MET-Targeting Anticancer Drugs—De Novo Design and Identification by Drug Repurposing
Abstract
1. Introduction
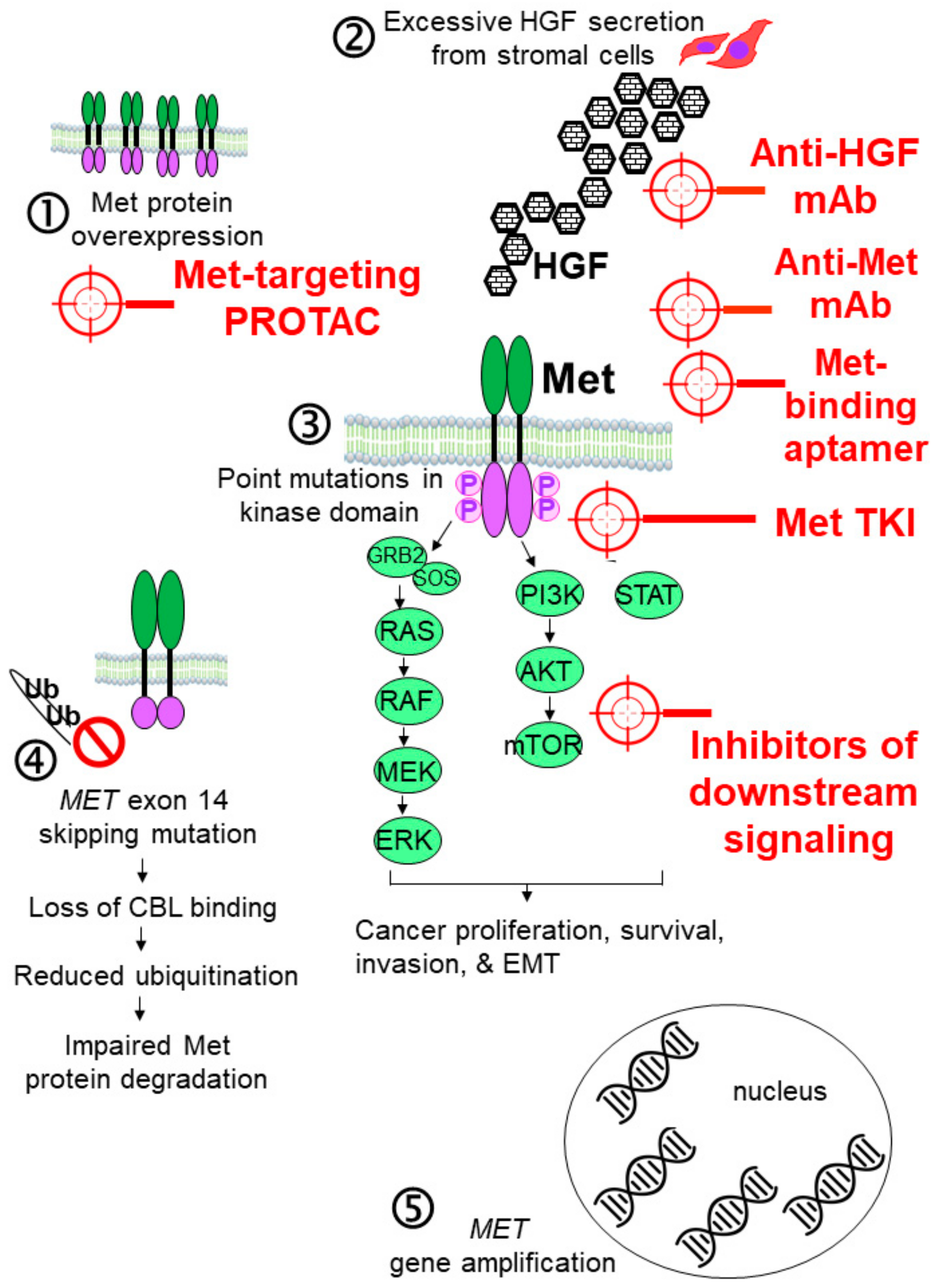
2. Recent Development of Met-Targeting Drug Candidates for Cancer Therapy
2.1. Antibody-Based Inhibitors of the HGF-MET Axis
2.1.1. Anti-HGF mAbs
2.1.2. Anti-Met mAbs
2.1.3. Bispecific Antibodies Simultaneously Targeting Met and Other Signaling Proteins
Simultaneously Targeting Met and EGFR
Simultaneously Targeting Met and VEGFR-2
Simultaneously Targeting Met and PD-1
2.1.4. Antibody–Drug Conjugates (ADC) Targeting Met
2.2. Inhibiting Met Dimerization
2.3. Small-Molecule Met Tyrosine Kinase Inhibitors (TKIs)
2.3.1. Type Ia Met TKIs
2.3.2. Type Ib Met TKIs
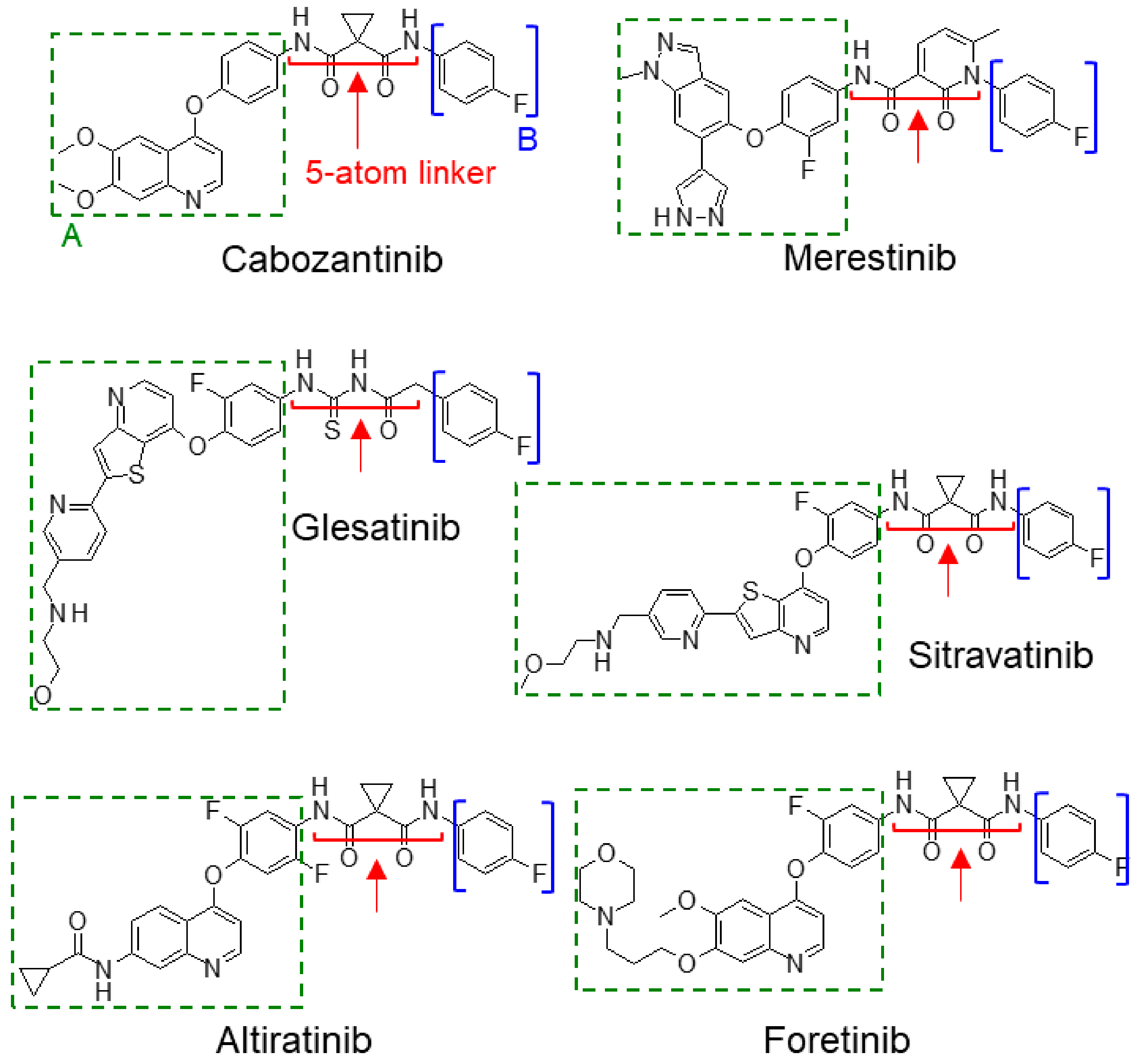
2.3.3. Type II Met TKIs
2.3.4. Type III Met TKIs
2.3.5. Novel Met TKIs with Distinct Binding Mode
2.3.6. Met Inhibitors Derived from Natural Sources
2.3.7. Met-Targeting PROTACs
3. Repurposing of Non-Oncology Drug as Met Inhibitors
3.1. High-Content-Analysis (HCA)-Based Screening for Met Inhibitors
3.2. In Silico Structure-Based Repurposing Screening for Met Inhibitors
3.3. Kinobeads Technology for Kinase Drug Repurposing
4. Combination of Met Inhibitors with Other Cancer Treatment Modalities to Overcome Drug Resistance
4.1. Use of Met TKIs to Overcome Drug Resistance to EGFR-Targeted TKIs
4.2. Use of Met Inhibitors to Potentiate Antitumor Response to Cancer Immunotherapy
4.3. Use of Met TKIs to Overcome Drug Resistance to Chemotherapy
5. Inherent and Acquired Resistance to Met TKIs
5.1. Inherent Resistance to Met TKIs
5.2. Acquired Resistance to Met TKIs
5.2.1. On-Target Resistance Mechanisms
5.2.2. Off-Target Resistance Mechanism
6. Further Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Giordano, S.; Ponzetto, C.; Di Renzo, M.F.; Cooper, C.S.; Comoglio, P.M. Tyrosine kinase receptor indistinguishable from the c-met protein. Nature 1989, 339, 155–156. [Google Scholar] [CrossRef]
- Naldini, L.; Vigna, E.; Narsimhan, R.P.; Gaudino, G.; Zarnegar, R.; Michalopoulos, G.K.; Comoglio, P.M. Hepatocyte growth factor (HGF) stimulates the tyrosine kinase activity of the receptor encoded by the proto-oncogene c-MET. Oncogene 1991, 6, 501–504. [Google Scholar] [PubMed]
- Bottaro, D.P.; Rubin, J.S.; Faletto, D.L.; Chan, A.M.; Kmiecik, T.E.; Vande Woude, G.F.; Aaronson, S.A. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 1991, 251, 802–804. [Google Scholar] [CrossRef] [PubMed]
- Linossi, E.M.; Estevam, G.O.; Oshima, M.; Fraser, J.S.; Collisson, E.A.; Jura, N. State of the structure address on MET receptor activation by HGF. Biochem. Soc. Trans. 2021, 49, 645–661. [Google Scholar] [CrossRef]
- Fu, J.; Su, X.; Li, Z.; Deng, L.; Liu, X.; Feng, X.; Peng, J. HGF/c-Met pathway in cancer: From molecular characterization to clinical evidence. Oncogene 2021, 40, 4625–4651. [Google Scholar] [CrossRef]
- Gentile, A.; Trusolino, L.; Comoglio, P.M. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008, 27, 85–94. [Google Scholar] [CrossRef]
- Chen, T.; You, Y.; Jiang, H.; Wang, Z.Z. Epithelial-mesenchymal transition (EMT): A biological process in the development, stem cell differentiation and tumorigenesis. J. Cell. Physiol. 2017, 232, 3261–3272. [Google Scholar] [CrossRef]
- Matsumoto, K.; Funakoshi, H.; Takahashi, H.; Sakai, K. HGF-Met pathway in regeneration and drug discovery. Biomedicines 2014, 2, 275–300. [Google Scholar] [CrossRef]
- Cooper, C.S.; Park, M.; Blair, D.G.; Tainsky, M.A.; Huebner, K.; Croce, C.M.; Vande Woude, G.F. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature 1984, 311, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xia, M.; Jin, K.; Wang, S.; Wei, H.; Fan, C.; Wu, Y.; Li, X.; Li, G.; Zeng, Z.; et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol. Cancer 2018, 17, 45. [Google Scholar] [CrossRef] [PubMed]
- Safi, D.; Hejleh, T.A.; Furqan, M. Narrative review: Mesenchymal-epithelial transition inhibitors-meeting their target. Transl. Lung Cancer Res. 2021, 10, 462–474. [Google Scholar] [CrossRef]
- Cappuzzo, F.; Marchetti, A.; Skokan, M.; Rossi, E.; Gajapathy, S.; Felicioni, L.; del Grammastro, M.; Sciarrotta, M.G.; Buttitta, F.; Incarbone, M.; et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J. Clin. Oncol. 2009, 27, 1667–1674. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Patil, T. MET alterations in advanced non-small cell lung cancer. Lung Cancer 2023, 178, 254–268. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Wu, W.; Wang, L.; Qu, J.; Han, Q.; Wang, H.; Song, S.; Liu, N.; Wang, Y.; Hou, H. Identification of MET fusions as novel therapeutic targets sensitive to MET inhibitors in lung cancer. J. Transl. Med. 2023, 21, 150. [Google Scholar] [CrossRef] [PubMed]
- Malik, R.; Mambetsariev, I.; Fricke, J.; Chawla, N.; Nam, A.; Pharaon, R.; Salgia, R. MET receptor in oncology: From Biomarker to therapeutic target. Adv. Cancer Res. 2020, 147, 259–301. [Google Scholar] [PubMed]
- Miranda, O.; Farooqui, M.; Siegfried, J.M. Status of agents targeting the HGF/c-Met axis in lung cancer. Cancers 2018, 10, 280. [Google Scholar] [CrossRef]
- Schoffski, P.; Gordon, M.; Smith, D.C.; Kurzrock, R.; Daud, A.; Vogelzang, N.J.; Lee, Y.; Scheffold, C.; Shapiro, G.I. Phase II randomized discontinuation trial of cabozantinib in patients with advanced solid tumors. Eur. J. Cancer 2017, 20, 296–304. [Google Scholar] [CrossRef]
- Scagliotti, G.; von Pawel, J.; Novello, S.; Ramlau, R.; Favaretto, A.; Barlesi, F.; Akerley, W.; Orlov, S.; Santoro, A.; Spigel, D.; et al. Phase III multinational, randomized, double-blind, placebo-controlled study of tivantinib (ARQ 197) plus erlotinib versus erlotinib alone in previously treated patients with locally advanced or metastatic nonsquamous non-small-cell lung cancer. J. Clin. Oncol. 2015, 33, 2667–2674. [Google Scholar] [CrossRef]
- Spigel, D.R.; Edelman, M.J.; O’byrne, K.; Paz-Ares, L.; Mocci, S.; Phan, S.; Shames, D.S.; Smith, D.; Yu, W.; Paton, V.E.; et al. Results from the phase III randomized trial of onartuzumab plus erlotinib versus erlotinib in previously treated stage IIIB or IV non-small-cell lung cancer: METLung. J. Clin. Oncol. 2017, 35, 412–420. [Google Scholar] [CrossRef]
- Lai, G.G.; Guo, R.; Drilon, A.; Tan, D.S. Refining patient selection of MET-activated non-small cell lung cancer through biomarker precision. Cancer Treat. Rev. 2022, 110, 102444. [Google Scholar] [CrossRef]
- Qin, K.; Hong, L.; Zhang, J.; Le, X. MET amplification as a resistance driver to TKI therapies in lung cancer: Clinical challenges and opportunities. Cancers 2023, 15, 612. [Google Scholar] [CrossRef]
- Santalahti, K.; Havulinna, A.; Maksimow, M.; Zeller, T.; Blankenberg, S.; Vehtari, A.; Joensuu, H.; Jalkanen, S.; Salomaa, V.; Salmi, M. Plasma levels of hepatocyte growth factor and placental growth factor predict mortality in a general population: A prospective cohort study. J. Intern. Med. 2017, 282, 340–352. [Google Scholar] [CrossRef] [PubMed]
- Matumoto, K.; Umitsu, M.; De Silva, D.M.; Roy, A.; Bottaro, D.P. Hepatocyte growth factor/MET in cancer progression and biomarker discovery. Cancer Sci. 2017, 108, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Doi, T.; Yamaguchi, K.; Komatsu, Y.; Muro, K.; Nishina, T.; Nakajima, T.E.; Tang, R.; Yang, H.; Zhang, Y.; Jung, A.S.; et al. A Phase 1/1b tolerability study of rilotumumab alone or in combination with cisplatin and capecitabine in Japanese patients with gastric cancer. Jpn. J. Clin. Oncol. 2017, 47, 1002–1009. [Google Scholar] [CrossRef]
- Iveson, T.; Donehower, R.C.; Davidenko, I.; Tjulandin, S.; Deptala, A.; Harrison, M.; Nirni, S.; Lakshmaiah, K.; Thomas, A.; Jiang, Y.; et al. Rilotumumab in combination with epirubicin, cisplatin, and capecitabine as first-line treatment for gastric or oesophagogastric junction adenocarcinoma: An open-label, dose de-escalation phase 1b study and a double-blind, randomized phase 2 study. Lancet Oncol. 2014, 15, 1007–1018. [Google Scholar] [CrossRef]
