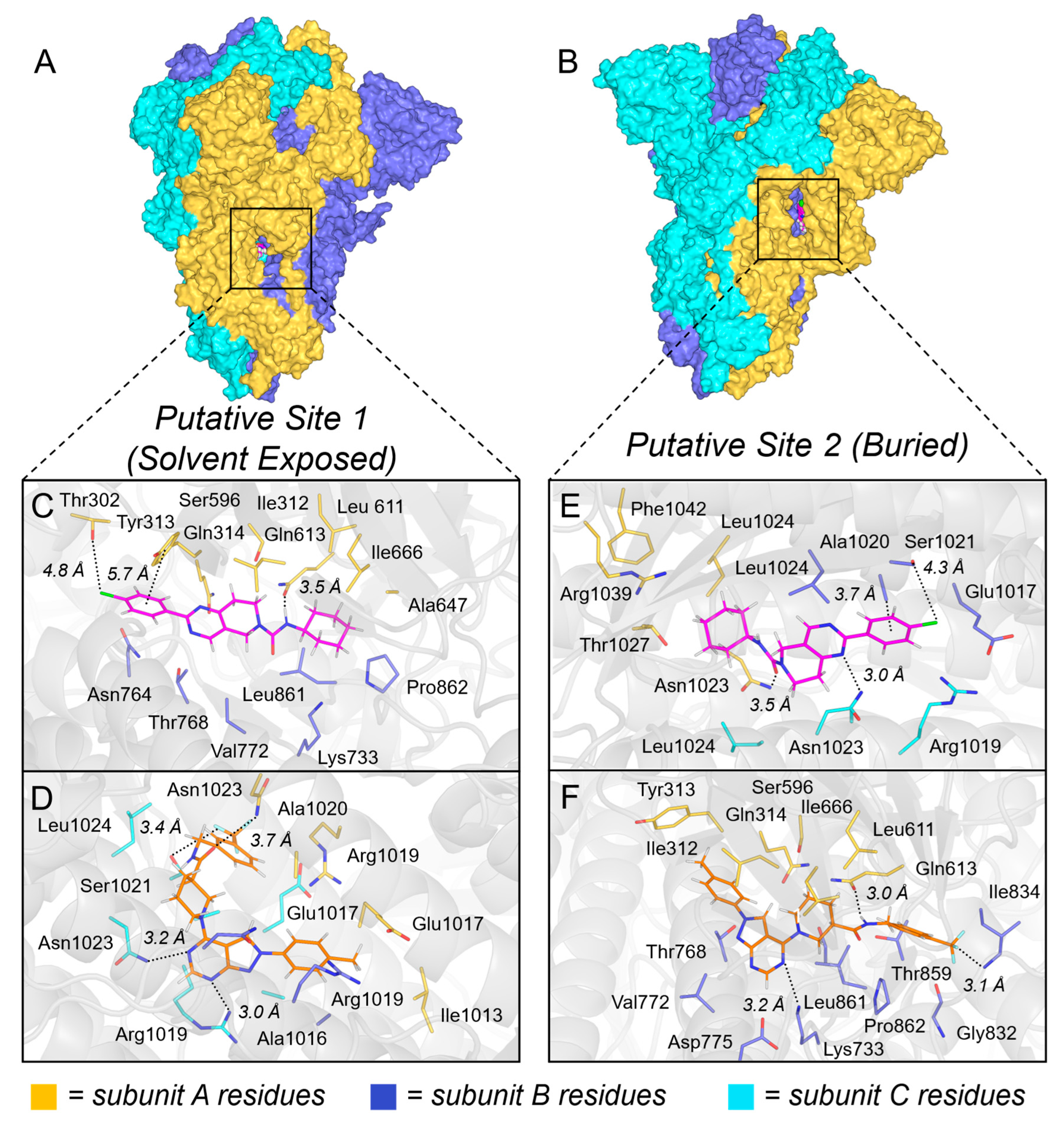
3.18. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sharma, A.; Ahmad Farouk, I.; Lal, S.K. COVID-19: A Review on the Novel Coronavirus Disease Evolution, Transmission, Detection, Control and Prevention. Viruses 2021, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.-W.; Yuan, S.; Chu, H.; Sridhar, S.; Yuen, K.-Y. COVID-19 drug discovery and treatment options. Nat. Rev. Microbiol. 2024, 22, 391–407. [Google Scholar] [CrossRef] [PubMed]
- Zmasek, C.M.; Lefkowitz, E.J.; Niewiadomska, A.; Scheuermann, R.H. Genomic evolution of the Coronaviridae family. Virology 2022, 570, 123–133. [Google Scholar] [CrossRef]
- Kesheh, M.M.; Hosseini, P.; Soltani, S.; Zandi, M. An overview on the seven pathogenic human coronaviruses. Rev. Med. Virol. 2022, 32, e2282. [Google Scholar] [CrossRef]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef]
- Wong, A.C.P.; Li, X.; Lau, S.K.P.; Woo, P.C.Y. Global Epidemiology of Bat Coronaviruses. Viruses 2019, 11, 174. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Yuan, S.; Kok, K.H.; To, K.K.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.; Poon, R.W.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef]
- Prévost, J.; Finzi, A. The great escape? SARS-CoV-2 variants evading neutralizing responses. Cell Host Microbe 2021, 29, 322–324. [Google Scholar] [CrossRef]
- Zhao, Y.; Huang, J.; Zhang, L.; Chen, S.; Gao, J.; Jiao, H. The global transmission of new coronavirus variants. Env. Res. 2022, 206, 112240. [Google Scholar] [CrossRef] [PubMed]
- Kustin, T.; Harel, N.; Finkel, U.; Perchik, S.; Harari, S.; Tahor, M.; Caspi, I.; Levy, R.; Leshchinsky, M.; Ken Dror, S.; et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2-mRNA-vaccinated individuals. Nat. Med. 2021, 27, 1379–1384. [Google Scholar] [CrossRef] [PubMed]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef]
- Santos, T.M.; Lisboa, A.B.P.; Rodrigues, W.; Gomes, H.; Abrahão, J.; Del-Bem, L.-E. Human variation in the protein receptor ACE2 affects its binding affinity to SARS-CoV-2 in a variant-dependent manner. J. Biomol. Struct. Dyn. 2023, 41, 2947–2955. [Google Scholar] [CrossRef]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Santaniello, A.; Perruolo, G.; Cristiano, S.; Agognon, A.L.; Cabaro, S.; Amato, A.; Dipineto, L.; Borrelli, L.; Formisano, P.; Fioretti, A.; et al. SARS-CoV-2 Affects Both Humans and Animals: What Is the Potential Transmission Risk? A Literature Review. Microorganisms 2023, 11, 514. [Google Scholar] [CrossRef]
