Design of a First-in-Class homoPROTAC to Induce ICP0 Degradation in Human Herpes Simplex Virus 1
Abstract
1. Introduction
2. Results
2.1. ICP0 Structural Analysis Reveals a Superior Druggable Pocket for Bivalent Ligand Design
2.2. Virtual Screening Identifies High-Affinity Core Scaffolds
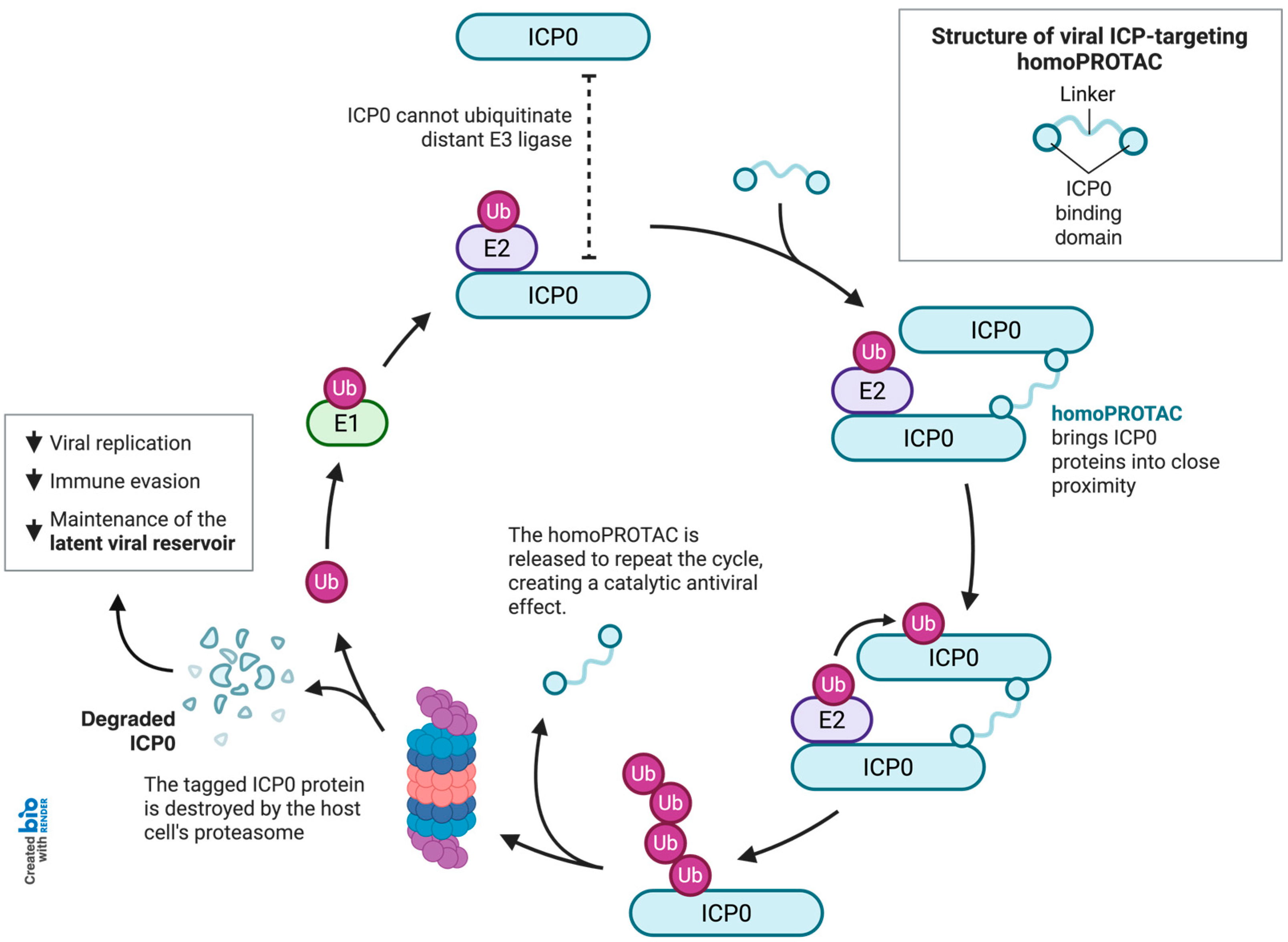
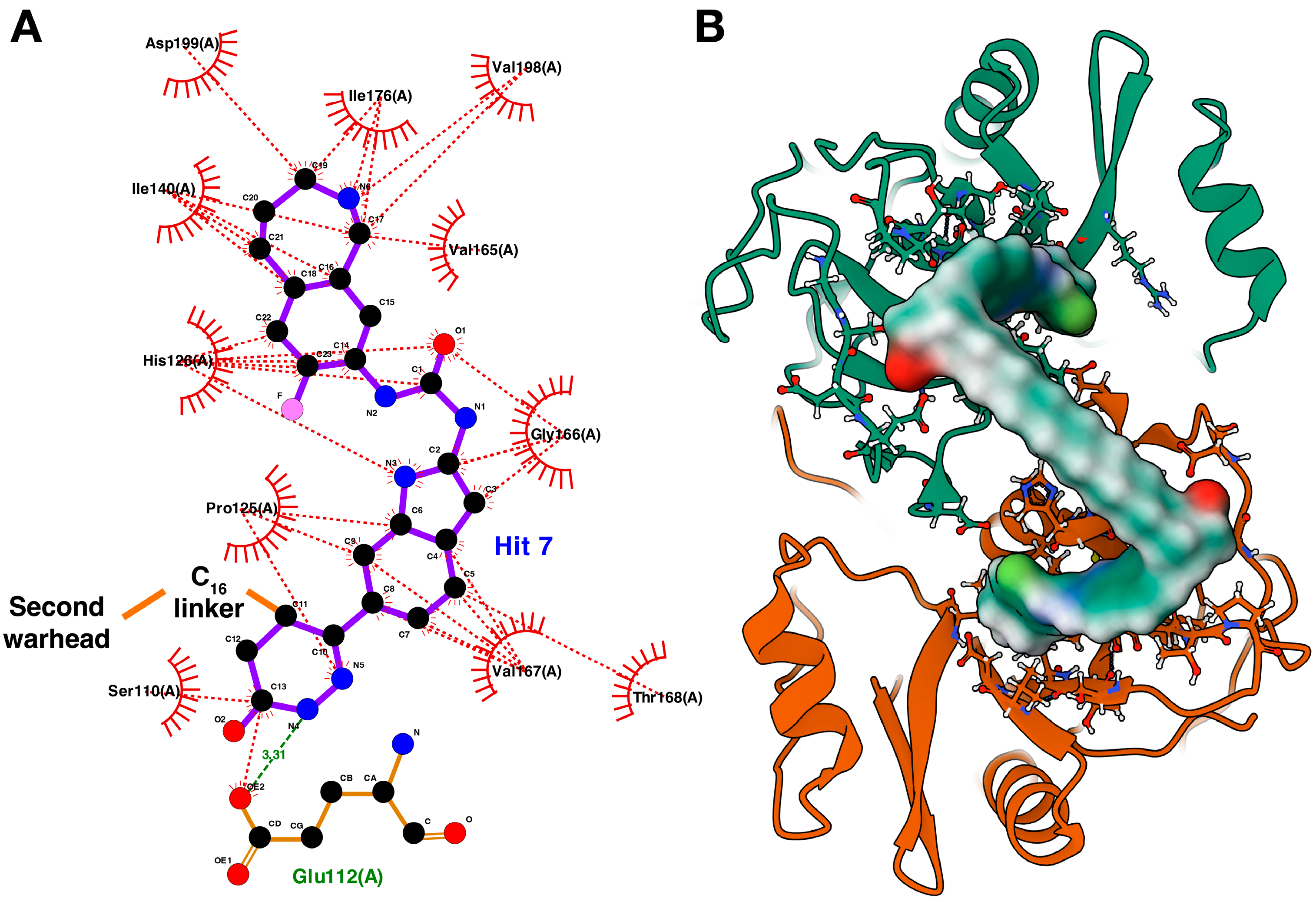
2.3. Rational Design of the Lead Bivalent Degrader ICP0-deg-01
3. Discussion
4. Materials and Methods
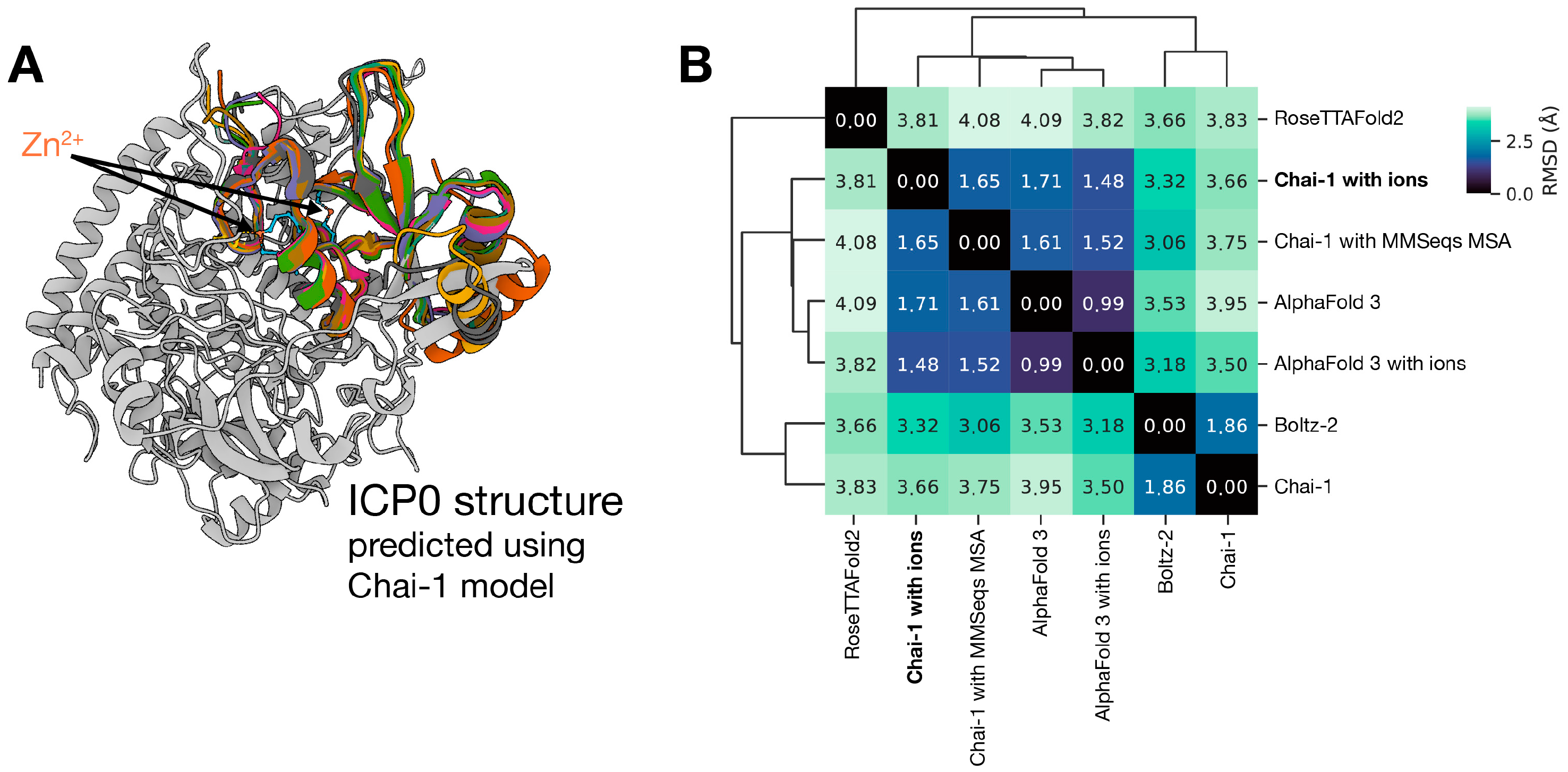
4.1. ICP0 Sequence Acquisition and AI-Driven Structural Prediction
4.2. Druggability Analysis and Site Selection
4.3. Small Molecule Library Preparation
4.4. Virtual Screening and Hit Identification
4.5. Hit Elaboration and Bivalent Degrader Design
4.6. ADMET Prediction
4.7. Structural Visualization
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| HSV-1 | Human Herpes Simplex Virus 1 |
| TPD | Targeted Protein Degradation |
| PROTAC | Proteolysis-Targeting Chimera |
| UPS | Ubiquitin–Proteasome System |
| ADMET | Absorption, Distribution, Metabolism, Excretion, and Toxicity |
| RMSD | Root Mean Square Deviation |
| ICP | Infected Cell Protein |
| CYP | Cytochrome P450 |
| pLDDT | predicted Local Distance Difference Test |
| pTM | predicted TM-score |
| ipTM | interface pTM |
| SA Score | Synthetic Accessibility Score |
| SASA | Solvent-Accessible Surface Area |
Appendix A
Appendix A.1

Appendix A.2. Structural Model Confidence
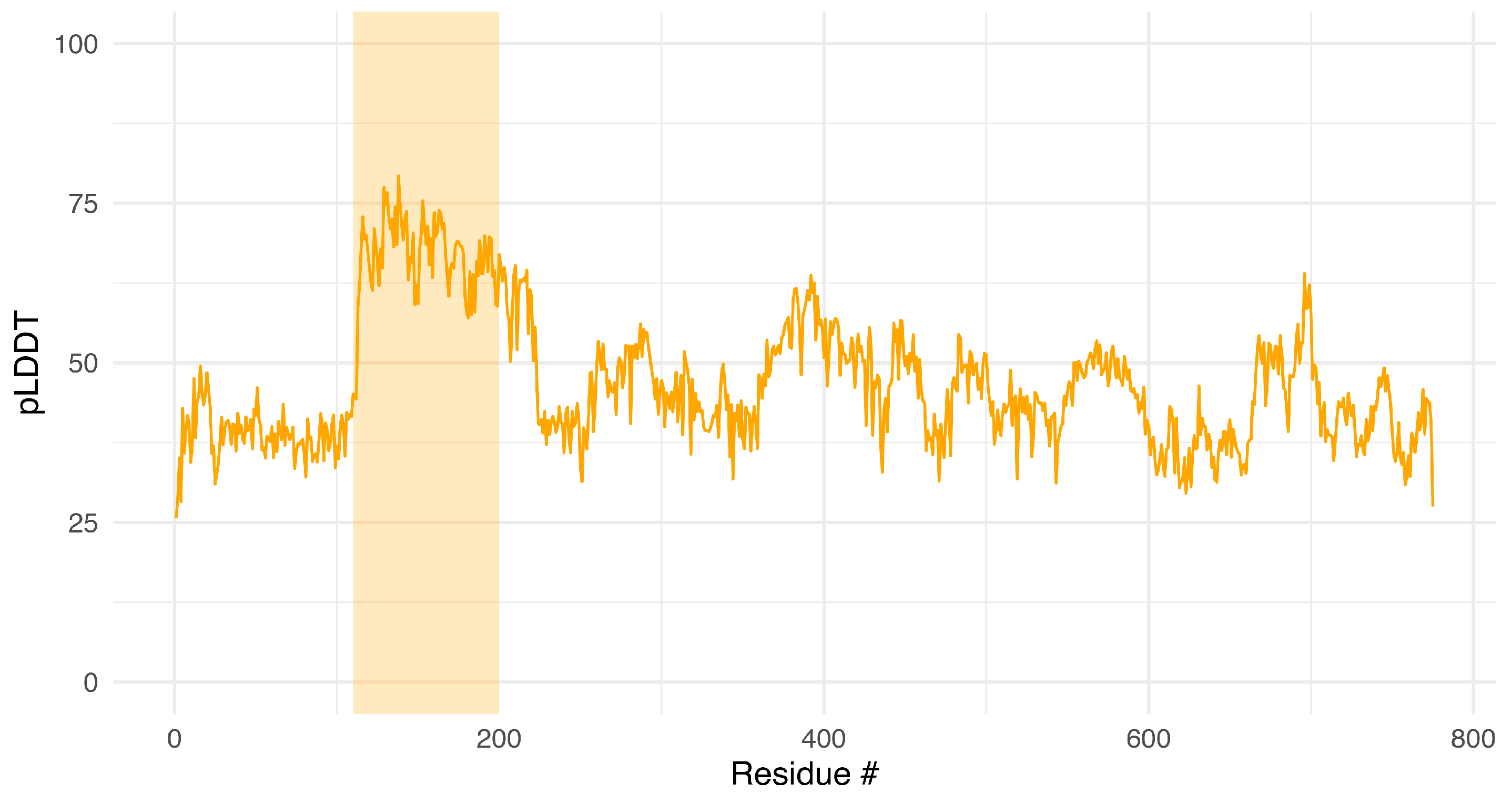
Appendix A.3. Predicted ADMET Profile of Lead Candidate ICP0-deg-01
| Property | Predicted Value |
|---|---|
| Molecular Weight (Da) | 1091.34 |
| Consensus LogP | 9.84 |
| Topological Polar Surface Area (Å2) | 220.82 |
| Rotatable Bonds | 27 |
| Aqueous Solubility (ESOL) | Insoluble (LogS = −12.5) |