- Cunningham, D.T.N.; Davidenko, I.; Murad, A.M. Phase III, randomized, double-blind, multicenter, placebo (P)-controlled trial of rilotumumab (R) plus epirubicin, cisplatin and capecitabine (ECX) as first-line therapy in patients (pts) with advanced MET-positive (pos) gastric or gastroesophageal junction (G/GEJ) cancer: RILOMET-1 study. J. Clin. Oncol. 2015, 33, 4000. [Google Scholar]
- Mok, T.S.K.; Geater, S.L.; Su, W.-C.; Tan, E.-H.; Yang, J.C.-H.; Chang, G.-C.; Han, M.; Komarnitsky, P.; Payumo, F.; Garrus, J.E.; et al. A randomized phase 2 study comparing the combination of ficlatuzumab and gefitinib with gefitinib alone in Asian patients with advanced stage pulmonary adenocarcinoma. J. Thorac. Oncol. 2016, 11, 1736–1744. [Google Scholar] [CrossRef]
- Taguchi, F.; Solomon, B.; Gregorc, V.; Roder, H.; Gray, R.; Kasahara, K.; Nishio, M.; Brahmer, J.; Spreafico, A.; Ludovini, V.; et al. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: A multicohort cross-institutional study. J. Natl. Cancer Inst. 2007, 99, 838–846. [Google Scholar] [CrossRef] [PubMed]
- Binz, H.K.; Bakker, T.R.; Phillips, D.J.; Cornelius, A.; Zitt, C.; Gottler, T.; Sigrist, G.; Fiedler, U.; Ekawardhani, S.; Dolado, I.; et al. Design and characterization of MP0250, a tri-specific anti-HGF/anti-VEGF DARPin® drug candidate. mAbs 2017, 9, 1262–1269. [Google Scholar] [CrossRef]
- Fiedler, U.; Cornelius, A.; Ekawardhani, S.; Dawson, K.M.; Gilboy, P.; Stumpp, M.T.; Dolado, I. Potency of bortezomib in combination with MP0250, a bispecific VEGF- and HGF-targeting darpin, in a preclinical multiple myeloma model. J. Clin. Oncol. 2014, 32, e19574. [Google Scholar] [CrossRef]
- Merchant, M.; Ma, X.; Maun, H.R.; Zheng, Z.; Peng, J.; Romero, M.; Huang, A.; Yang, N.-Y.; Nishimura, M.; Greve, J.; et al. Monovalent antibody design and mechanism of action of onartuzumab, a MET antagonist with anti-tumor activity as a therapeutic agent. Proc. Natl. Acad. Sci. USA 2013, 110, E2987–E2996. [Google Scholar] [CrossRef]
- Spigel, D.R.; Ervin, T.J.; Ramlau, R.A.; Daniel, D.B.; Goldschmidt, J.H., Jr.; Blumenschein, G.R., Jr.; Krzakowski, M.J.; Robinet, G.; Godbert, B.; Barlesi, F.; et al. Randomized phase II trial of onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. J. Clin. Oncol. 2013, 31, 4105–4114. [Google Scholar] [CrossRef] [PubMed]
- Koeppen, H.; Yu, W.; Zha, J.; Pandita, A.; Penuel, E.; Rangell, L.; Raja, R.; Mohan, S.; Patel, R.; Desai, R.; et al. Biomarker analyses from a placebo-controlled phase II study evaluating erlotinib + onartuzumab in advanced non-small cell lung cancer: MET expression levels are predictive of patient benefit. Clin. Cancer Res. 2014, 20, 4488–4498. [Google Scholar] [CrossRef] [PubMed]
- Brower, V. Onartuzumab ineffective in non-small-cell lung cancer. Lancet Oncol. 2017, 18, e66. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zeng, W.; Wortinger, M.A.; Yan, S.B.; Cornwell, P.; Peek, V.L.; Stephens, J.R.; Tetreault, J.W.; Xia, J.; Manro, J.R.; et al. LY2875358, a neutralizing and internalizing anti-MET bivalent antibody, inhibits HGF-dependent and HGF-independent MET activation and tumor growth. Clin. Cancer Res. 2014, 20, 6059–6070. [Google Scholar] [CrossRef] [PubMed]
- Camidge, D.R.; Moran, T.; Demedts, I.; Grosch, H.; Mileham, K.; Molina, J.; Juan-Vidal, O.; Bepler, G.; Goldman, J.W.; Park, K.; et al. A randomized, open-label phase II study evaluating emibetuzumab plus erlotinib and emibetuzumab monotherapy in MET immunohistochemistry positive NSCLC patients with acquired resistance to erlotinib. Clin. Lung Cancer 2022, 23, 300–310. [Google Scholar] [CrossRef]
- Hultberg, A.; Morello, V.; Huyghe, L.; De Jonge, N.; Blanchetot, C.; Hanssens, V.; De Boeck, G.; Silence, K.; Festjens, E.; Heukers, R.; et al. Depleting MET-expressing tumor cells by ADCC provides a therapeutic advantage over inhibiting HGF/MET signaling. Cancer Res. 2015, 75, 3373–3383. [Google Scholar] [CrossRef]
- Aftimos, P.G.; Barthelemy, P.; Rolfo, C.D.; Hanssens, V.; De Jonge, N.; Dreier, K.S.T.; De Haard, H.; Peeters, M.; Thibault, A.; Awada, A. A phase I, first-in-human study of argx-111, a monoclonal antibody targeting c-met in patients with solid tumors. J. Clin. Oncol. 2017, 33, 2580. [Google Scholar] [CrossRef]
- Lee, J.M.; Kim, B.; Lee, S.B.; Jeong, Y.; Oh, Y.M.; Song, Y.-J.; Jung, S.; Choi, J.; Cheong, K.H.; Kim, D.U.; et al. Cbl-independent degradation of Met: Ways to avoid agonism of bivalent Met-targeting antibody. Oncogene 2014, 33, 34–43. [Google Scholar] [CrossRef]
- Oh, Y.M.; Song, Y.-J.; Lee, S.B.; Jeong, Y.; Kim, B.; Kim, G.W.; Kim, K.E.; Lee, J.M.; Cho, M.-Y.; Choi, J.; et al. A new anti-c-met antibody selected by a mechanism-based dual-screening method: Therapeutic potential in cancer. Mol. Cell 2012, 34, 523–529. [Google Scholar] [CrossRef]
- Lee, J.; Kim, S.T.; Park, S.; Lee, S.; Park, S.H.; Park, J.O.; Lim, H.Y.; Ahn, H.; Bok, H.; Kim, K.-M.; et al. Trial of anti-MET monoclonal antibody in MET-overexpressed refractory cancer. Clin. Color. Cancer 2018, 17, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Goetsch, L.; Tucker, L.; Zhang, Q.; Gonzalez, A.; Vaidya, K.S.; Oleksijew, A.; Boghaert, E.; Song, M.; Sokolova, I.; et al. Anti-c-met monoclonal antibody ABT-700 breaks oncogene addiction in tumors with MET amplification. BMC Cancer 2016, 16, 105. [Google Scholar] [CrossRef]
- Stickler, J.H.; LoRusso, P.; Salgia, R.; Kang, Y.K.; Yen, C.J.; Lin, C.C.; Ansell, P.; Motwani, M.; Wong, S.; Yue, H.; et al. Phase I dose-escalation and –expansion study of telisotuzumab (ABT-700), an anti-c-Met antibody, in patients with advanced solid tumors. Mol. Cancer Ther. 2020, 19, 1210–1217. [Google Scholar] [CrossRef]
- Wang, J.; Anderson, M.G.; Oleksijew, A.; Vaidya, K.S.; Boghaert, E.R.; Tucker, L.; Zhang, Q.; Han, E.K.; Palma, J.P.; Naumovski, L.; et al. ABBV-399, a c-Met antibody-drug conjugate that targets both MET-amplified and c-Met-overexpressing tumors, irrespective of MET pathway dependence. Clin. Cancer Res. 2017, 23, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Grandal, M.M.; Havrylov, S.; Poulsen, T.T.; Koefoed, K.; Dahlman, A.; Galler, G.R.; Conrotto, P.; Collins, S.; Eriksen, K.W.; Kaufman, D.; et al. Simultaneous targeting of two distinct epitopes on MET effectively inhibits MET- and HGF-driven tumor growth by multiple mechanisms. Mol. Cancer Ther. 2017, 16, 2780–2791. [Google Scholar] [CrossRef] [PubMed]
- Pollmann, S.E.; Calvert, V.S.; Rao, S.; Boca, S.M.; Madhavan, S.; Horak, I.D.; Kjaer, A.; Petricoin, E.F.; Kragh, M.; Poulsen, T.T. Acquired resistance to a MET antibody in vivo can be overcome by the MET antibody mixture Sym015. Mol. Cancer Ther. 2018, 17, 1259–1270. [Google Scholar] [CrossRef]
- Basilico, C.; Modica, C.; Maione, F.; Vigna, E.; Comoglio, P.M. Targeting the MET oncogene by concomitant inhibition of receptor and ligand via an antibody-“decoy” strategy. Int. J. Cancer 2018, 143, 1774–1785. [Google Scholar] [CrossRef]
- Lindsey, S.; Langhans, S.A. Crosstalk of oncogenic signaling pathways during epithelial-mesenchymal transition. Front. Oncol. 2014, 4, 358. [Google Scholar] [CrossRef]
- Peng, K.C.; Su, J.W.; Xie, Z.; Wang, H.M.; Fang, M.M.; Li, W.F.; Chen, Y.-Q.; Guan, X.-H.; Su, J.; Yan, H.-H.; et al. Clinical outcomes of EGFR+/METamp+ vs. EGFR+/METamp- untreated patients with advanced non-small cell lung cancer. Thorac. Cancer 2022, 13, 1619–1630. [Google Scholar] [CrossRef]
- Castoldi, R.; Ecker, V.; Wiehle, L.; Majety, M.; Busl-Schuller, R.; Asmussen, M.; Nopora, A.; Jucknischke, U.; Osl, F.; Kobold, S.; et al. A novel bispecific EGFR/met antibody blocks tumor-promoting phenotypic effects induced by resistance to EGFR inhibition and has potent antitumor activity. Oncogene 2013, 32, 5593–5601. [Google Scholar] [CrossRef]