- Tan, C.C.S.; Lam, S.D.; Richard, D.; Owen, C.J.; Berchtold, D.; Orengo, C.; Nair, M.S.; Kuchipudi, S.V.; Kapur, V.; van Dorp, L.; et al. Transmission of SARS-CoV-2 from humans to animals and potential host adaptation. Nat. Commun. 2022, 13, 2988. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef]
- Hale, V.L.; Dennis, P.M.; McBride, D.S.; Nolting, J.M.; Madden, C.; Huey, D.; Ehrlich, M.; Grieser, J.; Winston, J.; Lombardi, D.; et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 2022, 602, 481–486. [Google Scholar] [CrossRef]
- Kuchipudi, S.V.; Surendran-Nair, M.; Ruden, R.M.; Yon, M.; Nissly, R.H.; Vandegrift, K.J.; Nelli, R.K.; Li, L.; Jayarao, B.M.; Maranas, C.D.; et al. Multiple spillovers from humans and onward transmission of SARS-CoV-2 in white-tailed deer. Proc. Natl. Acad. Sci. USA 2022, 119, e2121644119. [Google Scholar] [CrossRef]
- Chandler, J.C.; Bevins, S.N.; Ellis, J.W.; Linder, T.J.; Tell, R.M.; Jenkins-Moore, M.; Root, J.J.; Lenoch, J.B.; Robbe-Austerman, S.; DeLiberto, T.J.; et al. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). Proc. Natl. Acad. Sci. USA 2021, 118, e2114828118. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Shan, K.-J.; Wang, W.; Zhang, S.; Huan, Q.; Qian, W. Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. J. Genet. Genom. 2021, 48, 1111–1121. [Google Scholar] [CrossRef]
- Farhud, D.D.; Mojahed, N. SARS-CoV-2 Notable Mutations and Variants: A Review Article. Iran. J. Public Health 2022, 51, 1494–1501. [Google Scholar] [CrossRef]
- Kumar, R.; Srivastava, Y.; Muthuramalingam, P.; Singh, S.K.; Verma, G.; Tiwari, S.; Tandel, N.; Beura, S.K.; Panigrahi, A.R.; Maji, S.; et al. Understanding Mutations in Human SARS-CoV-2 Spike Glycoprotein: A Systematic Review & Meta-Analysis. Viruses 2023, 15, 856. [Google Scholar] [CrossRef]
- Magazine, N.; Zhang, T.; Wu, Y.; McGee, M.C.; Veggiani, G.; Huang, W. Mutations and Evolution of the SARS-CoV-2 Spike Protein. Viruses 2022, 14, 640. [Google Scholar] [CrossRef] [PubMed]