| GI Absorption | Low |
| Blood–Brain Barrier Permeation | No |
| P-glycoprotein (Pgp) Substrate | Yes |
| CYP1A2 Inhibitor | No |
| CYP2C19 Inhibitor | No |
| CYP2C9 Inhibitor | No |
| CYP2D6 Inhibitor | No |
| CYP3A4 Inhibitor | No |
| Lipinski Rule of 5 Violations | 4 |
| Bioavailability Score | 0.17 |
| PAINS Alerts | 0 |
| Synthetic Accessibility Score | 8.7 |
References
- James, C.; Harfouche, M.; Welton, N.J.; Turner, K.M.; Abu-Raddad, L.J.; Gottlieb, S.L.; Looker, K.J. Herpes Simplex Virus: Global Infection Prevalence and Incidence Estimates, 2016. Bull. World Health Organ. 2020, 98, 315–329. [Google Scholar] [CrossRef]
- Su, D.; Han, L.; Shi, C.; Li, Y.; Qian, S.; Feng, Z.; Yu, L. An Updated Review of HSV-1 Infection-Associated Diseases and Treatment, Vaccine Development, and Vector Therapy Application. Virulence 2024, 15, 2425744. [Google Scholar] [CrossRef]
- Riley, G.; Cloete, E.; Walls, T. Challenges of Timely Investigation and Treatment of Neonatal Herpes Simplex Virus Infection. J. Paediatr. Child Health 2023, 59, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.J.; Jaggi, U.; Tormanen, K.; Wang, S.; Hirose, S.; Ghiasi, H. The Anti-Apoptotic Function of HSV-1 LAT in Neuronal Cell Cultures but Not Its Function during Reactivation Correlates with Expression of Two Small Non-Coding RNAs, sncRNA1&2. PLoS Pathog. 2024, 20, e1012307. [Google Scholar] [CrossRef] [PubMed]
- Dochnal, S.A.; Krakowiak, P.A.; Whitford, A.L.; Cliffe, A.R. Physiological Oxygen Concentration during Sympathetic Primary Neuron Culture Improves Neuronal Health and Reduces HSV-1 Reactivation. Microbiol. Spectr. 2024, 12, e02031-24. [Google Scholar] [CrossRef] [PubMed]