- Patnaik, A.; Gordon, M.; Tsai, F.; Papadopoulos, K.P.; Rasco, D.; Beeram, M.; Fu, S.; Janku, F.; Hynes, S.M.; Gundala, S.R.; et al. A phase I study of LY3164530, a bispecific antibody targeting MET and EGFR, in patients with advanced or metastatic cancer. Cancer Chemother. Pharmacol. 2018, 82, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.; Gordon, E.; Claesson-Welsh, L. Mechanisms and regulation of endothelial VEGF receptor signaling. Nat. Rev. Mol. Cell Biol. 2016, 17, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.; Hurwitz, H.I.; Sandler, A.B.; Miles, D.; Coleman, R.L.; Deurloo, R.; Chinot, O.L. Bevacizumab (Avastin) in cancer treatment: A review of 15 years of clinical experience and future outlook. Cancer Treat. Rev. 2020, 86, 102017. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Kim, Y.J.; Lee, S.; Kim, Y.S. A heterodimeric Fc-based bispecific antibody simultaneously targeting VEGFR-2 and Met exhibits potent antitumor activity. Mol. Cancer Ther. 2013, 12, 2748–2759. [Google Scholar] [CrossRef]
- Sun, Z.-J.; Wu, Y.; Hou, W.-H.; Wang, Y.-X.; Yuan, Q.-Y.; Wang, H.-J.; Yu, M. A novel bispecific c-MET/PD-1 antibody with therapeutic potential in solid cancer. Oncotarget 2017, 8, 29067–29079. [Google Scholar] [CrossRef]
- Wu, Y.; Yu, M.; Sun, Z.; Hou, W.; Wang, Y.; Yuan, Q.; Mo, W. Generation and characterization of bispecific antibody targeting both PD-1 and c-Met. Protein Pept. Lett. 2018, 24, 1105–1112. [Google Scholar] [CrossRef]
- Hou, W.; Yuan, Q.; Yuan, X.; Wang, Y.; Mo, W.; Wang, H.; Yu, M. A novel tetravalent bispecific antibody targeting programmed death 1 and tyrosine-protein kinase Met for treatment of gastric cancer. Investig. New Drugs 2019, 37, 876–889. [Google Scholar] [CrossRef]
- Strickler, J.H.; Weekes, C.D.; Nemunaitis, J.; Ramanathan, R.K.; Heist, R.S.; Morgensztern, D.; Angevin, E.; Bauer, T.M.; Yue, H.; Motwani, M.; et al. First-in-human phase I, dose-escalation and -expansion study of telisotuzumab vedotin, an antibody-drug conjugate targeting c-Met, in patients with advanced solid tumors. J. Clin. Oncol. 2018, 36, 3298–3306. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Wang, L.; Sun, X.; Tang, M.; Quan, H.-T.; Zhang, L.-S.; Lou, L.-G.; Gou, S.-H. SHR-A1403, a novel c-Met antibody-drug conjugate, exerts encouraging anti-tumor activity in c-Met-overexpressing models. Acta Pharmacol. Sin. 2019, 40, 971–979. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, X.; Sun, X.; Li, J.; Wang, W.; Zhang, L.; Gou, S. Preclinical pharmacokinetics of a novel anti-c-Met antibody-drug conjugate, SHR-A1403, in rodents and non-human primates. Xenobiotica 2019, 49, 1097–1105. [Google Scholar] [CrossRef]
- Gymnopoulos, M.; Betancourt, O.; Blot, V.; Fujita, R.; Galvan, D.; Lieuw, V.; Nguyen, S.; Snedden, J.; Stewart, C.; Villicana, J.; et al. TR1801-ADC: A highly potent cMet antibody-drug conjugate with high activity in patient-derived xenograft models of solid tumors. Mol. Oncol. 2020, 14, 54–68. [Google Scholar] [CrossRef]
- Fujita, R.; Blot, V.; Wong, E.; Stewart, C.; Lieuw, V.; Richardson, R.; Banah, A.; Villicana, J.; Timmer, A.; Coronella, J.; et al. A novel no-agonist c-Met antibody drug conjugate with superior potency over a c-Met tyrosine kinase inhibitor in c-Met amplified and non-amplified cancers. Cancer Biol. Ther. 2020, 21, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Sellmann, C.; Doerner, A.; Knuehl, C.; Rasche, N.; Sood, V.; Krah, S.; Rhiel, L.; Messemer, A.; Wesolowski, J.; Schuette, M.; et al. Balancing selectivity and efficacy of bispecific epidermal growth factor receptor (EGFR) x c-MET antibodies and antibody-drug conjugates. J. Biol. Chem. 2016, 291, 25106–25119. [Google Scholar] [CrossRef] [PubMed]
- Prusty, D.K.; Adam, V.; Zadegan, R.M.; Irsen, S.; Famulok, M. Supramolecular aptamer nano-constructs for receptor-mediated targeting and light-triggered release of chemotherapeutics into cancer cells. Nat. Commun. 2018, 9, 535. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, L.; Tian, J.; Zhou, Z.; Li, J.; Yang, H. Nongenetic engineering strategies for regulating receptor oligomerization in living cells. Chem. Soc. Rev. 2020, 49, 1545–1568. [Google Scholar] [CrossRef] [PubMed]
- Ueki, R.; Sando, S. A DNA aptamer to c-Met inhibits cancer cell migration. Chem. Commun. 2014, 50, 13131–13134. [Google Scholar] [CrossRef]
- Piater, B.; Doerner, A.; Guenther, R.; Kolmar, H.; Hock, B. Aptamers binding to c-Met inhibiting tumor cell migration. PLoS ONE 2015, 10, e0142412. [Google Scholar] [CrossRef]
- Mogaki, R.; Okuro, K.; Ueki, R.; Sando, S.; Aida, T. Molecular glue that spatiotemporally turns on protein-protein interactions. J. Am. Chem. Soc. 2019, 141, 8035–8040. [Google Scholar] [CrossRef]
- Schiering, N.; Knapp, S.; Marconi, M.; Flocco, M.M.; Cui, J.; Perego, R.; Rusconi, L.; Cristiani, C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc. Natl. Acad. Sci. USA 2003, 100, 12654–12659. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.J. Targeting receptor tyrosine kinase MET in cancer: Small molecule inhibitors and clinical progress. J. Med. Chem. 2014, 57, 4427–4453. [Google Scholar] [CrossRef]
- Underiner, T.L.; Herbertz, T.; Miknyoczki, S.J. Discovery of small molecule c-met inhibitors: Evolution and profiles of clinical candidates. Anticancer Agents Med. Chem. 2010, 10, 7–27. [Google Scholar] [CrossRef] [PubMed]
- Lv, P.C.; Yang, Y.S.; Wang, Z.C. Recent progress in the development of small molecule c-Met inhibitors. Curr. Top. Med. Chem. 2019, 19, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Parikh, P.K.; Ghate, M.D. Recent advances in the discovery of small molecule c-Met kinase inhibitors. Eur. J. Med. Chem. 2017, 143, 1103–1138. [Google Scholar] [CrossRef] [PubMed]
- Modi, V.; Dunbrack, R.L. Defining a new nomenclature for the structures of active and inactive kinases. Proc. Natl. Acad. Sci. USA 2019, 116, 6818–6827. [Google Scholar] [CrossRef]
- Vijayan, R.S.K.; He, P.; Modi, V.; Duong-Ly, K.C.; Ma, H.; Peterson, J.R.; Dunbrack, R.L.; Levy, R.M. Conformational analysis of the DFG-out kinase motif and biochemical profiling of structurally validated type II inhibitors. J. Med. Chem. 2015, 58, 466–479. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacol. Res. 2016, 103, 26–48. [Google Scholar] [CrossRef] [PubMed]
- Nolen, B.; Taylor, S.; Ghosh, G. Regulation of protein kinases: Controlling activity through activation segment conformation. Mol. Cell 2004, 15, 661–675. [Google Scholar] [CrossRef]
- Eathiraj, S.; Palma, R.; Volckova, E.; Hirschi, M.; France, D.S.; Ashwell, M.A.; Chan, T.C. Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197. J. Biol. Chem. 2011, 286, 20666–20676. [Google Scholar] [CrossRef]
- Cui, J.J.; Tran-Dube, M.; Shen, H.; Nambu, M.; Kung, P.P.; Pairish, M.; Jia, L.; Meng, J.; Funk, L.; Botrous, I.; et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-Met) kinase and anaplastic lymphoma kinase (ALK). J. Med. Chem. 2011, 54, 6342–6363. [Google Scholar] [CrossRef]
- Drilon, A.E.; Camidge, D.R.; Ou, S.H.; Clark, J.W.; Socinski, M.A.; Weiss, J.; Riely, G.J.; Winter, M.; Wang, S.C.; Monti, K.; et al. Efficacy and safety of crizotinib in patients (PTS) with advanced MET exon 14-altered non-small cell drug cancer (NSCLC). J. Clin. Oncol. 2016, 34, 108. [Google Scholar] [CrossRef]
- Drilon, A.; Clark, J.W.; Weiss, J.; Ou, S.-H.I.; Camidge, D.R.; Solomon, B.J.; Otterson, G.A.; Villaruz, L.C.; Riely, G.J.; Heist, R.S.; et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat. Med. 2020, 26, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Kwak, E.L.; Bang, Y.-J.; Camidge, D.R.; Shaw, A.T.; Solomon, B.; Maki, R.G.; Ou, S.-H.I.; Dezube, B.J.; Jänne, P.A.; Costa, D.B.; et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010, 363, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Network NCC. National Comprehensive Cancer Network: Non-Small Cell Lung Cancer. 2022. Available online: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf (accessed on 1 May 2023).