- Zieneldien, T.; Kim, J.; Cao, J.; Cao, C. COVID-19 Vaccines: Current Conditions and Future Prospects. Biology 2021, 10, 960. [Google Scholar] [CrossRef] [PubMed]
- Manicassamy, B.; Wang, J.; Jiang, H.; Rong, L. Comprehensive Analysis of Ebola Virus GP1 in Viral Entry. J. Virol. 2005, 79, 4793–4805. [Google Scholar] [CrossRef]
- Guo, Y.; Tisoncik, J.; McReynolds, S.; Farzan, M.; Prabhakar, B.S.; Gallagher, T.; Rong, L.; Caffrey, M. Identification of a new region of SARS-CoV S protein critical for viral entry. J. Mol. Biol. 2009, 394, 600–605. [Google Scholar] [CrossRef]
- Wang, J.; Cheng, H.; Ratia, K.; Varhegyi, E.; Hendrickson, W.G.; Li, J.; Rong, L. A comparative high-throughput screening protocol to identify entry inhibitors of enveloped viruses. J. Biomol. Screen. 2014, 19, 100–107. [Google Scholar] [CrossRef]
- Rumschlag-Booms, E.; Zhang, H.; Soejarto, D.D.; Fong, H.H.; Rong, L. Development of an antiviral screening protocol: One-stone-two-birds. J. Antivir. Antiretrovir. 2011, 3, 8–10. [Google Scholar] [CrossRef]
- He, J.; Choe, S.; Walker, R.; Di Marzio, P.; Morgan, D.O.; Landau, N.R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 1995, 69, 6705–6711. [Google Scholar] [CrossRef]
- Xia, S.; Liu, M.; Wang, C.; Xu, W.; Lan, Q.; Feng, S.; Qi, F.; Bao, L.; Du, L.; Liu, S.; et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020, 30, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Iwanaga, N.; Cooper, L.; Rong, L.; Maness, N.J.; Beddingfield, B.; Qin, Z.; Crabtree, J.; Tripp, R.A.; Yang, H.; Blair, R.; et al. ACE2-IgG1 fusions with improved in vitro and in vivo activity against SARS-CoV-2. iScience 2022, 25, 103670. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Lipinski, C.A. Rule of five in 2015 and beyond: Target and ligand structural limitations, ligand chemistry structure and drug discovery project decisions. Adv. Drug Deliv. Rev. 2016, 101, 34–41. [Google Scholar] [CrossRef]
- Schafer, A.; Xiong, R.; Cooper, L.; Nowar, R.; Lee, H.; Li, Y.; Ramirez, B.E.; Peet, N.P.; Caffrey, M.; Thatcher, G.R.J.; et al. Evidence for distinct mechanisms of small molecule inhibitors of filovirus entry. PLoS Pathog. 2021, 17, e1009312. [Google Scholar] [CrossRef]
