Abstract
Insect-transmitted plant viruses are a major threat to global agricultural crop production. Receptors play a prominent role in the interplay between host-pathogen and vector interaction. The virus–vector relationship involves both viral and vector receptors. Receptors-like kinases (RLKs) and receptor-like proteins play a crucial role in plant immunity, which acts as a basal defense. Pathogens can evade or block host recognition by their effector proteins to inhibit pathogen recognition receptor (PRR)-mediated signaling. Intriguingly, RLKs are also known to interact with viral proteins and impact plant susceptibility against viruses, while the endocytic receptors in vectors assist in the binding of the virus to the vectors. Unlike other receptors of fungi and bacteria which have three different domains located from extracellular or intracellular to perceive a multitude of molecular patterns, the characterization of viral receptors is quite complex and limited since the virus is directly injected into plant cells by insect vectors. Little is known about these receptors. Unraveling the receptors involved in virus entry and transmission within the vector will provide vital information in virus–vector interactions. This review focuses on efforts undertaken in the identification and characterization of receptors of plant viruses within the host and vector. This will lead to a better understanding of the cellular mechanism of virus transmission and spread, and further suggests new alternative tools for researchers to develop an integrated approach for the management of viral diseases and associated vectors.
Keywords:
aphids; thrips; whiteflies; plant and leaf hoppers; plant immunity; RLKs; virus transmission 1. Introduction
In general, receptors are proteins on the cellular surface that transmit a signal upon binding with the respective extracellular molecule, signaling molecule, and ligands. The ligands can be hormones, growth factors, nutrients, etc. However, in viruses, the receptors are not signaling molecules, but they are essential for the infectious cycle of viruses within their host and transmission by their vectors. In the case of animal viruses, viruses gain entry through the host receptors. In other words, receptors on the cell surface of the host are the principal determinant of the infection process, as they act like a lock to access the cell. The viral infection happens through the binding of viral capsid proteins to cellular receptors of the host cell resulting in penetration of the viral genome [1]. Unlike animal viruses, plant receptors are not the principal determinant of the infection because the virus entry into the plant cells happens through mechanical damage with or without the insect vectors. So, the receptors have a different purpose. The receptors we discuss in the context of plant viruses have different features and understanding than animal viruses. This review discusses two groups of receptors in two major sections. One is the receptors that are associated with the insect vectors that have a huge role in determining the specificity of vectors while the other section discusses receptors present on the cellular surface of plants that sense and trigger an immune response. To the best of our knowledge, there is no comprehensive review that covers the receptors in the infectious cycle of plant viruses. This review aims to summarize progress on the identification and characterization of viral receptors that are critical for transmission and in plants that sense and mount defense against viruses.
2. Viral Receptors within the Vector
Most of the plant viruses that cause extensive losses to agriculture are transmitted by sap-sucking insect vectors that include aphids, leafhoppers, planthoppers, thrips, and whiteflies. These insect vectors transmit more than 70% of the plant viruses [2]. The uptake and transmission of viruses by their vector requires a tight association between viral proteins and vector-associated proteins, usually referred to as receptors (Table 1). The identification of these receptors is a key factor in understanding the mechanism of virus transmission and opens avenues to study virus–vector relationships and restrict virus spread. To date, several vector proteins interacting with viral proteins have been identified. However, of these identified proteins only a few virus-interacting proteins have been functionally characterized (Figure 1). Here, we summarize and discuss the vector determinants involved in virus uptake, retention, and transmission of plant viruses.
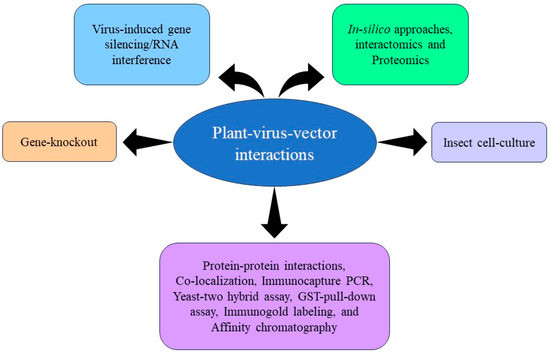
Figure 1.
Schematic representation of some of the common techniques used for the identification and functional characterization of proteins associated with plant–virus–vector pathosystem.
2.1. Aphid-Associated Viral Receptors
Aphids are phloem-feeding insects, well known as major pests in agriculture. In addition to weakening the plants by feeding, aphids also transmit several plant viruses [3]. Various strategies have been employed to identify receptors of viruses within aphid vectors. Virus overlay and immunoblot assays of English grain aphid as a vector (Sitobion avenae) and corn-leaf aphid as a non-vector (Rhopalosiphum maidis) showed that two accessory salivary gland (ASG)-associated proteins (SaM35 and SaM50) act as receptors for barley yellow dwarf virus (BYDV)-MAV isolate [4] (Figure 2). In addition, 50 kDa protein (P50) particles extracted from wheat aphid (Schizaphis graminum) and grain aphid (Sitobion avenae) exhibited specific binding to purified virus particles of BYDV-GAV isolate. A significant reduction in transmission efficiencies of BYDV-GAV by both the aphid vectors was observed upon antiserum feeding.