- Mathieu, L.N.; Larkins, E.; Akinboro, O.; Roy, P.; Amatya, A.K.; Fiero, M.H.; Mishra-Kalyani, P.S.; Helms, W.S.; Myers, C.E.; Skinner, A.M.; et al. FDA approval summary: Capmatinib and tepotinib for the treatment of metastatic NSCLC harboring MET exon 14 skipping mutations or alterations. Clin. Cancer Res. 2022, 28, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Q.; Yang, G.; Marando, C.; Koblish, H.K.; Hall, L.M.; Fridman, J.S.; Behshad, E.; Wynn, R.; Li, Y.; et al. A novel kinase inhibitor, INCB28060, blocks c-MET-dependent signaling, neoplastic activities, and cross-talk with EGFR and HER-3. Clin. Cancer Res. 2011, 17, 7127–7138. [Google Scholar] [CrossRef]
- Markham, A. Tepotinib: First approval. Drugs 2020, 80, 829–833. [Google Scholar] [CrossRef]
- Wolf, J.; Seto, T.; Han, J.Y.; Reguart, N.; Garon, E.B.; Groen, H.J.; Tan, D.S.W.; Hida, T.; de Jonge, M.; Orlov, S.V.; et al. Capmatinib in MET exon 4-mutated or MET-amplified non-small-cell lung cancer. N. Engl. J. Med. 2020, 383, 944–957. [Google Scholar] [CrossRef]
- Paik, P.K.; Felip, E.; Veillon, R.; Sakai, H.; Cortot, A.B.; Garassino, M.C.; Mazieres, J.; Viteri, S.; Senellart, H.; Van Meerbeeck, J.; et al. Tepotinib in non-small-cell lung cancer with MET exom 14 skipping mutations. N. Engl. J. Med. 2020, 383, 931–943. [Google Scholar] [CrossRef]
- Zhu, X.; Lu, Y.; Lu, S. Landscape of savolitinib development for the treatment of non-small cell lung cancer with MET alteration—A narrative review. Cancers 2022, 14, 6122. [Google Scholar] [CrossRef]
- Lu, S.; Fang, J.; Cao, L.; Li, X.; Guo, Q.; Zhou, J.; Cheng, Y.; Jiang, L.; Chen, Y.; Zhang, H.; et al. Abstract CT031: Preliminary efficacy and safety results of savolitinib treating patients with pulmonary sarcomatoid carcinoma (PSC) and other types of non-small cell lung cancer (NSCLC) harboring MET exon 14 skipping mutations. Cancer Res. 2019, 79, CT031. [Google Scholar] [CrossRef]
- Ahn, M.; Cantarini, M.; Frewer, P.; Hawkins, G.; Peters, J.; Howarth, P.; Ahmed, G.; Sahota, T.; Hartmaier, R.; Li-Sucholeiki, X.; et al. P1.01-134 SAVANNAH: Phase II trial of osimertinib + savolitinib in EGFR-mutant, MET-driven advanced NSCLC, following prior osimertinib. J. Thorac. Oncol. 2019, 14, S415–S416. [Google Scholar] [CrossRef]
- Yu, H.; Goldberg, S.; Le, X.; Piotrowska, Z.; Smith, P.; Mensi, I.; Kirova, B.; Chmielecki, J.; Li-Sucholeicki, X.; Szekeres, P.; et al. P2.01-22 ORCHARD: A phase II platform study in patients with advanced NSCLC who have progressed on first-line osimertinib therapy. J. Thorac. Oncol. 2019, 14, S647. [Google Scholar] [CrossRef]
- Ugolini, A.; Kenigsberg, M.; Rak, A.; Vallee, F.; Houtmann, J.; Lowinski, M.; Capdevila, C.; Khider, J.; Albert, E.; Martinet, N.; et al. Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo [4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844). J. Med. Chem. 2016, 59, 7066–7074. [Google Scholar] [CrossRef] [PubMed]
- Angevin, E.; Spitaleri, G.; Rodon, J.; Dotti, K.; Isambert, N.; Salvagni, S.; Moreno, V.; Assadourian, S.; Gomez, C.; Harnois, M.; et al. A first-in-human phase I study of SAR125844, a selective MET tyrosine kinase inhibitor, in patients with advanced solid tumors with MET amplification. Eur. J. Cancer 2017, 87, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Shih, J.; Zhong, B.; Shi, H.; Xue, D.; Choy, G.S.; Redkar, S. Abstract 2096: Bozitinib, a highly selective inhibitor of cMet, demonstrates robust activity in gastric, lung, hepatic and pancreatic in vivo models. Cancer Res. 2017, 77, 2096. [Google Scholar] [CrossRef]
- Hu, H.; Mu, Q.; Bao, Z.; Chen, Y.; Liu, Y.; Chen, J.; Wang, K.; Wang, Z.; Nam, Y.; Jiang, B.; et al. Mutational landscape of secondary glioblastoma guides MET-targeted trial in brain tumor. Cell 2018, 175, 1665–1678. [Google Scholar] [CrossRef]
- Berthou, S.; Aebersold, D.M.; Schmidt, L.S.; Stroka, D.; Heigl, C.; Streit, B.; Stalder, D.; Gruber, G.; Liang, C.; Howlett, A.R.; et al. The Met kinase inhibitor SU11274 exhibits a selective inhibition pattern toward different receptor mutated variants. Oncogene 2004, 23, 5387–5393. [Google Scholar] [CrossRef]
- Felip, E.; Sakai, H.; Patel, J.; Horn, L.; Veillon, R.; Griesinger, F.; Bruns, R.; Scheele, J.; Paik, P. OA12.01 phase II data for the MET inhibitor tepotinib in patients with advanced NSCLC and MET exon 14-skipping mutations. J. Thorac. Oncol. 2018, 13, S347. [Google Scholar] [CrossRef]
- Wolf, J.; Seto, T.; Han, J.Y.; Reguart, N.; Garon, E.B.; Groen, H.J.M.; Tan, D.S.-W.; Hida, T.; De Jonge, M.J.; Orlov, S.V.; et al. Capmatinib (INC280) in METΔex14-mutated advanced non-small cell lung cancer (NSCLC): Efficacy data from the phase II GEOMETRY mono-1 study. J. Clin. Oncol. 2019, 37, 9004. [Google Scholar] [CrossRef]
- Landi, L.; Chiari, R.; Tiseo, M.; Dlnca, F.; Dazzi, C.; Chella, A.; Delmonte, A.; Bonanno, L.; Giannarelli, D.; Cortinovise, D.L.; et al. Crizotinib in MET deregulated or ROS1 rearranged pretreated non-small-cell lung cancer (METROS): A phase II, prospective, multicenter, two-arms trial. Clin. Cancer Res. 2019, 25, 7312–7319. [Google Scholar] [CrossRef]
- Paik, P.K.; Veillon, R.; Cortot, A.B.; Felip, E.; Sakai, H.; Mazieres, J.; Griesinger, F.; Horn, L.; Senellart, H.; Van Meerbeeck, J.P.; et al. Phase II study of tepotinib in NSCLC patients with METex14 mutations. J. Clin. Oncol. 2019, 37, 9005. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, Q.; Zhang, L.; Hu, S.; Chen, T.; Li, H.F.; Chen, Y.; Xu, Y.; Lu, T. Discovery, optimization and biological evaluation for novel c-Met kinase inhibitors. Eur. J. Med. Chem. 2018, 143, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, L.-S.; Xu, H.-C.; Wang, M.-S.; Zhao, X.-E.; Ming, Z.-H.; Zhu, X.-L.; Huang, W.; Yang, G.-F. 2,7-naphthyridinone-based MET kinase inhibitors: A promising novel scaffold for antitumor drug development. Eur. J. Med. Chem. 2019, 178, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Boezio, A.A.; Copeland, K.W.; Rex, K.; Albrecht, B.K.; Bauer, D.; Bellon, S.F.; Boezio, C.; Broome, M.A.; Choquette, D.; Coxon, A.; et al. Discovery of (R)-6-(1-(8-Fluoro-6-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl)ethyl)-3-(2-methoxyethoxy)-1,6-naphthyridin-5(6H)-one (AMG 337), a Potent and Selective Inhibitor of MET with High Unbound Target Coverage and Robust In Vivo Antitumor Activity. J. Med. Chem. 2016, 59, 2328–2342. [Google Scholar] [PubMed]
- Dussault, I.; Bellon, S.F. C-Met inhibitors with different binding modes: Two is better than one. Cell Cycle 2008, 7, 1157–1160. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Miyamoto, N.; Hirayama, T.; Oki, H.; Okada, K.; Tawada, M.; Iwata, H.; Nakamura, K.; Yamasaki, S.; Miki, H.; et al. Structure-based design, synthesis, and evaluation of imidazo[1,2-b]pyridazine and imidazo[1,2-a]pyridine derivatives as novel dual c-Met and VEGFR2 kinase inhibitors. Bioorg. Med. Chem. 2013, 21, 7686–7698. [Google Scholar] [CrossRef]
- Qi, B.; Yang, Y.; Gong, G.; He, H.; Yue, X.; Xu, X.; Hu, Y.; Li, J.; Chen, T.; Wan, X.; et al. Discovery of N1-(4-((7-(3-(4-ethylpiperazin-1-yl)propoxy)-6-methoxyquinolin-4-yl)oxy)-3,5-difluorophenyl)-N3-(2-(2,6-difluorophenyl)-4-oxothiazolidin-3-yl)urea as a multi-tyrosine kinase inhibitor for drug-sensitive and drug-resistant cancers treatment. Eur. J. Med. Chem. 2019, 163, 10–27. [Google Scholar] [CrossRef]
- Puccini, A.; Marín-Ramos, N.I.; Bergamo, F.; Schirripa, M.; Lonardi, S.; Lenz, H.-J.; Loupakis, F.; Battaglin, F. Safety and tolerability of c-MET inhibitors in cancer. Drug Saf. 2019, 42, 211. [Google Scholar] [CrossRef]
- D’angelo, N.D.; Bellon, S.F.; Booker, S.K.; Cheng, Y.; Coxon, A.; Dominguez, C.; Fellows, I.; Hoffman, D.; Hungate, R.; Kaplan-Lefko, P.; et al. Design, synthesis, and biological evaluation of potent c-Met inhibitors. J. Med. Chem. 2008, 51, 5766–5779. [Google Scholar] [CrossRef]
- Zhao, S.J.; Zhang, Y.; Zhou, H.Y.; Xi, S.C.; Zou, B.; Bao, G.L.; Wang, L.; Wang, J.; Zeng, T.; Gong, P.; et al. Synthesis and biological evaluation of 4-(2-fluorophenoxy)-3,3′-bipyridine derivatives as potential c-met inhibitors. Eur. J. Med. Chem. 2016, 120, 37–50. [Google Scholar] [CrossRef]
- Wang, L.X.; Xu, S.; Liu, X.B.; Chen, X.Y.; Xiong, H.H.; Hou, S.S.; Zou, W.; Tang, Q.; Zheng, P.; Zhu, W. Discovery of thinopyrimidine-triazole conjugates as c-Met targeting and apoptosis inducing agents. Bioorg. Chem. 2018, 77, 370–380. [Google Scholar] [CrossRef]
- Nan, X.; Li, H.J.; Fang, S.B.; Li, Q.Y.; Wu, Y.C. Structure-based discovery of novel 4-(2-fluorophenoxy)quinolone derivatives as c-Met inhibitors using isocyanide-involved multicomponent reactions. Eur. J. Med. Chem. 2020, 193, 112241. [Google Scholar] [CrossRef]