- Grams, T.R.; Edwards, T.G.; Bloom, D.C. A Viral lncRNA Tethers HSV-1 Genomes at the Nuclear Periphery to Establish Viral Latency. J. Virol. 2023, 97, e01438-23. [Google Scholar] [CrossRef]
- Bautista, L.; Sirimanotham, C.; Espinoza, J.; Cheng, D.; Tay, S.; Drayman, N. A Drug Repurposing Screen Identifies Decitabine as an HSV-1 Antiviral. Microbiol. Spectr. 2024, 12, e01754-24. [Google Scholar] [CrossRef]
- Gosavi, A.A.; Thorat, P.A.; Mulla, J.A.S. Formulation and Evaluation of Acyclovir Loaded Transferosomal Gel for Transdermal Drug Delivery. J. Drug Deliv. Ther. 2024, 14, 122–130. [Google Scholar] [CrossRef]
- Spataro, F.; Ria, R.; Choul, N.; Vacca, A.; Solimando, A.G.; Girolamo, A.D. Acyclovir Desensitization. A Case Report and a Review of Desensitization Strategies. J. Infect. Dev. Ctries. 2025, 19, 174–180. [Google Scholar] [CrossRef]
- Kawsar, S.M.A.; Munia, N.S.; Saha, S.; Ozeki, Y. In Silico Pharmacokinetics, Molecular Docking and Molecular Dynamics Simulation Studies of Nucleoside Analogs for Drug Discovery—A Mini Review. Mini-Rev. Med. Chem. 2024, 24, 1070–1088. [Google Scholar] [CrossRef]
- Liu, S.; Knafels, J.D.; Chang, J.S.; Waszak, G.A.; Baldwin, E.T.; Deibel, M.R.; Thomsen, D.R.; Homa, F.L.; Wells, P.A.; Tory, M.C.; et al. Crystal Structure of the Herpes Simplex Virus 1 DNA Polymerase*. J. Biol. Chem. 2006, 281, 18193–18200. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, D.M.; Castillo, E.; Duarte, L.F.; Arriagada, J.; Corrales, N.; Farías, M.A.; Henríquez, A.; Agurto-Muñoz, C.; González, P.A. Current Antivirals and Novel Botanical Molecules Interfering with Herpes Simplex Virus Infection. Front. Microbiol. 2020, 11, 139. [Google Scholar] [CrossRef] [PubMed]