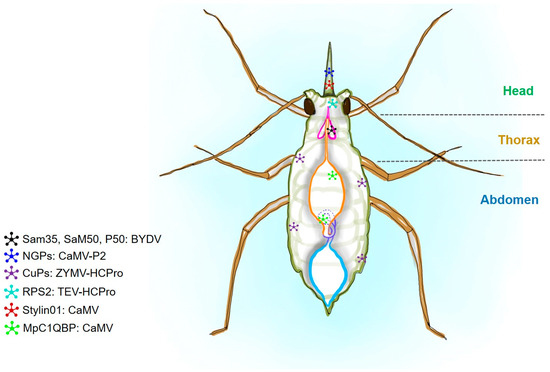
Figure 2.
A schematic diagram of an aphid showing the putative proteins corresponding to viral interactive partners. Barley yellow dwarf virus (BYDV), cauliflower mosaic virus (CaMV), zucchini yellow mosaic virus (ZYMV), and tobacco etch virus (TEV) are shown in combination with their putative partners. Host proteins are represented as non-glycosylated proteins (NGPs), Cuticular proteins (CuPs), ribosomal protein S2 (RPS2), and complement component 1Q subcomponent-binding protein (C1QBP). The presence of the viral receptors in aphids is shown with star shapes and different colors.
Immunogold labeling showed that the P50 protein is located at the plasma membrane surrounding the ASG in the head tissues of S. graminum. All these studies suggested that P50, SaM35, and SaM50 are associated with virus transmission [5]. Seddas and co-workers employed SDS-PAGE and 2D electrophoresis (2DE) and showed that three green-peach aphid (Myzus persicae) proteins, Rack-1, GAPDH3, and actin, may be involved in transcytosis of beet western yellows virus (BWYV) particles in the aphid vector [6]. P2 protein encoded by DNA viruses such as cauliflower mosaic virus (CaMV) acts as a receptor for a non-glycosylated protein deeply embedded in the chitin matrix of aphids. These receptors were found in the tip of the aphid’s maxillary stylets [7]. Cuticular proteins (CuPs) are major components of insect cuticles and have been identified as receptors of viruses in several insects including aphids. In aphids, CuPs of M. persicae interact with the helper-component protease (HCPro) of the Zucchini yellow mosaic virus [8]. To identify the aphid receptors, tobacco etch virus (TEV) (genus, potyvirus)-encoded HCPro was used as bait to select interacting proteins among the proteins extracted from aphid head tissues. Among the various proteins identified, ribosomal protein S2 (RPS2) was selected for further analysis. Cloning and heterologous expression of the corresponding M. persicae gene confirmed specific interactions between TEV-HCPro and RPS2. Further investigations suggested that RPS2 is involved in virus transmission [9] in insect vectors. Genomics and proteomics were applied to identify the wheat aphid (S. graminum) proteins mediating virus transmission. Of the identified proteins, co-immunoprecipitation (Co-IP) and mass spectrometry (MS) analysis showed that cyclophilin A and B proteins interact with the RPV strain of cereal yellow dwarf virus (CYDV-RPV) and may mediate virus transport from the hindgut lumen into the hemocoel [10]. Using the Far-Western blot technique, Linz and co-workers identified that membrane alanyl aminopeptidase N (APN) acts as a receptor for pea enation mosaic virus (PEMV)-encoded coat protein (CP) in the gut of the pea aphid (Acyrthosiphon pisum) [11]. A study conducted by Liang and Gao (2017) showed that the cuticular protein MPCP4 of the green peach aphid (M. persicae) binds with the CP of cucumber mosaic virus (CMV) [12].
RNAi-mediated silencing of MPCP4 suppressed the ability of M. persicae to acquire CMV. All these lines of evidence indicate that MPCP4 is a putative receptor of CMV and helps in virus acquisition. RNAi-mediated silencing of stylin-01 and stylin-02 showed that stylin-01 might act as a receptor of CaMV in the stylet of pea aphids (A. pisum) and M. persicae [13]. A novel aphid protein, membrane-bound Ephrin receptor (Eph), was found to be involved in the transmission of the Turnip yellows virus (TuYV) by M. persicae. A significant reduction in TuYV accumulation and transmission by M. persicae was observed after in planta feeding of dsRNA (dsRNAEph)-targeting Eph-mRNA [14]. Interestingly, CuPs MPCP2 was found to interact with potato virus Y (PVY). However, silencing of MPCP2 through oral dsRNA feeding resulted in a 47% reduction in the virus transmission efficiency of M. persicae, indicating its potential role in virus transmission [15]. Affinity purification coupled with high-resolution mass spectrometry of M. persicae resulted in the identification of 11 putative proteins, suggesting higher interaction probability with structural proteins of potato leafroll virus (PLRV). Yeast-two hybrid (YTH) showed the physical interaction of three of these vector proteins with PLRV-encoded structural proteins, in addition to a few other luteoviruses. Immunoprecipitation (IP) of complement component 1Q subcomponent-binding protein (C1QBP) showed the partial co-localization of MpC1QBP with PLRV in cytoplasmic puncta along the periphery of aphid gut epithelial cells. Artificial delivery of chemical inhibitors of C1QBP to aphids resulted in increased PLRV acquisition and transmission by aphids, supporting the role of C1QBP in PLRV acquisition and transmission by green peach aphids [16]. A combined mRNA and protein analysis of M. persicae infected with CMV enabled the identification of several viral putative regulators, including ribosomal proteins cytochrome P450 enzymes [17]. To date, several viral receptor proteins in aphids have been identified and their specific role has not yet been fully understood [18] (Figure 2). Further, research will pave the way toward a safe alternative to insecticides used for managing aphids and lowering the damage caused by transmitted viruses.