- Qi, B.H.; Yang, Y.; He, H.; Yue, X.P.; Zhou, Y.T.; Zhou, X.; Chen, Y.; Liu, M.; Zhang, A.; Wei, F. Identification of novel N1-(2-aryl-1,3-thiazolidin-4-one)-N3-aryl ureas showing potent multi-tyrosine kinase inhibitory activities. Eur. J. Med. Chem. 2018, 146, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.D.; Duan, Y.L.; Xiong, H.H.; Chen, T.; Xiao, Z.; Wang, L.X.; Xiao, Y.; Huang, S.; Xiong, Y.; Zhu, W.; et al. Synthesis and antiproliferative activity of 6,7-disubstituted-4-phenoxyquinoline derivatives bearing the 1,8-naphthyridin-2-one moiety. Eur. J. Med. Chem. 2018, 158, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.B.; Peek, V.L.; Ajamie, R.; Buchanan, S.G.; Graff, J.R.; Heidler, S.A.; Hui, Y.-H.; Huss, K.L.; Konicek, B.W.; Manro, J.R.; et al. LY2801653 is an orally bioavailable multi-kinase inhibitor with potent activity against MET, MST1R, and other oncoproteins, and displays anti-tumor activities in mouse xenograft models. Investig. New Drugs 2013, 31, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Konicek, B.W.; Capen, A.R.; Credille, K.M.; Ebert, P.J.; Falcon, B.L.; Heady, G.L.; Patel, B.K.; Peek, V.L.; Stephens, J.R.; Stewart, J.A.; et al. Merestinib (LY2801653) inhibits neurotrophic receptor kinase (NTRK) and suppresses growth of NTRK fusion bearing tumors. Oncotarget 2018, 9, 13796–13806. [Google Scholar] [CrossRef]
- Recondo, G.; Bahcall, M.; Spurr, L.F.; Che, J.; Ricciuti, B.; Leonardi, G.C.; Lo, Y.-C.; Li, Y.Y.; Lamberti, G.; Nguyen, T.; et al. Molecular mechanisms of acquired resistance to MET tyrosine kinase inhibitors in patients with MET exon 14 mutant NSCLC. Clin. Cancer Res. 2020, 26, 2615–2625. [Google Scholar] [CrossRef]
- Engstrom, L.D.; Aranda, R.; Lee, M.; Tovar, E.A.; Essenburg, C.J.; Madaj, Z.; Chiang, H.; Briere, D.; Hallin, J.; Lopez-Casas, P.P.; et al. Glesatinib exhibits antitumor activity in lung cancer models and patients harboring MET exon 14 mutations and overcomes mutation-mediated resistance to type I MET inhibitors in nonclinical models. Clin. Cancer Res. 2017, 23, 6661–6672. [Google Scholar] [CrossRef]
- Besterman, J.M.; Fournel, M.; Dupont, I.; Bonfils, C.; Dubay, M.; Ste-Croix, H.; Maroun, C.R. Potent preclinical antitumor activity of MGCD265, an oral Met/VEGFR kinase inhibitor in phase II clinical development, in combination with taxanes or erlotinib. J. Clin. Oncol. 2010, 28, e13595. [Google Scholar] [CrossRef]
- Kollmannsberger, C.; Hurwitz, H.; Bazhenova, L.; Cho, B.C.; Hong, D.; Park, K.; Reckamp, K.L.; Sharma, S.; Der-Torossian, H.; Christensen, J.G.; et al. Phase I study evaluating glesatinib (MGCD265), an inhibitor of MET and AXL, in patients with non-small cell lung cancer and other advanced solid tumors. Target. Oncol. 2023, 18, 105–118. [Google Scholar] [CrossRef]
- Peters, S.; Paz-Ares, L.; Herbst, R.S.; Reck, M. Addressing CPI resistance in NSCLC: Targeting TAM receptors to modulate the tumor microenvironment and future prospects. J. Immunother. Cancer 2022, 10, e004863. [Google Scholar] [CrossRef]
- He, K.; Berz, D.; Gadgeel, S.M.; Iams, W.T.; Bruno, D.S.; Blakely, C.M.; Spira, A.I.; Patel, M.R.; Waterhouse, D.M.; Richards, D.A.; et al. MRTX-500 phase 2 trial: Sitravatinib with nivolumab in patients with nonsquamous NSCLC progressing on or after checkpoint inhibitor therapy or chemotherapy. J. Thorac. Oncol. 2023, 18, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yu, X.; Huang, D.; Ma, Z.; Gao, B.; Cui, J.; Chu, Q.; Zhou, Q.; Sun, M.; Day, D.; et al. SAFERON-103: A phase 1b study of the safety and efficacy of sitravatinib combined with tislelizumab in patients with locally advanced or metastatic non-small cell lung cancer. J. Immunother. Cancer 2023, 11, e006055. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.D.; Kaufman, M.D.; Leary, C.B.; Turner, B.A.; Wise, S.C.; Ahn, Y.M.; Booth, R.J.; Caldwell, T.M.; Ensinger, C.L.; Hood, M.M.; et al. Altiratinib inhibits tumor growth, invasion, angiogenesis, and microenvironment-mediated drug resistance via balanced inhibition of MET, TIE2, and VEGFR2. Mol. Cancer Ther. 2015, 14, 2023–2034. [Google Scholar] [CrossRef]
- Piao, Y.; Park, S.Y.; Henry, V.; Smith, B.D.; Tiao, N.; Flynn, D.L.; De Groot, J.F. Novel MET/TIE2/VEGFR2 inhibitor altiratinib inhibits tumor growth and invasiveness in bevacizumab-resistant glioblastoma mouse models. Neuro Oncol. 2016, 18, 1230–1241. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, Y.; Mukohara, Y.; Tomioka, H.; Funakoshi, Y.; Kiyota, N.; Fujiwara, Y.; Yashiro, M.; Hirakawa, K.; Hirai, M.; Minami, H. Foretinib (GSK1363089), a multi-kinase inhibitor of MET and VEGFRs, inhibits growth of gastric cancer cell lines by blocking inter-receptor tyrosine kinase networks. Investig. New Drugs 2012, 30, 1352–1360. [Google Scholar] [CrossRef] [PubMed]
- Davare, M.A.; Saborowski, A.; Eide, C.A.; Tognon, C.; Smith, R.L.; Elferich, J.; Agarwal, A.; Tyner, J.W.; Shinde, U.P.; Lowe, S.W.; et al. Foretinib is a potent inhibitor of oncogenic ROS1 fusion proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 19519–19524. [Google Scholar] [CrossRef]
- Bergethon, K.; Shaw, A.T.; Ou, S.-H.I.; Katayama, R.; Lovly, C.M.; McDonald, N.T.; Massion, P.P.; Siwak-Tapp, C.; Gonzalez, A.; Fang, R.; et al. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 2012, 30, 863–870. [Google Scholar] [CrossRef]
- Fujino, T.; Mitsudomi, T. Acquired resistance mechanism for MET tyrosine kinase inhibitor. JTO Clin. Res. Rep. 2021, 2, 100134. [Google Scholar] [CrossRef]
- Fujino, T.; Suda, K.; Koga, T.; Hamada, A.; Ohara, S.; Chiba, M.; Shimoji, M.; Takemoto, T.; Soh, J.; Mitsudomi, T. Foretinib can overcome common on-target resistance mutations after capmatinib/tepotinib treatment in NSCLCs with MET exon 14 skipping mutation. J. Hematol. Oncol. 2022, 15, 79. [Google Scholar] [CrossRef]
- Shah, M.A.; Wainberg, Z.A.; Catenacci, D.V.; Hochster, H.S.; Ford, J.; Kunz, P.; Lee, F.-C.; Kallender, H.; Cecchi, F.; Rabe, D.C.; et al. Phase II study evaluating 2 dosing schedules of oral foretinib (GSK1363089), cMET/VEGFR2 inhibitor, in patients with metastatic gastric cancer. PLoS ONE 2013, 8, e54014. [Google Scholar] [CrossRef]
- Rimassa, L.; Personeni, N.; Simonelli, M.; Santoro, A. Tivantinib: A new promising mesenchymal-epithelial transition factor inhibitor in the treatment of hepatocellular carcinoma. Future Oncol. 2013, 9, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Munshi, N.; Jeay, S.; Li, Y.; Chen, C.-R.; France, D.S.; Ashwell, M.A.; Hill, J.; Moussa, M.M.; Leggett, D.S.; Li, C.J. ARQ197, a novel and selective inhibitor of the human c-Met receptor tyrosine kinase with antitumor activity. Mol. Cancer Ther. 2010, 9, 1544–1553. [Google Scholar] [CrossRef] [PubMed]
- Katayama, R.; Aoyama, A.; Yamori, T.; Qi, J.; Oh-Hara, T.; Song, Y.; Engelman, J.A.; Fujita, N. Cytotoxic activity of tivantinib (ARQ197) is not due solely to c-Met inhibition. Cancer Res. 2013, 73, 3087–3096. [Google Scholar] [CrossRef] [PubMed]
- Basilico, C.; Pennacchietti, S.; Vigna, E.; Chiriaco, C.; Arena, S.; Bardelli, A.; Valdembri, D.; Serini, G.; Michieli, P. Tivantinib (ARQ197) displays cytotoxic activity that is independent of its ability to bind Met. Clin. Cancer Res. 2013, 19, 2381–2392. [Google Scholar] [CrossRef]
- Weekes, C.D.; Clark, J.W.; Zhu, A.X. Tivantinib for advanced hepatocellular carcinoma: Is MET still a viable target? Lancet Oncol. 2018, 19, 591–592. [Google Scholar] [CrossRef]
- Santoro, A.; Rimassa, L.; Borbath, I.; Daniele, B.; Salvagni, S.; Van Laethem, J.L.; Van Vlierberghe, H.; Trojan, J.; Kolligs, F.T.; Weiss, A.; et al. Tivantinib for second-line treatment of advanced hepatocellular carcinoma: A randomized, placebo-controlled phase 2 study. Lancet Oncol. 2013, 14, 55–63. [Google Scholar] [CrossRef]
- Collie, G.W.; Barlind, L.; Bazzaz, S.; Börjesson, U.; Dale, I.L.; Disch, J.S.; Habeshian, S.; Jetson, R.; Khurana, P.; Madin, A.; et al. Discovery of a selective c-Met inhibitor with a novel binding mode. Bioorg. Med. Chem. Lett. 2022, 75, 128948. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, L.; Peng, J.; Ward, R.; Hao, P.; Wang, J.; Zhang, N.; Yang, Y.; Guo, X.; Xiang, C.; et al. Dictamnine, a novel c-Met inhibitor, suppresses the proliferation of lung cancer cells by downregulating the PI3K/AKT/mTOR and MAPK signaling pathways. Biochem. Pharmacol. 2022, 195, 114864. [Google Scholar] [CrossRef]
- An, S.; Yu, J.; Xu, T.; Hao, P. Application of dictamnine in preparation of c-Met inhibitor for treating cancer. Chinese Patent No. CN110755435, 2020. [Google Scholar]