- Hsieh, C.L.; Goldsmith, J.A.; Schaub, J.M.; DiVenere, A.M.; Kuo, H.C.; Javanmardi, K.; Le, K.C.; Wrapp, D.; Lee, A.G.; Liu, Y.; et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 2020, 369, 1501–1505. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, T.; Zhang, Y.; Yang, E.S.; Schramm, C.A.; Shi, W.; Pegu, A.; Oloniniyi, O.K.; Henry, A.R.; Darko, S.; et al. Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science 2021, 373, eabh1766. [Google Scholar] [CrossRef]
- Sharanya, C.S.; Sabu, A.; Haridas, M. Potent phytochemicals against COVID-19 infection from phyto-materials used as antivirals in complementary medicines: A review. Future J. Pharm. Sci. 2021, 7, 113. [Google Scholar] [CrossRef]
- Shuster, A.; Pechalrieu, D.; Jackson, C.B.; Abegg, D.; Choe, H.; Adibekian, A. Clinical Antiviral Drug Arbidol Inhibits Infection by SARS-CoV-2 and Variants through Direct Binding to the Spike Protein. ACS Chem. Biol. 2021, 16, 2845–2851. [Google Scholar] [CrossRef] [PubMed]
- Freidel, M.R.; Vakhariya, P.A.; Sardarni, S.K.; Armen, R.S. The Dual-Targeted Fusion Inhibitor Clofazimine Binds to the S2 Segment of the SARS-CoV-2 Spike Protein. Viruses 2024, 16, 640. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Halgren, T. New method for fast and accurate binding-site identification and analysis. Chem. Biol. Drug Des. 2007, 69, 146–148. [Google Scholar] [CrossRef]
- Ye, G.; Liu, B.; Li, F. Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain. Nat. Commun. 2022, 13, 1214. [Google Scholar] [CrossRef] [PubMed]
- Sander, T.; Freyss, J.; von Korff, M.; Reich, J.R.; Rufener, C. OSIRIS, an entirely in-house developed drug discovery informatics system. J. Chem. Inf. Model. 2009, 49, 232–246. [Google Scholar] [CrossRef]
- Salunke, S.; O’Brien, F.; Cheng Thiam Tan, D.; Harris, D.; Math, M.-C.; Ariën, T.; Klein, S.; Timpe, C. Oral drug delivery strategies for development of poorly water soluble drugs in paediatric patient population. Adv. Drug Deliv. Rev. 2022, 190, 114507. [Google Scholar] [CrossRef]
- Lipinski, C.A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 2000, 44, 235–249. [Google Scholar] [CrossRef]
- Zhang, L.; Narayanan, K.K.; Cooper, L.; Chan, K.K.; Skeeters, S.S.; Devlin, C.A.; Aguhob, A.; Shirley, K.; Rong, L.; Rehman, J.; et al. An ACE2 decoy can be administered by inhalation and potently targets omicron variants of SARS-CoV-2. EMBO Mol. Med. 2022, 14, e16109. [Google Scholar] [CrossRef]