2.2. Planthopper and Leafhopper-Associated Virus Receptors
Identification of receptors of plant viruses in leafhoppers and planthoppers is crucial for understanding the transmission mechanism. Researchers are continuously working in this area and identifying novel proteins taking advantage of several molecular and biological tools. Guoying and co-workers showed that a 32 kDa membrane-associated protein of rice brown planthopper (Nilaparvata lugens) acts as a potential receptor of rice ragged stunt oryzavirus (RRSV) (genus, Oryzavirus) [19]. Further, small brown planthopper (SBPH, Laodelphax striatellus Fallén) proteins separated by 2D-gel electrophoresis were screened for rice stripe virus (RSV)-binding molecules using a virus overlay assay of protein blots. Wherein, mass spectrometry was employed and five proteins that bound to purified RSV particles in vitro were identified. The virus-binding capabilities of these proteins were elucidated further using in-vitro assays. Of the five putative proteins, a receptor for activated protein C kinase (RACK) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH3) were found to be involved in epithelial transcytosis of virus particles and three ribosomal proteins (RPL5, RPL7a, and RPL8) presumptively involved in infection and propagation of RSV in vector cells [20] (Figure 3). Huo and co-workers (2014) revealed that the vitellogenin receptor of SBPH is required for transovarial transmission of RSV [21].
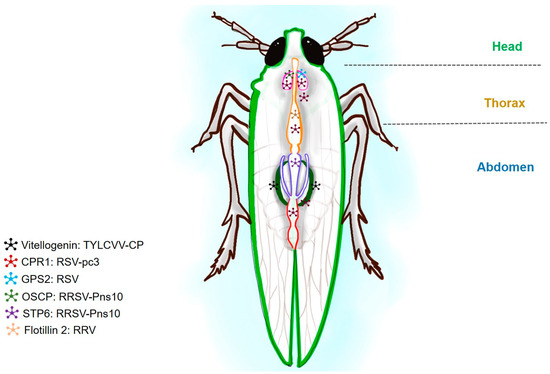
Figure 3.
A schematic diagram of planthopper showing the putative proteins corresponding to viral interactive partners. Tomato yellow leaf curl virus (TYLCV), rice stripe virus (RSV), and rice ragged stunt oryzavirus (RRSV) are shown in combination with their putative partners. Insect proteins such as vitellogenin (Vg), G-protein Pathway Suppressor 2 (GPS2), oligomycin-sensitivity conferral protein (OSCP), sugar transporter 6 (STP), and flotillin are shown in combination with their respective viruses. The presence of the viral receptors in the planthopper is shown with star shapes and different colors.
Another study by Wang and co-workers showed that the capsid protein of RSV binds to the G-protein Pathway Suppressor 2 (GPS2) of SBPH and activates the c-Jun N-terminal kinase (JNK) signaling pathway that promotes RSV replication in the vector [22]. In addition, a cuticular protein, CPR1 from SBPH, interacts with the nucleocapsid protein (pc3) of RSV both in vivo and in vitro and co-localizes with RSV in the hemocytes of SBPH. The protein aids viral survival in the hemolymph [23]. The Pns10 encoded by rice dwarf virus (RDV) was found to specifically interact with cytoplasmic actin of rice green leafhopper (Nephotettix cincticeps) but not with zigzag leafhopper (Recilia dorsalis), emphasizing the role of actin in virus transmission and virus–vector specificity [24]. A non-structural protein (Pns10) of rice ragged stunt virus (RRSV) interacting with host oligomycin-sensitivity conferral protein (OSCP) of BPH was recently identified. The interaction between BPH OSCP and RRSV Pns10 was verified using GST pull-down assay. This was the first evidence of direct interaction of RRSV protein with mitochondria. Suppressing the OSCP gene significantly reduced the viral load in RRSV-infected BPHs, revealing the role of mitochondrial protein in virus proliferation [25]. Thirteen different proteins of SBPH interacting with the nucleocapsid (N) protein of RSV were identified using the GAL4-based YTH system. Among these, the interaction of RPL18 protein was further validated and downregulation of RPL18 dramatically reduced viral protein expression, indicating the requirement of RPL18 in RSV translation and replication [26]. A sugar transporter 6 of L. striatellus was found to be involved in the entry of RSV to midgut epithelial cells and is also involved in virus transmission [27]. Proteomic analysis of viruliferous SBPH revealed that α-tubulin 2 interacts with nonstructural protein 3 (NS3) of RSV and is involved in the passage of RSV through midgut and salivary glands and leads to successful horizontal transmission [28]. A plasma membrane protein, flotillin 2, was identified to facilitate the infection of RSV in its vector, SBPH [29]. Interaction studies further revealed that in corn planthoppers (Peregrinus maidis), 107 proteins interact with glycoproteins of the maize mosaic virus (MMV). Further, the interaction of Cyclophilin A and apolipophorin III with MMV glycoproteins was validated in an insect cell line study [30]. In addition, voltage-dependent anion channel 2 (VDAC2) of SBPH showed interaction with RSV-encoded RNA-dependent RNA polymerase (RdRP). These interactions facilitated the accumulation of RSV in SBPH [31]. In silico analysis of interactions between glycoprotein of MMV and Syntaxin-18 (PmStx18) of corn planthopper (P. maidis) revealed PmStx18 as a putative receptor of MMV [32] (Figure 2). These receptor proteins will provide new clues for studies of the complicated relationship between viruses and vectors by planthoppers.