- Aliebrahimi, S.; Kouhsari, S.M.; Arab, S.S.; Shadboorestan, A.; Ostad, S.N. Phytochemicals, withaferin A and carnosol, overcome pancreatic cancer stem cells as c-Met inhibitors. Biomed. Pharmacother. 2018, 106, 1527–1536. [Google Scholar] [CrossRef]
- Kelm, J.M.; Pandey, D.S.; Malin, E.; Kansou, H.; Arora, S.; Kumar, R.; Gavande, N.S. PROTAC’ing oncoproteins: Targeted protein degradation for cancer therapy. Mol. Cancer 2023, 22, 62. [Google Scholar] [CrossRef]
- Smith, B.E.; Wang, S.L.; Jaime-Figueroa, S.; Harbin, A.; Wang, J.; Hamman, B.D.; Crews, C.M. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat. Commun. 2019, 10, 131. [Google Scholar] [CrossRef] [PubMed]
- Bondeson, D.P.; Smith, B.E.; Burslem, G.M.; Buhimschi, A.D.; Hines, J.; Jaime-Figueroa, S.; Wang, J.; Hamman, B.D.; Ishchenko, A.; Crews, C.M. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem. Biol. 2018, 25, 78–87.e5. [Google Scholar] [CrossRef]
- Burslem, G.M.; Smith, B.E.; Lai, A.C.; Jaime-Figueroa, S.; McQuaid, D.C.; Bondeson, D.P.; Toure, M.; Dong, H.; Qian, Y.; Wang, J.; et al. The advantages of targeted protein degradation over inhibition: An RTK case study. Cell Chem. Biol. 2018, 25, 67–77.e3. [Google Scholar] [CrossRef] [PubMed]
- Sachkova, A.A.; Andreeva, D.V.; Tikhomirov, A.S.; Scherbakov, A.M.; Salnikova, D.I.; Sorokin, D.V.; Bogdanov, F.B.; Rysina, Y.D.; Shchekotikhin, A.E.; Shchegravina, E.S.; et al. Design, synthesis and in vitro investigation of cabozantinib-based PROTACs to target c-Met kinase. Pharmaceutics 2022, 14, 2829. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.; Cho, W.C. Drug repurposing for cancer therapy in the era of precision medicine. Curr. Mol. Pharmacol. 2022, 15, 895–903. [Google Scholar] [CrossRef]
- Denner, P.; Schmalowsky, J.; Prechtl, S. High-content analysis in preclinical drug discovery. Comb. Chem. High Throughput Screen. 2008, 11, 216–230. [Google Scholar] [CrossRef]
- Oh, J.W.; Oh, Y.J.; Han, S.; Her, N.G.; Nam, D.H. High-content analysis-based sensitivity prediction and novel therapeutics screening for c-Met-addicted glioblastoma. Cancers 2021, 13, 372. [Google Scholar] [CrossRef]
- Cutinho, P.F.; Venkataramana, C.H.S.; Suma, B.V. In silico hit identification, drug repurposing, pharmacokinetic and toxicity prediction of c-Met kinase inhibitors for cancer therapy. In Proceedings of the Conference on Drug Design and Discovery Technologies, Bengaluru, India, 21–22 November 2019; Murahari, M., Sundar, L., Chaki, S., Poongavanam, V., Bhat, P., Nayak, U.Y., Eds.; Royal Society of Chemistry: London, UK, 2019; pp. 54–59. [Google Scholar]
- Klaeger, S.; Heinzlmeir, S.; Wilhelm, M.; Polzer, H.; Vick, B.; Koenig, P.-A.; Reinecke, M.; Ruprecht, B.; Petzoldt, S.; Meng, C.; et al. The target landscape of clinical kinase drugs. Science 2017, 358, eaan4368. [Google Scholar] [CrossRef]
- Bantscheff, M.; Eberhard, D.; Abraham, Y.; Bastuck, S.; Boesche, M.; Hobson, S.; Mathieson, T.; Perrin, J.; Raida, M.; Rau, C.; et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 2007, 25, 1035–1044. [Google Scholar] [CrossRef]
- Fernandes, M.; Jamme, P.; Cortot, A.B.; Kherrouche, Z.; Tulasne, D. When the MET receptor kicks in to resist targeted therapies. Oncogene 2021, 40, 4061–4078. [Google Scholar] [CrossRef]
- Bean, J.; Brennan, C.; Shih, J.-Y.; Riely, G.; Viale, A.; Wang, L.; Chitale, D.; Motoi, N.; Szoke, J.; Broderick, S.; et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc. Natl. Acad. Sci. USA 2007, 104, 20932–20937. [Google Scholar] [CrossRef] [PubMed]
- Engelman, J.A.; Zejnullahu, K.; Mitsudomi, T.; Song, Y.; Hyland, C.; Park, J.O.; Lindeman, N.; Gale, C.-M.; Zhao, X.; Christensen, J.; et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007, 316, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef]
- Ramalingam, S.S.; Cheng, Y.; Zhou, C.; Ohe, Y.; Imamura, F.; Cho, B.C.; Lin, M.-C.; Majem, M.; Shah, R.; Rukazenkov, Y.; et al. LBA50 Mechanisms of acquired resistance to first-line osimertinib: Preliminary data from the phase III FLAURA study. Ann. Oncol. 2018, 29, 740. [Google Scholar] [CrossRef]
- Cooper, A.J.; Sequist, L.V.; Lin, J.J. Third-generation EGFR and ALK inhibitors: Mechanisms of resistance and management. Nat. Rev. Clin. Oncol. 2022, 19, 499–514. [Google Scholar] [CrossRef]
- Suzawa, K.; Offin, M.; Schoenfeld, A.J.; Plodkowski, A.J.; Odintsov, I.; Lu, D. Acquired MET exon 14 alteration drives secondary resistance to epidermal growth factor tyrosine kinase inhibitor in EGFR-mutated lung cancer. JCO Precis. Oncol. 2019, 3, PO.19.00011. [Google Scholar] [CrossRef]
- Le, X.; Puri, S.; Negrao, M.V.; Nilsson, M.B.; Robichaux, J.; Boyle, T.; Hicks, J.K.; Lovinger, K.L.; Roarty, E.; Rinsurongkawong, W.; et al. Landscape of EGFR-dependent and –independent resistance mechanisms to osimertinib and continuation therapy beyond progression in EGFR-mutant NSCLC. Clin. Cancer Res. 2018, 24, 6195–6203. [Google Scholar] [CrossRef]
- Shi, P.; Oh, Y.-T.; Zhang, G.; Yao, W.; Yue, P.; Li, Y.; Kanteti, R.; Riehm, J.; Salgia, R.; Owonikoko, T.K.; et al. Met gene amplification and protein hyperactivation is a mechanism of resistance to both first and third generation EGFR inhibitors in lung cancer treatment. Cancer Lett. 2016, 380, 494–504. [Google Scholar] [CrossRef]
- Deng, L.; Kiedrowski, L.A.; Ravera, E.; Cheng, H.; Halmos, B. Response to dual crizotinib and osimertinib treatment in a lung cancer patient with MET amplification detected by liquid biopsy who acquired secondary resistance to EGFR tyrosine kinase inhibition. J. Thorac. Oncol. 2018, 13, e169–e172. [Google Scholar] [CrossRef]
- Zhu, V.W.; Schrock, A.B.; Ali, S.M.; Ou, S.I. Differential response to a combination of full-dose osimertinib and crizotinib in a patient with EGFR-mutant non-small cell lung cancer and emergent MET amplification. Lung Cancer 2019, 10, 21–26. [Google Scholar] [CrossRef]
- Sequist, L.V.; Han, J.Y.; Ahn, M.J.; Cho, B.C.; Yu, H.; Kim, S.W.; Yang, J.C.-H.; Lee, J.S.; Su, W.-C.; Kowalski, D.; et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: Interim results from a multicenter, open-label, phase 1b study. Lancet Oncol. 2020, 21, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Smit, E.F.; Dooms, C.; Raskin, J.; Nadal, E.; Tho, L.M.; Le, X.; Mazieres, J.; Hin, H.S.; Morise, M.; Zhu, V.W.; et al. INSIGHT2: A phase II study of tepotinib plus osimertinib in MET-amplified NSCLC and first-line osimertinib resistance. Future Oncol. 2022, 18, 1039–1054. [Google Scholar] [CrossRef] [PubMed]
- Bauml, J.; Cho, B.C.; Park, K.; Lee, K.H.; Cho, E.K.; Kim, D.W.; Kim, S.-W.; Haura, E.B.; Sabari, J.K.; Sanborn, R.E.; et al. Amivantamab in combination with lazertinib for the treatment of osimertinib-relapsed, chemotherapy-naïve EGFR mutant (EGFRm) non-small cell lung cancer (NSCLC) and potential biomarkers for response. J. Clin. Oncol. 2021, 39, 9006. [Google Scholar] [CrossRef]
- Park, K.; Haura, E.B.; Leighl, N.B.; Mitchell, P.; Shu, C.A.; Girard, N.; Viteri, S.; Han, J.-Y.; Kim, S.-W.; Lee, C.K.; et al. Amivantamab in EGFR exon 20 insertion-mutated non-small-cell lung cancer progressing on platinum chemotherapy: Initial results from the CHRYSALIS phase I study. J. Clin. Oncol. 2021, 39, 3391–3402. [Google Scholar] [CrossRef]
- Saigi, M.; Alburquerque-Bejar, J.J.; Mc Leer-Florin, A.; Pereira, C.; Pros, E.; Romero, O.A.; Baixeras, N.; Esteve-Codina, A.; Nadal, E.; Brambilla, E.; et al. MET-oncogenic and JAK2-inactivating alterations are independent factors that affect regulation of PD-L1 expression in lung cancer. Clin. Cancer Res. 2018, 24, 4579–4587. [Google Scholar] [CrossRef]
- Mazieres, J.; Drilon, A.; Lusque, A.B.; Mhanna, L.; Cortot, A.B.; Mezquita, L.; Thai, A.A.; Mascaux, C.; Couraud, S.; Veillon, R.; et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: Results from the IMMUNOTARGET registry. Ann. Oncol. 2019, 30, 1321–1328. [Google Scholar] [CrossRef]
- Mack, P.C.; Klein, M.I.; Ayers, K.L.; Zhou, X.; Guin, S.; Fink, M.; Rossi, M.; Ai-Kateb, H.; O’connell, T.; Hantash, F.M.; et al. Targeted next-generation sequencing reveals exceptionally high rates of molecular driver mutations in never-smokers with lung adenocarcinoma. Oncologist 2022, 27, 476–486. [Google Scholar] [CrossRef]
- Awad, M.M.; Oxnard, G.R.; Jackman, D.M.; Savukoski, D.O.; Hall, D.; Shivdasani, P.; Heng, J.C.; Dahlberg, S.E.; Jänne, P.A.; Verma, S.; et al. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J. Clin. Oncol. 2016, 34, 721–730. [Google Scholar] [CrossRef]
- Schrock, A.B.; Frampton, G.M.; Suh, J.; Chalmers, Z.R.; Rosenzweig, M.; Erlich, R.L.; Halmos, B.; Goldman, J.; Forde, P.; Leuenberger, K.; et al. Characterization of 298 patients with lung cancer harboring MET exon 14 skipping alterations. J. Thorac. Oncol. 2016, 11, 1493–1502. [Google Scholar] [CrossRef]