- Markgren, P.O.; Hamalainen, M.; Danielson, U.H. Kinetic analysis of the interaction between HIV-1 protease and inhibitors using optical biosensor technology. Anal. Biochem. 2000, 279, 71–78. [Google Scholar] [CrossRef]
- Argade, M.D.; Anirudhan, V.; Bradley, S.P.; Tomorowicz, L.; Bott, R.; Sownthirarajan, B.; Zielinski, C.A.; Sloan, J.P.; Nikolic, D.S.; Gaisin, A.M.; et al. Refinement of imidazo[2,1-a]pyrimidines in pursuit of potential drug candidates against group 2 influenza A viruses. Eur. J. Med. Chem. 2025, 292, 117679. [Google Scholar] [CrossRef] [PubMed]
- Argade, M.D.; Achi, J.G.; Bott, R.; Morsheimer, K.M.; Owen, C.D.; Zielinski, C.A.; Gaisin, A.M.; Alvarez, M.; Moore, T.W.; Bu, F.; et al. Guardians at the Gate: Optimization of Small Molecule Entry Inhibitors of Ebola and Marburg Viruses. J. Med. Chem. 2025, 68, 135–155. [Google Scholar] [CrossRef] [PubMed]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.; Honig, B.; Shaw, D.E.; Friesner, R.A. A hierarchical approach to all-atom protein loop prediction. Proteins 2004, 55, 351–367. [Google Scholar] [CrossRef]
- Johnston, R.C.; Yao, K.; Kaplan, Z.; Chelliah, M.; Leswing, K.; Seekins, S.; Watts, S.; Calkins, D.; Chief Elk, J.; Jerome, S.V.; et al. Epik: pK(a) and Protonation State Prediction through Machine Learning. J. Chem. Theory Comput. 2023, 19, 2380–2388. [Google Scholar] [CrossRef]
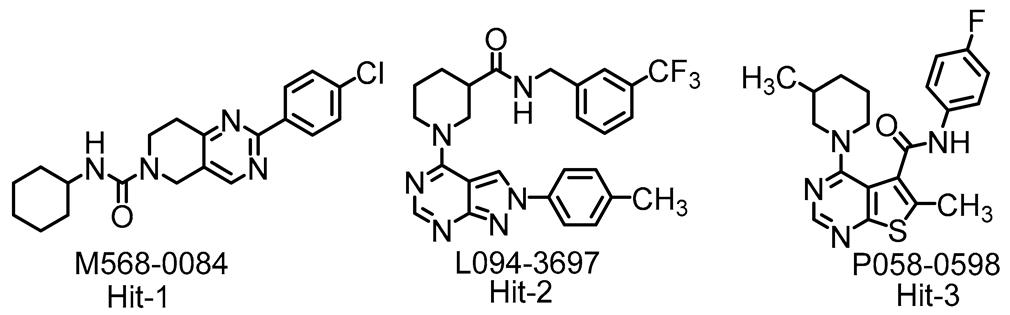 | ||||||
| Variant | Hit-1 | Hit-2 | Hit-3 | |||
|---|---|---|---|---|---|---|
| EC50 (µM) a | SI b | EC50 (µM) a | SI b | EC50 (µM) a | SI b | |
| A (Wuhan) | 0.31 ± 0.09 | 210 | 4.93 ± 0.47 | 20 | 10.63 ± 0.69 | 9 |
| B.1.1.7 (alpha) | 0.54 ± 0.07 | 120 | 4.27 0.53 | 23 | 12.69 ± 1.03 | 8 |
| B.1.351 (beta) | 0.32 ± 0.05 | 203 | 3.18 1.04 | 31 | 13.03 ± 1.65 | 8 |
| P.1 (gamma) | 0.35 ± 0.04 | 186 | 3.11 0.41 | 32 | 15.53 ± 0.91 | 6 |
| B.1.617.2 (Delta) | 0.21 ± 0.01 | 310 | 5.36 ± 0.05 | 19 | 10.75 ± 0.35 | 9 |
| B.1.1.529 BA.1 (Omicron) | 0.98 ± 0.07 | 66 | 7.23 ± 0.46 | 14 | 29.53 ± 0.84 | 3 |
| B.1.1.529 BA.2 | 0.43 ± 0.01 | 232 | 7.63 ± 0.35 | 13 | ND | - |
| B.1.1.529 BA.4/5 | 0.23 ± 0.03 | 434 | 7.31 ± 1.02 | 14 | ND | - |