2.3. Thrips-Associated Virus Receptors
Thrips-transmitted viruses cause severe losses to various crop plants worldwide [33,34]. Young thrips larvae acquire the virus while feeding on infected hosts and are transmitted by adults throughout their lifespan. Adults cannot acquire the virus while young larvae are unable to transmit the virus [35,36]. Viruses infect the gut epithelium of thrips and reach the salivary glands from where the virus is transmitted to other hosts. During this viral movement, several uncharacterized vector proteins (receptors) are involved [37]. Few studies have been undertaken to identify the thrips proteins involved in virus acquisition and transmission. The very initial study for the identification of receptors of viruses in thrips was carried out in 1998 by Bandla and co-workers where a midgut protein (50-kDa) was identified as a potential receptor of tomato spotted wilt virus (TSWV) glycoproteins in western flower thrips (Frankliniella occidentalis) [38]. Similarly, another 94-kDa protein of F. occidentalis and cotton thrips (Thrips tabaci) was found to interact with the envelope glycoprotein (G2) of TSWV [39]. In silico analysis was performed to identify receptors of groundnut bud necrosis virus (GBNV) glycoprotein (GN) in melon thrips (T. palmi), suggesting that C-type lectin is the primary cellular receptor to interact with GBNV-GN [40] (Figure 4).
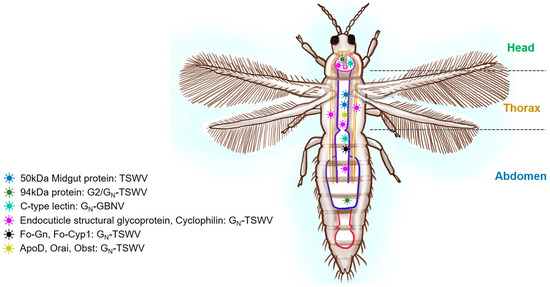
Figure 4.
A schematic diagram of thrips showing the putative proteins corresponding to viral interactive partners. Tomato spotted wilt virus (TSWV) with midgut proteins, endocuticle structural glycoprotein (Fo-GN), cyclophilin (Fo-Cyp1), apolipoprotein-D (ApoD), orai-2-like (Orai), and obstructor-E-like isoform X2 (Obst). Major segments like the head, thorax, and abdomen are labeled. The presence of the viral receptors in thrips is shown with different colors and shapes.
Receptors of TSWV glycoprotein GN were identified using gel overlay assay. The study identified six TSWV-interacting proteins (TIPS) from F. occidentalis. Further validation showed that two TIPS, an endocuticle structural glycoprotein, and cyclophilin, interacted with virus-encoded enveloped glycoprotein (GN). These proteins were found to be involved in virus entry or facilitate other virus infection processes in thrips [41]. Zheng and co-workers screened the F. occidentalis-TSWV YTH library and identified 74 thrips proteins, including ubiquitin-related proteins interacting with TSWV [42]. Later interaction of ubiquitin-related protein UBR7 with TSWV GN in F. occidentalis was validated using surface plasma resonance and GST pull-down assays [43]. The interaction of endocuticle structural glycoprotein (Fo-GN) and cyclophilin (Fo-Cyp1) in TSWV and F. occidentalis was also confirmed by employing immunoblotting and proteomic analysis [44]. To further understand the TSWV transmission mechanism by western flower thrips species, a split-ubiquitin membrane-based assay was employed to identify the potential vector proteins involved in virus transmission. Out of 67 identified proteins, 3 proteins, apolipoprotein-D (ApoD), orai-2-like (Orai), and obstructor-E-like isoform X2 (Obst), were selected for further validation. Protein Obst was found to be overexpressed in viruliferous thrips whereas silencing Obst resulted in decreased virus acquisition in larvae and transmission by adults, indicating the possible role of Obst in TSWV acquisition and transmission in F. occidentalis [45]. At present, most of our understanding of thrips viral receptors is based on F. occidentalis and TSWV. Further research is needed with other viruses and thrips vectors for a better understanding of virus transmission by thrips.
2.4. Whiteflies-Associated Virus Receptors
Plant viruses transmitted by whiteflies (Bemisia tabaci) are causative agents of many serious diseases of crop plants [46]. The coat protein (CP) is a structural protein involved in the movement of the virus in the host [47]. Various B. tabaci proteins undergo protein–protein interactions, act as viral receptors, and are involved in virus acquisition and transmission. B. tabaci heat shock proteins (BtHSP16 and BtHSP70) interact with the tomato yellow leaf curl Sardinia virus (TYLCSV)- CP and are involved in protection against other begomoviruses while translocating in the whitefly (Figure 5) [48,49].