- Birkmann, A.; Bonsmann, S.; Kropeit, D.; Pfaff, T.; Rangaraju, M.; Sumner, M.; Timmler, B.; Zimmermann, H.; Buschmann, H.; Ruebsamen-Schaeff, H. Discovery, Chemistry, and Preclinical Development of Pritelivir, a Novel Treatment Option for Acyclovir-Resistant Herpes Simplex Virus Infections. J. Med. Chem. 2022, 65, 13614–13628. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Zhou, L.; Wu, J.; Cheng, J.; Duan, Y.; Qian, W. Anti-HSV-1 Agents: An Update. Front. Pharmacol. 2025, 15, 1451083. [Google Scholar] [CrossRef]
- Frejborg, F.; Kalke, K.; Hukkanen, V. Current Landscape in Antiviral Drug Development against Herpes Simplex Virus Infections. Smart Med. 2022, 1, e20220004. [Google Scholar] [CrossRef] [PubMed]
- Kuo, J.-Y.; Yeh, C.-S.; Wang, S.-M.; Chen, S.-H.; Wang, J.-R.; Chen, T.-Y.; Tsai, H.-P. Acyclovir-Resistant HSV-1 Isolates among Immunocompromised Patients in Southern Taiwan: Low Prevalence and Novel Mutations. J. Med. Virol. 2023, 95, e28985. [Google Scholar] [CrossRef]
- Zhong, G.; Chang, X.; Xie, W.; Zhou, X. Targeted Protein Degradation: Advances in Drug Discovery and Clinical Practice. Signal Transduct. Target. Ther. 2024, 9, 308. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, F.; Pal, S.; Hu, Q. Proteolysis-Targeting Drug Delivery System (ProDDS): Integrating Targeted Protein Degradation Concepts into Formulation Design. Chem. Soc. Rev. 2024, 53, 9582–9608. [Google Scholar] [CrossRef]
- Xue, Y.; Bolinger, A.A.; Zhou, J. Novel Approaches to Targeted Protein Degradation Technologies in Drug Discovery. Expert Opin. Drug Discov. 2023, 18, 467–483. [Google Scholar] [CrossRef]
- Ambrozkiewicz, M.C.; Cuthill, K.J.; Harnett, D.; Kawabe, H.; Tarabykin, V. Molecular Evolution, Neurodevelopmental Roles and Clinical Significance of HECT-Type UBE3 E3 Ubiquitin Ligases. Cells 2020, 9, 2455. [Google Scholar] [CrossRef]