- Sabari, J.K.; Leonardi, G.C.; Shu, C.A.; Umeton, R.; Montecalvo, J.; Ni, A.; Chen, R.; Dienstag, J.; Mrad, C.; Bergagnini, I.; et al. PD-L1 expression, tumor mutational burden, and response to immunotherapy in patients with MET exon 14 altered lung cancers. Ann. Oncol. 2018, 29, 2085–2091. [Google Scholar] [CrossRef]
- Glodde, N.; Bald, T.; van den Boorn-Konijnenberg, D.; Nakamura, K.; O’Donnell, J.S.; Szczepanski, S.; Brandes, M.; Eickhoff, S.; Das, I.; Shridhar, N.; et al. Reactive neutrophil responses dependent on the receptor tyrosine kinase c-MET limit cancer immunotherapy. Immunity 2017, 47, 789–802.e9. [Google Scholar] [CrossRef] [PubMed]
- Martin, V.; Chiriaco, C.; Modica, C.; Acquadro, A.; Cortese, M.; Galimi, F.; Perera, T.; Gammaitoni, L.; Aglietta, M.; Comoglio, P.M.; et al. Met inhibition revokes IFNγ-induction of PD-1 ligands in MET-amplified tumors. Br. J. Cancer 2019, 120, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Liang, Q.; Sun, Z.; Yuan, X.; Hou, W.; Wang, Y.; Wang, H.; Yu, M. Development of bispecific anti-c-Met/PD-1 diabodies for the treatment of solid tumors and the effect of c-Met binding affinity on efficacy. OncoImmunology 2021, 10, 1914954. [Google Scholar] [CrossRef] [PubMed]
- Choueiri, T.K.; Powles, T.; Burotto, M.; Escudier, B.; Bourlon, M.T.; Zurawski, B.; Oyervides Juárez, V.M.; Hsieh, J.J.; Basso, U.; Shah, A.Y.; et al. Nivolumab plus cabozantinib versus sunitinib for advanced renal-cell carcinoma. N. Engl. J. Med. 2021, 384, 829–841. [Google Scholar] [CrossRef]
- George, D.J.; Lee, C.H.; Heng, D. New approaches to first-line treatment of advanced renal cell carcinoma. Ther. Adv. Med. Oncol. 2021, 13, 17588359211034708. [Google Scholar] [CrossRef]
- Wood, G.E.; Hockings, H.; Hilton, D.M.; Kermorgant, S. The role of MET in chemotherapy resistance. Oncogene 2021, 40, 1927–1941. [Google Scholar] [CrossRef]
- To, K.K.; Cho, W.C. Mesenchymal epithelial transition (MET): A key player tin chemotherapy resistance and an emerging target for potentiating cancer immunotherapy. Curr. Cancer Drug Targets 2022, 22, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Lasagna, N.; Fantappiè, O.; Solazzo, M.; Morbidelli, L.; Marchetti, S.; Cipriani, G.; Ziche, M.; Mazzanti, R. Hepatocyte growth factor and inducible nitric oxide synthase are involved in multidrug resistance-induced angiogenesis in hepatocellular carcinoma cell lines. Cancer Res. 2006, 66, 2673–2682. [Google Scholar] [CrossRef]
- Ozasa, H.; Oguri, T.; Maneno, K.; Takakuwa, O.; Kunii, E.; Yagi, Y.; Uemura, T.; Kasai, D.; Miyazaki, M.; Niimi, A. Significance of c-Met overexpression in cytotoxic anticancer drug-resistant small cell lung cancer cells. Cancer Sci. 2014, 105, 1032–1039. [Google Scholar] [CrossRef]
- Hung, T.-H.; Li, Y.-H.; Tseng, C.-P.; Lan, Y.-W.; Hsu, S.-C.; Chen, Y.-H.; Huang, T.-T.; Lai, H.-C.; Chen, C.-M.; Choo, K.-B.; et al. Knockdown of c-Met induced apoptosis in ABCB1-overexpressed multidrug-resistance cancer cell lines. Cancer Gene Ther. 2015, 22, 262–270. [Google Scholar] [CrossRef]
- Deying, W.; Feng, G.; Shumei, L.; Hui, Z.; Ming, L.; Hongqing, W. CAF-derived HGF promotes cell proliferation and drug resistance by up-regulating the c-Met/PI3K/Akt and GRP78 signaling in ovarian cancer cells. Biosci. Rep. 2017, 37, BSR20160470. [Google Scholar] [CrossRef] [PubMed]
- Rotow, J.K.; Gui, P.; Wu, W.; Raymond, V.M.; Lanman, R.B.; Kaye, E.J.; Peled, N.; de la Cruz, F.F.; Nadres, B.; Corcoran, R.B.; et al. Co-occurring alterations in the RAS-MAPK pathway limit response to MET inhibitor treatment in MET exon 14 skipping mutation-positive lung cancer. Clin. Cancer Res. 2020, 26, 439–449. [Google Scholar] [CrossRef]
- Jamme, P.; Fernandes, M.; Copin, M.C.; Descarpentries, C.; Escande, F.; Morabito, A.; Grégoire, V.; Jamme, M.; Baldacci, S.; Tulasne, D.; et al. Alterations in the PI3K pathay drive resistance to MET inhibitors in NSCLC harboring MET exon 14 skipping mutations. J. Thorac. Oncol. 2020, 15, 741–751. [Google Scholar] [CrossRef]
- Guo, R.; Offin, M.; Brannon, A.R.; Chang, J.; Chow, A.; Delasos, L.; Somwar, R.; Wilkins, O.; Scott, K.; Tian, Y.; et al. MET inhibitor resistance in patients with MET exon 14-altered lung cancers. J. Clin. Oncol. 2019, 37, 9006–9007. [Google Scholar] [CrossRef]
- Fujino, T.; Kobayashi, Y.; Suda, K.; Koga, T.; Nishino, M.; Ohara, S.; Chiba, M.; Shimoji, M.; Tomizawa, K.; Takemoto, T.; et al. Sensitivity and resistance of MET exon 14 mutations in lung cancer to eight MET tyrosine kinase inhibitors in vitro. J. Thorac. Oncol. 2019, 14, 1753–1765. [Google Scholar] [CrossRef] [PubMed]
- Riedel, R.; Fassunke, J.; Tumbrink, H.L.; Scheel, A.H.; Heydt, C.; Hieggelke, L.; Scheffler, M.; Heimsoeth, A.; Nogova, L.; Michels, S.; et al. Resistance to MET inhibition in MET-dependent NSCLC and therapeutic activity after switching from type I to type II MET inhibitors. Eur. J. Cancer 2023, 179, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Suzawa, K.; Offin, M.; Lu, D.; Kurzatkowski, C.; Vojnic, M.; Smith, R.S.; Sabari, J.K.; Tai, H.; Mattar, M.; Khodos, I.; et al. Activation of KRAS mediates resistance to targeted therapy in MET exon 14-mutant non-small cell lung cancer. Clin. Cancer Res. 2019, 25, 1248–1260. [Google Scholar] [CrossRef]
- Zhang, Y.; Yin, J.; Peng, F. Acquired resistance to crizotinib in advanced lung adenocarcinoma with MET exon 14 skipping. Lung Cancer 2017, 113, 69–71. [Google Scholar] [CrossRef]
- Nilsson, M.B.; Sun, H.; Robichaux, J.; Pfeifer, M.; McDermott, U.; Travers, J.; Diao, L.; Xi, Y.; Tong, P.; Shen, L.; et al. A YAP/FOXM1 axis mediates EMT-associated EGFR inhibitor resistance and increased expression of spindle assembly checkpoint components. Sci. Transl. Med. 2020, 12, eaaz4589. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Yang, J.C.-H.; Yu, H.; Kim, S.-W.; Saka, H.; Horn, L.; Goto, K.; Ohe, Y.; Mann, H.; Thress, K.S.; et al. TATTON: A multi-arm, phase Ib trial of osimertinib combined with selumetinib, savolitinib, or durvalumab in EGFR-mutant lung cancer. Ann. Oncol. 2020, 31, 507–516. [Google Scholar] [CrossRef]
- Nie, N.; Li, J.; Zhang, J.; Dai, J.; Liu, Z.; Ding, Z.; Wang, Y.; Zhu, M.; Hu, C.; Han, R.; et al. First-line osimertinib in patients with EGFR-mutated non-small cell lung cancer: Effectiveness, resistance mechanisms, and prognosis of different subsequent treatments. Clin. Med. Insights Oncol. 2022, 16, 11795549221134735. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.; Adjei, A.A. MET: A promising anticancer therapeutic target. Nat. Rev. Clin. Oncol. 2012, 9, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Matsubara, D.; Goto, A.; Ota, S.; Sachiko, O.; Ishikawa, S.; Aburatani, H.; Miyazawa, K.; Fukayama, M.; Niki, T. Constitutive activation of c-Met is correlated with c-Met overexpression and dependent on cell-matrix adhesion in lung adenocarcinoma cell lines. Cancer Sci. 2008, 99, 14–22. [Google Scholar] [CrossRef]
- Friedlaender, A.; Drilon, A.; Banna, G.L.; Peters, S.; Addeo, A. The METeoric rise of MET in lung cancer. Cancer 2020, 126, 4826–4837. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Navas, T.; Herrick, W.G.; Hollingshead, M.G.; Bottaro, D.P.; Doroshow, J.H.; Parchment, R.E. Effective implementation of novel MET pharmacodynamic assays in translational studies. Ann. Transl. Med. 2017, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Frampton, G.M.; Ali, S.M.; Rosenzweig, M.; Chmielecki, J.; Lu, X.; Bauer, T.M.; Akimov, M.; Bufill, J.A.; Lee, C.; Jentz, D.; et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015, 5, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Sabari, J.K.; Santini, F.; Bergagnini, I.; Lai, W.V.; Arbour, K.C.; Drilon, A. Changing the therapeutic landscape in non-small cell lung cancers: The evolution of comprehensive molecular profiling improves access to therapy. Curr. Oncol. Rep. 2017, 19, 24. [Google Scholar] [CrossRef]
- Yin, W.; Cheng, J.; Tang, Z.; Toruner, G.; Hu, S.; Guo, M.; Robinson, M.; Medeiros, L.J.; Tang, G. MET amplification (MET/CEP7 ratio > 1.8) is an independent poor prognostic marker in patients with treatment-naïve non-small cell lung cancer. Clin. Lung Cancer 2021, 22, e512–e518. [Google Scholar] [CrossRef]
- Guo, M.Z.; Marron, K.A.; Spira, A.; Waterhouse, D.M.; Scott, S.C. Targeted treatment of non-small cell lung cancer: Focus on capmatinib with companion diagnostics. OncoTargets Ther. 2021, 14, 5321–5331. [Google Scholar] [CrossRef]