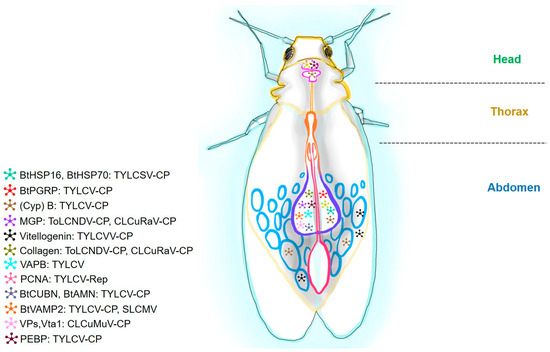
Figure 5.
A schematic diagram of a whitefly showing the putative proteins corresponding to viral interactive partners. Tomato yellow leaf curl virus (TYLCV), tomato leaf curl New Delhi virus (ToLCVNDV), cotton leaf curl Rajasthan virus (CLCuV-Ra), cotton leaf curl Multan virus (CLCuMuV), and tomato yellow leaf curl Sardinia virus (TYLCSV). Insect proteins are heat-shock proteins (BtHSP16 and BtHSP70), B. tabaci peptidoglycan recognition protein (BtPGRP), Cyclophilin (Cyp) B, midgut protein (MGP), vesicle-associated membrane protein-associated protein B (VAPB), proliferating cell nuclear antigen (PCNA), cubilin (BtCUBN), aminoless (BtAMN), B. tabaci vesicle-associated membrane protein 2 (BtVAMP2), vacuolar protein (Vps), sorting-associated protein twenty-associated 1 (Vta1), and phosphatidylethanolamine-binding protein (PEBP). Major segments like the head, thorax, and abdomen are labeled.
Wang and co-workers identified that peptidoglycan recognition protein encoded by the B. tabaci PGRP gene (BtPGRP) acts as a binding site for tomato yellow leaf curl virus (TYLCV) [50]. In vitro interactions were detected between BtPGRP and TYLCV by immunocapture PCR. Immunocapture PCR and co-localization were also used to identify the role of Cyclophilin (Cyp) B protein in the transmission of TYLCV [51]. In the begomovirus-whitefly system, cyp-encoded proteins might be required to refold virion particles and facilitate the virus movement across the membrane barriers in the vector. cDNA expression library screening against the CP of tomato leaf curl New Delhi virus (ToLCVNDV) and cotton leaf curl Rajasthan virus (CLCuV-Ra) was used using infected gut tissues [52]. Upon screening, midgut protein (MGP) was identified as a putative receptor for begomovirus in whiteflies. Also, an interaction study was carried out to decipher the potential role of vitellogenin (Vg; an egg yolk precursor protein) in virus transmission. Vg is a multifunctional protein expressed in a tissue-specific manner in insects. Recent studies suggest that the Vg receptor is essential for the transovarial transmission of TYLCV by B. tabaci [53]. In monopartite begomoviruses chili leaf curl virus (ChiLCV), silencing of B. tabaci hsp70 and fasciclin 2 (fas2) facilitate the virus infection. In addition, differential analysis of B. tabaci in response to ChiLCV further revealed an association of innate immunity-related genes such as Toll-like receptor 3 (TLR3), a transducer of erbB2.1 (TOB1), and GMP reductase. Silencing of TLR3 and TOB1 significantly reduces ChiLCV transmission, suggesting their negative regulatory role in virus pathogenesis [54].