- Pan, B.; Mountford, S.J.; Kiso, M.; Anderson, D.E.; Papadakis, G.; Jarman, K.E.; Tilmanis, D.R.; Maher, B.; Tran, T.; Shortt, J.; et al. Targeted Protein Degraders of SARS-CoV-2 Mpro Are More Active than Enzymatic Inhibition Alone with Activity against Nirmatrelvir Resistant Virus. Commun. Med. 2025, 5, 140. [Google Scholar] [CrossRef] [PubMed]
- Borges, P.H.O.; Ferreira, S.B.; Silva, F.P. Recent Advances on Targeting Proteases for Antiviral Development. Viruses 2024, 16, 366. [Google Scholar] [CrossRef]
- McCloskey, E.; Kashipathy, M.; Cooper, A.; Gao, P.; Johnson, D.K.; Battaile, K.P.; Lovell, S.; Davido, D.J. HSV-1 ICP0 Dimer Domain Adopts a Novel β-Barrel Fold. Proteins Struct. Funct. Bioinform. 2024, 92, 830–841. [Google Scholar] [CrossRef]
- Perusina Lanfranca, M.; van Loben Sels, J.M.; Ly, C.Y.; Grams, T.R.; Dhummakupt, A.; Bloom, D.C.; Davido, D.J. A 77 Amino Acid Region in the N-Terminal Half of the HSV-1 E3 Ubiquitin Ligase ICP0 Contributes to Counteracting an Established Type 1 Interferon Response. Microbiol. Spectr. 2022, 10, e00593-22. [Google Scholar] [CrossRef]
- Harrell, T.L.; Davido, D.J.; Bertke, A.S. Herpes Simplex Virus 1 (HSV-1) Infected Cell Protein 0 (ICP0) Targets of Ubiquitination during Productive Infection of Primary Adult Sensory Neurons. Int. J. Mol. Sci. 2023, 24, 2931. [Google Scholar] [CrossRef]
- Jan Fada, B.; Guha, U.; Zheng, Y.; Reward, E.; Kaadi, E.; Dourra, A.; Gu, H. A Novel Recognition by the E3 Ubiquitin Ligase of HSV-1 ICP0 Enhances the Degradation of PML Isoform I to Prevent ND10 Reformation in Late Infection. Viruses 2023, 15, 1070. [Google Scholar] [CrossRef]