- Xu, Z.; Li, H.; Dong, Y.; Cheng, P.; Luo, F.; Fu, S.; Gao, M.; Kong, L.; Che, N. Incidence and PD-L1 expression of MET 14 skipping in Chinese population: A non-selective NSCLC cohort study using RNA-based sequencing. OncoTargets Ther. 2020, 13, 245–6253. [Google Scholar] [CrossRef]
- Paik, P.K.; Drilon, A.; Fan, P.-D.; Yu, H.; Rekhtman, N.; Ginsberg, M.S.; Borsu, L.; Schultz, N.; Berger, M.F.; Rudin, C.M.; et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 2015, 5, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Piper-Vallillo, A.J.; Kobayashi, S.S.; Costa, D.B. Differential pattern of resistance and sensitivity to difference classes of MET inhibitors for MET-amplified tumors with MET-D1228X or MET-Y1230X mutations. JTO Clin. Res. Rep. 2021, 2, 100133. [Google Scholar] [PubMed]
- Kang, J.; Chen, H.-J.; Wang, Z.; Liu, J.; Li, B.; Zhang, T.; Yang, Z.; Wu, Y.-L.; Yang, J.-J. Osimertinib and cabozantinib combinatorial therapy in an EGFR-mutant lung adenocarcinoma patient with multiple MET secondary-site mutations after resistance to crizotinib. J. Thorac. Oncol. 2018, 13, E49–E53. [Google Scholar] [CrossRef] [PubMed]


| Type | Name | Molecular Target(s) * | Approval Status (Indications) | Representative Clinical Trials |
|---|---|---|---|---|
| Ia | Crizotinib (PF-02341066) | ROS1 > Met > ALK | Approved (indicated for advanced NSCLC with ALK or ROS1; breakthrough therapy for advanced NSCLC with MET exon 14 skipping as second-line therapy) | Phase 1 (NCT00585195)—first-in-class drug candidate demonstrating clinical efficacy in NSCLC patients bearing MET-exon-14-skipping mutations Phase 2 (NCT02465060) Phase 2 (NCT02499614) Phase 2 (NCT02034981) |
| Ib | Capmatinib (INCB28060) | Met | Approved (indicated for advanced NSCLC with MET exon 14 skipping) | Phase 2 (NCT02414139, GEOMETRY mono-1)—pivotal trial demonstrating substantial antitumor efficacy of capmatinib in NSCLC patients with MET-exon-14-skipping mutations Phase Ib/II (NCT01610336)—combination with geftinib Phase Ib/II (NCT02468661)—combination with erlotinib |
| Tepotinib (EMD1214063) | Met | Approved (breakthrough therapy for advanced NSCLC with MET exon 14 skipping as first-line therapy) | Phase 2 (NCT02864992, VISION)—pivotal trial demonstrating favorable ORR and mPFS from tepotinib in NSCLC patients with MET-exon-14-skipping mutations Phase Ib/II (NCT01982955, INSIGHT)—combination with gefitinib Phase II (NCT03940703, INSIGHT 2)—combination with osimertinib | |
| Savolitinib (AZD6094, volitinib) | Met | Conditionally approved in China for advanced NSCLC with MET exon 14 skipping mutations | Phase 2 (NCT02897479)—pivotal trial demonstrating favorable clinical outcome and safety profile from single-agent savolitinib in Chinese patients with PSC, brain metastasis, and NSCLC patients with MET-exon-14-skipping mutations. Savolitinib was conditionally approved in China. Phase Ib (NCT02143466, TATTON), Phase 2 (NCT03778229; SAVANNAH) and Phase 2 (NCT03944772; ORCHARD)—demonstrating clinical efficacy of savolitinib–osimertinib combination in acquired resistance setting in advanced NSCLC with MET alterations. | |
| APL-101 (Bozitinib) | Met | Under clinical investigation | Phase 1 (NCT03175224) | |
| SAR125844 | Met | Under clinical investigation | Phase 1/2 (NCT01391533) Phase 2 (NCT02435121) | |
| II | Cabozantinib (XL-184, BMS-907351) | VEGFR2 > Met > Ret > Kit > Flt-1/2/3/4 > AXL > Tie2 | Approved (indicated for renal cell carcinoma and advanced metastatic medullary thyroid carcinoma) | Phase 3 (NCT01865747)—cabozantinib improved PFS compared to everolimus in RCC patients who progressed after VEGFR-targeted therapy Phase 2 (NCT01639508)—cabozantinib showing favorable clinical efficacy in patients with RET-rearranged lung cancer |
| Merestinib (LY2801653) | DDR1 > Met ~ AXL > MKNK1/2 > FLT3 > DDR2 > MERTK > MST1R > ROS1 | Under clinical investigation | Phase 1 (NCT03027284) Phase 2 (NCT02711553) Phase 2 (NCT02920996) | |
| Glesatinib (MGCD265) | Met > RON > VEGFR1/2 /3 > Tie-2 | Under clinical investigation | Phase 1 (NCT00697632) Phase 2 (NCT02544633) | |
| Sitravatinib (MGCD516) | TAM receptors (Axl, Mer) > VEGFR2 > KIT > Met | Under clinical investigation | Phase 2 (NCT03606174)—combination with PD-1 checkpoint inhibitor Phase 3 (NCT03906071)—combination with PD-1 checkpoint inhibitor in metastatic NSCLC | |
| Altiratinib (DCC-2701) | Met > Tie2 > VEGFR2 | Under clinical investigation | Phase 1 (NCT02228811) | |
| Foretinib (GSK1363089) | Met > AXL > RON > VEGFRs | Product development terminated by sponsor company in 2015 | Phase 2 (NCT00726323) Phase 2 (NCT02034097)—product development terminated by sponsor | |
| III | Tivantinib (ARQ197) | Met > RON | Under clinical investigation | Phase 2 (NCT00988741) Phase 2 (NCT01892527)—combination with cetuximab in resistant MET high subjects Phase 2 (NCT01519414) |
| Combination | Cancer Type | clinicatrials.gov Identifier (Phase) | Status |
|---|---|---|---|
| Cabozantinib (Met TKI) + nivolumab (anti-PD-1 mAb) versus Sunitinib (multitargeted TKI) | Previously untreated advanced RCC | NCT03141177 (Phase 3) | Completed; nivolumab + cabozantinib had significant benefits over sunitinib with respect to PFS and OS. |
| cabozantinib (Met TKI) + Nivolumab (anti-PD-1 mAb) with or without ipilimumab (anti-CTLA mAb) | Metastatic genitourinary tumors | NCT02496208 (Phase 1) | Active, not recruiting (last update posted 26 April 2023) |
| APL-101 (Met TKI) + genolimzumab/nivolumab (anti-PD-1 mAb) | Locally advanced or metastatic HCC or RCC | NCT03655613 (Phase 1/2) | Terminated (due to administrative reasons; status update on 6 May 2022) |
| Capmatinib (Met TKI) + pembrolizumab (anti-PD-1 mAb) | NSCLC with PD-L1 expression > 50% and no EGFR mutation or ALK rearrangement | NCT04139317 (Phase 2) | Terminated (due to toxicity in the drug combination arm; status update on 27 February 2023) |
| Cabozantinib (Met/VEGFR TKI) + nivolumab (anti-PD-1 mAb) + ipilimumab (anti-CTLA mAb) | Metastatic soft-tissue sarcoma | NCT04551430 (Phase 2) | Active, not recruiting (last update posted 31 March 2023) |
| Cabozantinib (Met/VEGFR TKI) + nivolumab (anti-PD-1 mAb) | Metastatic microsatellite-stable colorectal cancer | NCT04963283 (Phase 2) | Recruiting (last update posted 15 December 2022) |
| Cabozantinib (Met/VEGFR TKI) + nivolumab (anti-PD-1 mAb) | Advanced HCC who progressed upon first-line therapy | NCT05039736 (Phase 2) | Not yet recruiting (estimated start date 24 February 2023) |
| Cabozantinib (Met/VEGFR TKI) + ipilimumab (anti-CTLA-4 mAb) + nivolumab (anti-PD-1 mAb) | Refractory cutaneous melanoma | NCT05200143 (Phase 2) | Recruiting (last update posted 13 June 2022) |
| Combination | Cancer Type | ClinicaTrials.gov Identifier (Phase) | Key Findings/Current Trial Status |
|---|---|---|---|
| Tivantinib (Met inhibitor) + cetuximab (EGFR mAb) + irinotecan (topoisomerase I inhibitor) | CRC | NCT01075048 (Phase 2) |
|
| Onartuzumab (anti-Met mAb) + FOLFOX + bevacizumab (anti-VEGF mAb) | CRC | NCT01418222 (Phase 2) |
|
| Rilotumumab (anti-HGF mAb) + epirubicin + Cisplatin + capecitabine | Gastric cancer | NCT00719550 (Phase 2) |
|
| Rilotumumab (anti-HGF mAb) + mitoxantrone + prednisone | Castration-resistant prostate cancer who had received previous taxane chemotherapy | NCT00770848 (Phase 2) |
|
| Crizotinib (Met/ALK inhibitor) + cyclophosphamide + topotecan | Refractory solid tumors or nnaplastic large-cell lymphoma | NCT01606878 (Phase 1) |
|
| Cabozantinib (Met/VEGFR inhibitor) + topotecan + cyclophosphamide | Refractory Ewing sarcoma or osteosarcoma | NCT04661852 (Phase 1) |
|
| Cabozantinib (Met/VEGFR inhibitor) + cisplatin / doxorubicin / methotrexate | Newly diagnosed osteosarcoma | NCT05691478 (Phase 2/3) |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
To, K.K.-W.; Leung, K.-S.; Cho, W.C.-S. MET-Targeting Anticancer Drugs—De Novo Design and Identification by Drug Repurposing. Drugs Drug Candidates 2023, 2, 591-623. https://doi.org/10.3390/ddc2030031
To KK-W, Leung K-S, Cho WC-S. MET-Targeting Anticancer Drugs—De Novo Design and Identification by Drug Repurposing. Drugs and Drug Candidates. 2023; 2(3):591-623. https://doi.org/10.3390/ddc2030031
Chicago/Turabian StyleTo, Kenneth Kin-Wah, Kwong-Sak Leung, and William Chi-Shing Cho. 2023. "MET-Targeting Anticancer Drugs—De Novo Design and Identification by Drug Repurposing" Drugs and Drug Candidates 2, no. 3: 591-623. https://doi.org/10.3390/ddc2030031
APA StyleTo, K. K.-W., Leung, K.-S., & Cho, W. C.-S. (2023). MET-Targeting Anticancer Drugs—De Novo Design and Identification by Drug Repurposing. Drugs and Drug Candidates, 2(3), 591-623. https://doi.org/10.3390/ddc2030031