Other begomoviruses such as ToLCNDV or CLCuV-encoded CP specifically interact with the collagen protein of the insects. Therefore, it will be interesting to understand how virus-encoded CPs take over collagen to escape the host immune response during virus transmission [55]. Moreover, ToLCNDV or CLCuV-CP was further used as bait against the total RNA of B. tabaci. The assay resulted in the identification of a thioredoxin-like protein (TLP) as a receptor of begomoviruses [56]. Split-ubiquitin-based YTH assay followed by GST pull-down and immunofluorescence were used to study the interactions between TYLCV and B. tabaci. Middle East Asia Minor 1 (MEAM1) whitefly species using a transcriptome database (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA407873; Accessed: 1 March 2024) suggests the abundance of vesicle-associated membrane protein-associated protein B (VAPB) in midgut tissues. A recent study showed that VAPB is involved in TYLCV-mediated virus transmission [57]. An in-depth study of TYLCV-encoded replication-associated protein (Rep) was found to be interacting with whitefly, proliferating cell nuclear antigen (PCNA), which recruits DNA Polδ and aids in virus replication within the vector [58]. In MEAM1, GST pull-down and LC-MS/MS were used to screen the binding partners of TYLCV-CP. The study identified two whitefly proteins, cubilin (BtCUBN) and aminoless (BtAMN), forming a receptor complex termed BtCubam (Figure 4). In this receptor complex, BtCUBN contributes a viral-binding region and BtAMN contributes to membrane anchorage, which facilitates the entry of begomoviruses into the vector midgut via clathrin-dependent endocytosis [59]. Furthermore, 50 whitefly proteins interacting with an intergenic region (IR) of tomato yellow leaf curl China virus (TYLCCNV) were identified. Dual luciferase analysis revealed that one of the identified proteins, hairy and enhancer of split homolog-1 (HES1), is specifically bound to the ‘CACGTG’ motif in the TYLCCNV-IR region. A decrease in viral transcription, accumulation, and transmission was observed after HES1 silencing. The findings of this study showed that interactions with whitefly proteins and the IR of TYLCCNV are involved in viral transcription in whiteflies. Proteomic interactions analyzed among TYLCV and B. tabaci showed the interaction of 15 putative whitefly proteins specifically with CP of TYLCV. Out of these 15 proteins, 1 protein tumorous imaginal disc (Tid) had stable interaction in in vitro assay, emphasizing that the DnaJ-C domain of Tid301-499aa was found to be the specific virus-binding site. Silencing of the target gene followed by the use of anti-Tid antibodies resulted in a higher quantity of TYLCV in the whitefly body, indicating its potential role in antiviral infection [60]. In addition, B. tabaci vesicle-associated membrane protein 2 (BtVAMP2) transcript levels were increased during TYLCV infection. Later, they found that TYLCV-CP was having physical interactions with BtVAMP2 [61]. Blocking of BtVAMP2 protein by feeding specific BtVAMP2 antibodies resulted in a significant reduction in virus titer in B. tabaci. Similar findings were observed in whiteflies infected with Sri Lankan cassava mosaic virus (SLCMV). Feeding BtVAMP2-linked antibodies showed reduced acquisition of SLCMV. These interactions demonstrated the presumptive role of BtVAMP2 in begomovirus acquisition by whiteflies. An interaction study was further used to identify 54 putative whitefly proteins involved in cotton leaf curl Multan virus (CLCuMuV) CP-mediated transmission. RNAi analysis showed that vacuolar protein (Vps), sorting-associated protein twenty-associated 1 (Vta1) is a positive regulator of CLCuMuV acquisition and transmission [62]. Similarly, screening of pepper whitefly-borne vein yellows virus (PeWBVYV) CP against the cDNA library of whitefly (MEAM1) resulted in the identification of C1QBP as an insect protein interacting with poleroviruses, suggesting C1QBP might be involved in virus transmission [63]. A recent study also found that tomato leaf curl Bangalore virus (ToLCBV) CP showed interactions with 102 distinct whitefly proteins (B. tabaci Asia 1). These proteins included HSP70, GroEL, enolase, nucleoproteins, lachesins, vitellogenin, succinate dehydrogenase, apolipophorins, salivary secreted proteins, 40s ribosomal proteins, tropomyosin, sorbitol dehydrogenase, GTP cyclohydrolase, dipeptidyl peptidase, annexin, E3 ubiquitin, and others [64]. Some of these proteins might be helpful for the virus and some favor the whiteflies. Another interesting study suggests a viable interaction between insect phosphatidylethanolamine-binding protein (PEBP) and TYLCV-CP wherein it downregulates the MAPK signaling cascade. This further activates apoptosis in whiteflies and increases viral titer [65].
Immunoprecipitation assay and DUAL membrane cDNA library screening technology were applied to understand interacting partners in RNA viruses. Cucurbit chlorotic yellows virus (CCYV)-encoded minor coat protein (CPm) interacts with several proteins of B. tabaci like tubulin beta chain (TUB), keratin type I cytoskeletal 9-like (KRT), and cytochrome-c-oxidase subunit 5A (COX). These proteins were found to be associated with virus retention within the vector and transmission of CCYV [66]. Transcription factors (TFs) involved in both old- and new-world begomovirus transmission were identified recently. A whitefly C2H2 zinc finger (ZF) protein, 100% identical to the vascular endothelial ZF-like (vezf) protein, was found to be interacting with the CP of the begomoviruses. Silencing of the vezf gene of B. tabaci led to the increased retention of mono or bipartite begomoviruses, TYLCV, cucurbit leaf crumple virus (CuLCrV), and sida golden mosaic virus (SiGMV), suggesting an inhibitory role of vezf during begomoviruses transmission [67]. Another transcription factor, zinc finger 330 (ZNF330), was involved in B. tabaci and a bipartite begomovirus ramie mosaic virus (RaMoV) pathogenesis [68]. Silencing ZNF300 resulted in a significant reduction in longevity and fecundity of RaMoV-infected female adults. The results demonstrated that ZNF300 is a negative regulator of RaMoV replication in B. tabaci Mediterranean (MED) species. Though several proteins interacting with viruses have been unraveled, other key viral receptors remain unknown. Further, research is needed to identify these receptors for a better understanding of virus transmission by whiteflies and for the implementation of novel strategies for managing them.

Table 1.
Vector receptor proteins are involved in viral replication, acquisition, and transmission.
Table 1.