- Hou, F.; Sun, Z.; Deng, Y.; Chen, S.; Yang, X.; Ji, F.; Zhou, M.; Ren, K.; Pan, D. Interactome and Ubiquitinome Analyses Identify Functional Targets of Herpes Simplex Virus 1 Infected Cell Protein 0. Front. Microbiol. 2022, 13, 856471. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Hembram, D. The Viral SUMO-Targeted Ubiquitin Ligase ICP0 Is Phosphorylated and Activated by Host Kinase Chk2. FASEB J. 2021, 35, 1952–1977. [Google Scholar] [CrossRef]
- Perusina Lanfranca, M.; Mostafa, H.H.; Davido, D.J. HSV-1 ICP0: An E3 Ubiquitin Ligase That Counteracts Host Intrinsic and Innate Immunity. Cells 2014, 3, 438–454. [Google Scholar] [CrossRef]
- Rodríguez, M.C.; Dybas, J.M.; Hughes, J.; Weitzman, M.D.; Boutell, C. The HSV-1 Ubiquitin Ligase ICP0: Modifying the Cellular Proteome to Promote Infection. Virus Res. 2020, 285, 198015. [Google Scholar] [CrossRef]
- Maillet, S.; Naas, T.; Crepin, S.; Roque-Afonso, A.-M.; Lafay, F.; Efstathiou, S.; Labetoulle, M. Herpes Simplex Virus Type 1 Latently Infected Neurons Differentially Express Latency-Associated and ICP0 Transcripts. J. Virol. 2006, 80, 9310–9321. [Google Scholar] [CrossRef]
- Zografou-Barredo, N.A.; Hallatt, A.J.; Goujon-Ricci, J.; Cano, C. A Beginner’s Guide to Current Synthetic Linker Strategies towards VHL-Recruiting PROTACs. Bioorg. Med. Chem. 2023, 88–89, 117334. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Passaro, S.; Corso, G.; Wohlwend, J.; Reveiz, M.; Thaler, S.; Somnath, V.R.; Getz, N.; Portnoi, T.; Roy, J.; Stark, H.; et al. Boltz-2: Towards Accurate and Efficient Binding Affinity Prediction. bioRxiv 2025. [Google Scholar] [CrossRef]
- Discovery, C.; Boitreaud, J.; Dent, J.; McPartlon, M.; Meier, J.; Reis, V.; Rogozhnikov, A.; Wu, K. Chai-1: Decoding the Molecular Interactions of Life. bioRxiv 2024. [Google Scholar] [CrossRef]
- Team, B.A.A.; Chen, X.; Zhang, Y.; Lu, C.; Ma, W.; Guan, J.; Gong, C.; Yang, J.; Zhang, H.; Zhang, K.; et al. Protenix—Advancing Structure Prediction Through a Comprehensive AlphaFold3 Reproduction. bioRxiv 2025. [Google Scholar] [CrossRef]