| SARS-CoV | 0.66 ± 0.02 | 99 | 15.14 ± 0.53 | 7 | 8.89 ± 0.30 | 11 |
| Compound | MW | cLogP | Solubility | Caco-2 | F % | Drug Score b |
|---|---|---|---|---|---|---|
| µg/mL | × 10−6 cm/s | |||||
| Hit-1 | 370 | 3.46 | 80 | 101 | 98 | 43 |
| Hit-2 | 494 | 3.6 | 4 | 96 | 23 | 26 |
| Hit-3 | 384 | 4.25 | 40 | 137 | 52 | 16 |
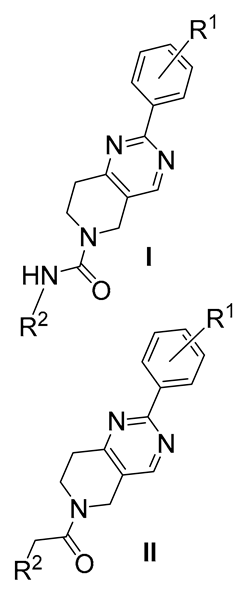 | Compound | R1 | R2 | EC50 (µM) a | SI b | |
|---|---|---|---|---|---|---|
| Hit-1 | I | 4-Cl | cyclohexyl- | 0.32 ± 0.03 | 210 | |
| 2 | I | 4-CF3 | cyclohexyl- | 5.20 ± 0.22 | 19 | |
| 3 | I | 3,4-di-F | cyclohexyl- | 0.93 ± 0.09 | 108 | |
| 4 | I | H | cyclohexyl- | 6.14 ± 0.33 | 16 | |
| 5 | I | H | 4-Cl-phenyl- | 2.53 ± 0.10 | 40 | |
| 6 | I | H | 4-F-phenyl- | 3.20 ± 0.13 | 31 | |
| 7 | I | H | 3,4-methylene dioxyphenyl- | 6.07 ± 0.25 | 16 | |
| 8 | I | 4-OMe | 3-F-4-Me-phenyl | 2.11 ± 0.33 | 47 | |
| 9 | II | 4-Cl | 2-Cl-phenyl- | 0.47 ± 0.02 | 207 | |
| 10 | II | 4-Cl | 4-Cl-phenyl- | 1.44 ± 0.14 | 69 | |
| 11 | II | 4-Cl | 3-MeO-phenyl- | 2.67 ± 0.16 | 28 | |
| 12 | II | 4-CF3 | 2-Cl-phenyl- | 7.13 ± 0.35 | 14 | |
| 13 | II | 3,4-di-F | 2-Cl-phenyl- | 0.23 ± 0.07 | 223 | |
| 14 | II | 3,4-di-F | 4-Cl-phenyl- | 0.68 ± 0.05 | 147 | |
| Compound | Metabolic Stability a | |||
|---|---|---|---|---|
| Plasma | Liver Microsome | |||
| 2 µM | 2 µM | 10 µM | No NADPH | |
| Human | Human | |||
| 1 | 99.5 | 78.7 | 96.6 | 100 |
| 3 | 100 | 79.4 | 98.1 | 100 |
| 7 | 100 | 67.3 | 100 | 100 |
| 10 | 100 | 31.1 | 84.5 | 76.5 |
| 11 | 100 | 23.1 | 72.5 | 100 |
| 13 | 100 | 28.3 | 74.2 | 100 |
| Verapamil | 17.8 | 64.1 | ||
| Mouse | Mouse | |||
| 1 | 100 | 50.6 | 88.4 | 100 |
| 3 | 99.1 | 65.9 | 89.6 | 100 |
| 7 | 100 | 81.3 | 95.1 | 100 |
| 10 | 100 | 28.9 | 95.1 | 100 |
| 11 | 100 | 16.4 | 63.6 | 100 |
| 13 | 100 | 23.0 | 45.9 | 100 |
| Verapamil | 17.9 | 59.1 | ||
| Dog | Dog | |||
| 1 | 100 | 81.4 | 98.0 | 100 |
| 3 | 100 | 85.5 | 98.9 | 100 |
| 7 | 100 | 94.6 | 100 | 100 |
| 10 | 100 | 71.5 | 57.9 | 100 |
| 11 | 100 | 31.1 | 68.1 | 100 |
| 13 | 100 | 72.7 | 82.1 | 100 |
| Verapamil | 21.1 | 69.9 | ||
| Monkey | Monkey | |||
| 1 | 100 | 54.4 | 94.9 | 100 |
| 3 | 100 | 70.5 | 97.7 | 97.9 |
| 7 | 100 | 72.6 | 91.8 | 95.4 |