Vector receptor proteins are involved in viral replication, acquisition, and transmission.
| Vector | Vector Protein | Protein Localization | Site of Interaction | Putative Role | Virus Protein | References |
|---|---|---|---|---|---|---|
| Aphids | SaM35 and SaM50 | - | - | Virus transmission | - | [4] |
| P50 | Plasma membrane surrounding the accessory salivary gland | - | Virus transmission | - | [5] | |
| Receptor for activated protein kinase 1 (Rack-1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH3), and actin | - | - | Epithelial transcytosis | - | [6] | |
| Cuticular proteins (CuPs) | - | - | Virus acquisition and transmission | Helper-component protease (HCPro) and CP | [8,12,15] | |
| Non-glycosylated protein | Chitin matrix | Maxillary stylet | - | Virus particles | [7] | |
| Ribosomal protein S2 (RPS2) | Cell membrane | - | Virus transmission | HCPro | [9] | |
| Cyclophilin A and B | - | - | Virus transport | - | [10] | |
| Aminopeptidase N (APN) | - | - | Receptor | CP | [11] | |
| Ephrin receptor (Eph) protein | - | - | Virus transmission | Minor coat protein (CPm) | [14] | |
| Stylin-01 | Maxillary stylets | - | Transmission | Helper component protein P2 | [13] | |
| Complement component 1 Q subcomponent-binding protein (C1QBP) | Gut epithelial cells | Cytoplasmic puncta and gut epithelial cells | Virus acquisition and transmission | Structural proteins | [16] | |
| Ribosomal proteins cytochrome P450 enzymes, and cuticle proteins | - | - | - | - | [17] | |
| Cuticle proteins and tubulins | - | - | - | - | [18] | |
| Leafhoppers | Actin | - | - | Virus–vector specificity | Nonstructural protein Pns10 | [24] |
| Planthoppers | 32-kDa membrane protein | - | - | - | - | [19] |
| RACK, GAPDH3, and ribosomal proteins (RPL5, RPL7a and RPL8) | - | - | Epithelial transcytosis, infection, and propagation of the virus | Nucleocapsid protein | [20] | |
| Vitellogenin | Ovary | Germarium | Transovarial transmission | Nucleocapsid protein (pc3) | [21] | |
| Cuticular protein (CPR1) | Hemolymph, salivary gland, gut, and ovary | Hemocytes | Virus transmission | pc3 | [23] | |
| G protein Pathway Suppressor 2 (GPS2) | Salivary gland cells | - | Viral replication | CP | [22] | |
| Host oligomycin-sensitivity conferral protein (OSCP) | Mitochondria | Cytoplasm of the salivary gland cells | Viral proliferation | Nonstructural protein Pns10 | [25] | |
| RPL18 | - | - | Virus accumulation | Nucleocapsid (N) protein | [26] | |
| Sugar transporter 6 | Midgut | Cell membrane | Viral entry into midgut epithelial cells | Nucleocapsid protein | [27] | |
| α-tubulin 2 | - | Midgut, hemocytes, and principal salivary glands | Horizontal transmission | Nonstructural protein 3 (NS3) | [28] | |
| Flotillin 2 | Plasma membrane of midgut epithelial cells | Gut microvilli | Virus entry in midgut epithelial cells | Nucleocapsid protein | [29] | |
| Cyclophilin A and Apolipophorin III | Insect cells | Glycoproteins | [30] | |||
| Voltage-dependent anion channel 2 (VDAC2) | - | - | Virus accumulation | RNA-dependent RNA polymerase | [31] | |
| Syntaxin-18 | - | - | - | Glycoprotein | [32] | |
| Thrips | Midgut protein (50 kDa; 94 kDa) | Midgut | Midgut | Translocation of virus in midgut | Glycoproteins | [38,39] |
| Endocuticle structural glycoprotein and cyclophilin | Midgut and salivary glands | Midgut and salivary glands | Virus entry | Glycoprotein (GN) | [41] | |
| C-type lectin | - | - | - | GN | [40] | |
| Glycoprotein (Fo-GN) and cyclophilin (Fo-Cyp1) | Midgut | Midgut | Virus entry | GN | [44] | |
| Apolipoprotein-D (ApoD), orai-2-like (Orai), and obstructor-E-like isoform X2 (Obst) | - | - | Virus acquisition and transmission | GN | [45] | |
| UBR7 | - | - | - | GN | [43] | |
| Whitefly (B. tabaci) | Heat shock proteins | Midgut | Midgut | Inhibits virus inside whitefly | CP | [48,49] |
| Peptidoglycan recognition protein | Midgut | Midgut | Whitefly immunity against the virus | CP | [50] | |
| Midgut protein | Midgut | Midgut | Translocation of virus in midgut | CP | [52] | |
| Cyclophilin B | Midgut, salivary gland, ovary | Midgut, salivary gland, and ovary | Helps suppress whitefly immune response | CP | [51] | |
| Vitellogenin | Ovary | Hemolymph and ovary | Viral entry into the ovary | CP | [53] | |
| Vesicle-associated membrane protein-associated protein B | Midgut | Midgut | Inhibits virus translocation across midgut | CP | [57] | |
| Collagen | Midgut | Midgut | Helps in viral adhesion and entry to the midgut epithelial cells | CP | [55] | |
| Thioredoxin like protein | - | - | - | CP | [56] | |
| Hairy and enhancer of split homolog-1 (HES1) | - | - | Viral transcription | Intergenic region | [69] | |