- Baek, M.; Anishchenko, I.; Humphreys, I.R.; Cong, Q.; Baker, D.; DiMaio, F. Efficient and Accurate Prediction of Protein Structure Using RoseTTAFold2. bioRxiv 2023. [Google Scholar] [CrossRef]
- Minibaeva, G.; Polishchuk, P. CReM-Dock: De Novo Design of Synthetically Feasible Compounds Guided by Molecular Docking. ChemRxiv 2024. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern Web App for 3D Visualization and Analysis of Large Biomolecular Structures. Nucleic Acids Res. 2021, 49, W431–W437. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
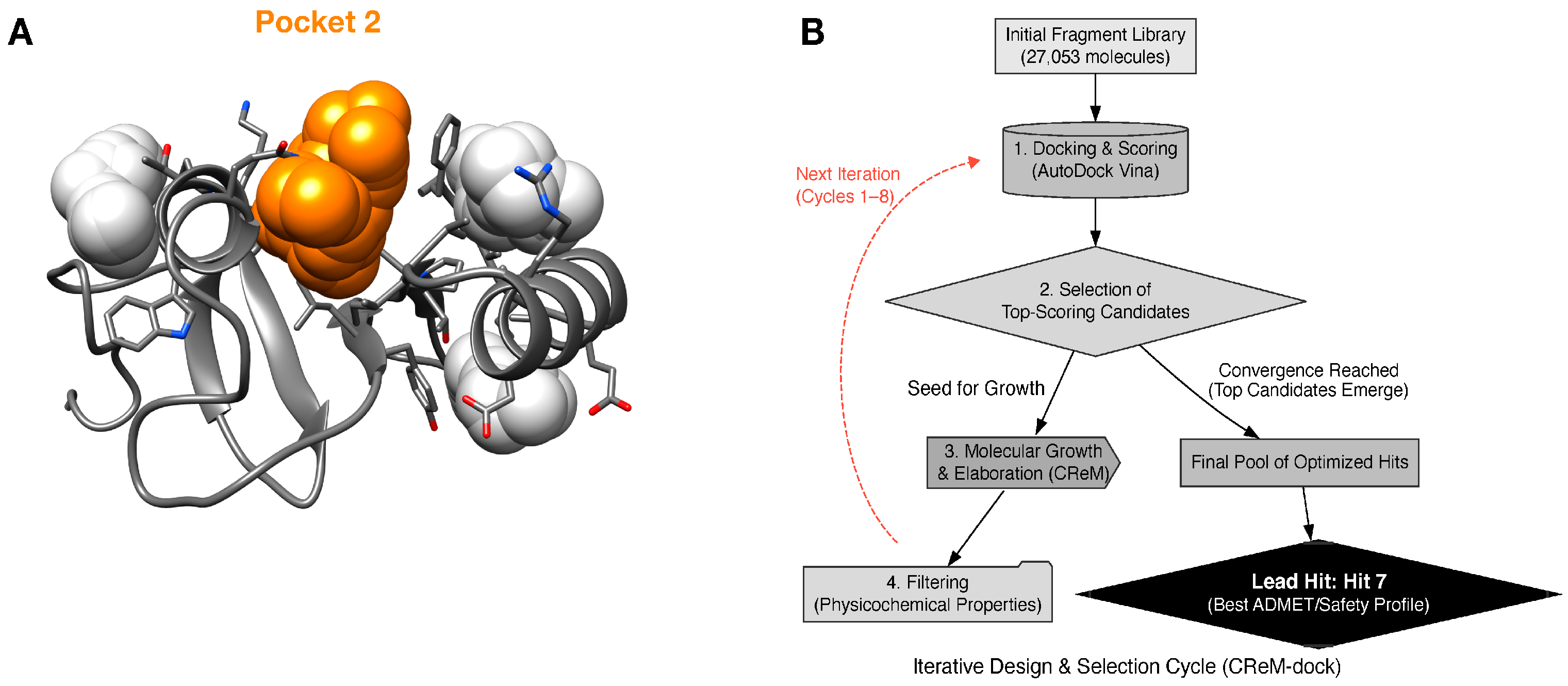
| Property | Pocket 1 | Pocket 2 (Target) | Pocket 3 | Pocket 4 |
|---|---|---|---|---|
| Druggability Score | 0.115 | 0.215 | 0.008 | 0.001 |
| Volume (Å3) | 303.173 | 617.16 | 249.952 | 290.706 |
| Number of Alpha Spheres | 35 | 59 | 16 | 17 |
| Total SASA (Å2) | 98.302 | 197.97 | 80.75 | 87.032 |
| Apolar SASA (Å2) | 71.25 | 130.424 | 73.249 | 59.174 |
| Hits | Structure | Predicted Affinity (kcal/mol) | MW (Da) | Consensus LogP | ESOL LogS | Predicted CYP Inhibition | SA Score |
|---|---|---|---|---|---|---|---|
| Hit 1 | 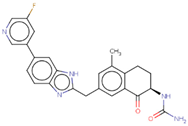 | −10.887 | 443.47 | 3.41 | −4.9 | 1A2, 2C19, 2C9, 3A4 | 3.66 |
| Hit 2 | 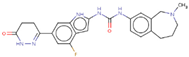 | −10.758 | 448.49 | 2.96 | −4.06 | 1A2, 2D6 | 3.78 |
| Hit 3 | 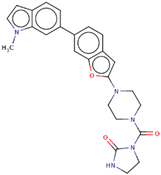 | −10.743 | 443.5 | 2.72 | −4.71 | 2C19, 2C9, 2D6, 3A4 | 3.65 |
| Hit 4 |  | −10.677 | 435.48 | 3.34 | −5.31 | 1A2, 2C19, 2C9, 2D6, 3A4 | 3.66 |
| Hit 5 | 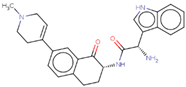 | −10.61 | 428.53 | 2.75 | −4.15 | 2C19, 2C9, 3A4 | 4.36 |