| 10 | 100 | 31.1 | 94.1 | 100 |
| 11 | 100 | 1.8 | 45.9 | 95.6 |
| 13 | 100 | 5.4 | 53.7 | 100 |
| Verapamil | 0.2 | 27.8 | ||
| Compound | CYP Inhibition * | |||
|---|---|---|---|---|
| CYP1A2 | CYP2C9 | CYP2D6 | CYP3A4 | |
| % Inhibition | ||||
| 1 | 13.82 | 78.50 | −0.57 | 68.77 |
| 3 | 2.58 | 73.02 | 10.09 | 53.56 |
| 7 | 32.03 | 65.44 | 3.22 | 81.20 |
| 10 | 9.31 | 44.23 | 50.68 | 63.14 |
| 11 | 33.05 | −5.89 | 48.48 | 81.24 |
| 13 | −0.15 | 51.66 | −1.74 | 50.22 |
| Parameter a | Unit | i.v. (5 mg/kg) b | p.o. (50 mg/kg) c |
|---|---|---|---|
| T1/2 | h | 2.35 | 4.0 |
| Tmax | h | 0.08 | 1.83 |
| Cmax | ng/mL | 2677 | 4440 |
| AUC(inf) | ng·h/mL | 5366 | 36,675 |
| MRT(inf) | h | 3.86 | 5.40 |
| Vz/F | mL/kg | 3172 | 4260 |
| Cl | mL/h/kg | 940 | 1376 |
| F | % | - | 68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bradley, S.P.; Galván Achi, J.M.; Cooper, L.; Argade, M.D.; Cheng, H.; Bott, R.; Zielinski, C.A.; Gaisin, A.M.; Jesikiewicz, L.T.; Villegas, J.A.; et al. Discovery and Characterization of 7,8-Dihydropyrido[4,3-d]pyrimidines as SARS-CoV-2 Entry Inhibitors. Drugs Drug Candidates 2025, 4, 47. https://doi.org/10.3390/ddc4040047
Bradley SP, Galván Achi JM, Cooper L, Argade MD, Cheng H, Bott R, Zielinski CA, Gaisin AM, Jesikiewicz LT, Villegas JA, et al. Discovery and Characterization of 7,8-Dihydropyrido[4,3-d]pyrimidines as SARS-CoV-2 Entry Inhibitors. Drugs and Drug Candidates. 2025; 4(4):47. https://doi.org/10.3390/ddc4040047
Chicago/Turabian StyleBradley, Sean P., Jazmin M. Galván Achi, Laura Cooper, Malaika D. Argade, Han Cheng, Ryan Bott, Christian A. Zielinski, Arsen M. Gaisin, Luke T. Jesikiewicz, José A. Villegas, and et al. 2025. "Discovery and Characterization of 7,8-Dihydropyrido[4,3-d]pyrimidines as SARS-CoV-2 Entry Inhibitors" Drugs and Drug Candidates 4, no. 4: 47. https://doi.org/10.3390/ddc4040047
APA StyleBradley, S. P., Galván Achi, J. M., Cooper, L., Argade, M. D., Cheng, H., Bott, R., Zielinski, C. A., Gaisin, A. M., Jesikiewicz, L. T., Villegas, J. A., Lee, H., Ratia, K., Peet, N. P., Rong, L., & Gaisina, I. N. (2025). Discovery and Characterization of 7,8-Dihydropyrido[4,3-d]pyrimidines as SARS-CoV-2 Entry Inhibitors. Drugs and Drug Candidates, 4(4), 47. https://doi.org/10.3390/ddc4040047