| Hit 6 | 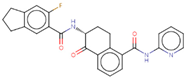 | −10.608 | 443.47 | 3.85 | −5.22 | 1A2, 2C19, 2C9, 2D6, 3A4 | 3.64 |
| Hit 7 |  | −10.557 | 434.47 | 2.73 | −3.69 | 1A2 | 3.62 |
| Hit 8 | 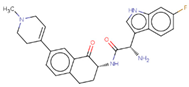 | −10.462 | 446.52 | 3.06 | −4.31 | 2C19, 2D6, 3A4 | 4.44 |
| Hit 9 |  | −10.433 | 446.56 | 3.55 | −5.18 | 1A2, 2D6, 3A4 | 3.39 |
| Hit 10 | 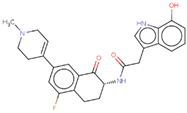 | −10.431 | 447.5 | 3.34 | −4.61 | 2C19, 2C9, 3A4 | 4.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salimova, L.; Sahin, A.; Ardicli, O.; Kurtoglu Babayev, F.H.; Sari, Z.B.; Sari, M.E.; Kurtoglu, M.G.; Ardicli, S.; Babayev, H. Design of a First-in-Class homoPROTAC to Induce ICP0 Degradation in Human Herpes Simplex Virus 1. Drugs Drug Candidates 2025, 4, 42. https://doi.org/10.3390/ddc4030042
Salimova L, Sahin A, Ardicli O, Kurtoglu Babayev FH, Sari ZB, Sari ME, Kurtoglu MG, Ardicli S, Babayev H. Design of a First-in-Class homoPROTAC to Induce ICP0 Degradation in Human Herpes Simplex Virus 1. Drugs and Drug Candidates. 2025; 4(3):42. https://doi.org/10.3390/ddc4030042
Chicago/Turabian StyleSalimova, Leyla, Ali Sahin, Ozge Ardicli, Fatima Hacer Kurtoglu Babayev, Zeynep Betul Sari, Muhammed Emin Sari, Muhammet Guzel Kurtoglu, Sena Ardicli, and Huseyn Babayev. 2025. "Design of a First-in-Class homoPROTAC to Induce ICP0 Degradation in Human Herpes Simplex Virus 1" Drugs and Drug Candidates 4, no. 3: 42. https://doi.org/10.3390/ddc4030042
APA StyleSalimova, L., Sahin, A., Ardicli, O., Kurtoglu Babayev, F. H., Sari, Z. B., Sari, M. E., Kurtoglu, M. G., Ardicli, S., & Babayev, H. (2025). Design of a First-in-Class homoPROTAC to Induce ICP0 Degradation in Human Herpes Simplex Virus 1. Drugs and Drug Candidates, 4(3), 42. https://doi.org/10.3390/ddc4030042