| Cubilin and aminoless | Midgut | Midgut | Virus acquisition and transmission | CP | [59] | |
| Proliferating cell nuclear antigen (PCNA) | - | Midgut and salivary gland | Helps with virus replication | Replication-associated protein (Rep) | [58] | |
| Tumorous imaginal discs (Tid) | - | - | Immune response | CP | [60] | |
| Vesicle-associated membrane protein 2 | - | - | Virus acquisition | CP | [61] | |
| Vacuolar protein sorting-associated protein (Vps) twenty-associated 1 (Vta1) | - | Midgut | Translocation of virus in midgut | CP | [62] | |
| C1QBP | - | - | Virus transmission | CP | [63] | |
| HSP70, GroEL, enolase, nucleoproteins, lachesins, vitellogenin, succinate dehydrogenase, apolipophorins, salivary secreted proteins, 40 s ribosomal proteins, tropomyosin, sorbitol dehydrogenase, GTP cyclohydrolase, dipeptidyl peptidase, annexin, E3 ubiquitin, and others | - | - | - | CP | [64] | |
| Phosphatidylethanolamine-binding protein (PEBP) | Midgut and salivary gland cell membrane | Cytoplasm | Regulation of autophagy and apoptosis | CP | [65] | |
| C2H2 zinc finger | - | - | Inhibits virus retention | CP | [67] | |
| Tubulin beta chain (TUB), keratin type I cytoskeletal 9-like (KRT), and cytochrome c oxidase subunit 5A (COX) | - | - | Virus retention and transmission | CPm | [66] | |
| Zinc finger 330 (ZNF330) | - | - | Antiviral response | CP | [68] |
CP = coat protein, and CPm = coat protein minor.
3. Viral Receptors in Plants
Plants are a well-evolved system that uses a variety of sophisticated immune mechanisms to combat many pathogens including fungi, bacteria, and viruses. The immune responses are switched on by the interaction of microbial signatures, either from pathogens or beneficial microbes, called Pathogen or Microbes-Associated Molecular Patterns (P/MAMP), or sometimes from molecules released by plants due to biotic or abiotic stresses referred to as damage-associated molecular patterns (DAMP) [70], with a cognate receptor called Pattern Recognition Receptors (PRR), located on the plant cell surface [71].
It becomes evident that plants have evolved two layers of defense viz., basal, or primary and secondary defense. The primary defense is better known as PAMP-triggered immunity (PTI), which involves the interaction of PAMP/DAMP molecules with its corresponding extracellular PRR, thereby conferring the first layer of defense against a broad range of pathogens. To bypass the PTI, virulent pathogens have evolved to produce effector molecules. Again, to counteract this virulent pathogen, plants activate a secondary defense, designated as effector-triggered immunity (ETI), which involves the interaction of specific pathogen effector/avirulent (Avr) protein with its cognate intracellular resistance (R) protein [72] leading to a highly specific restriction against the invading pathogen(s). Upon perception of signaling molecules, the plant activates downstream signaling and defense responses, including structural and biochemical changes, which enable the plant to combat invading pathogens [73,74]. On the other hand, pathogens have also evolved to overcome the non-viral PTI and cause successful infection. Consequently, plants have also evolved by producing R protein to protect themselves from infecting viruses, which is often referred to as the boom-and-bust cycle or zig-zag model [75].
Unlike fungal and bacterial pathogens that adventure through active penetration of host cells, plant viruses solely rely on vector transmission or opportunistic mechanical wounds for entry into the plant cells, provoking all essential viral proteins (coat protein (CP); replication protein (RP); movement protein (MP)) encoded by the viral genome to be translated in the plant cytoplasm itself. These translated proteins serve as Avr factors for defense activation [76]. Due to the direct nature of viral entry in plant systems, it has been arduous for the scientific community to characterize and understand the molecular mechanism of interaction of viral-PAMP (VAMP) with its cognate PRR. This review conferred the mechanisms of either direct or indirect interaction of viral protein with R protein and non-viral receptors involved in virus resistance and categorized the receptor proteins (R protein and non-viral coreceptor) based on interacting protein domains with its cognate-interacting partners (Figure 6).
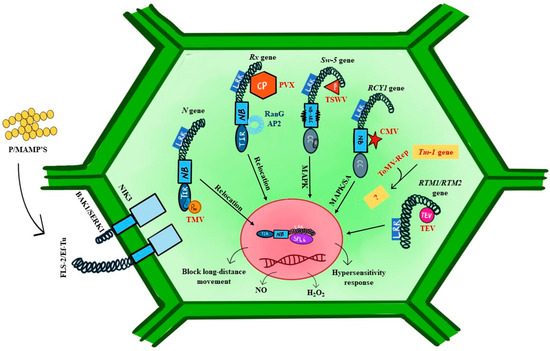
Figure 6.
A simplified diagram of virus and non-virus-associated receptors in plants. N-gene, Rx gene, Sw-5b gene, RCY1 gene, Tm-1, RTM1/RTM2 gene show the response against viruses. Tobacco mosaic virus (TMV), potato virus X (PVX), tomato spotted wilt virus (TSWV), cucumber mosaic virus (CMV), tomato mosaic virus (ToMV), and tobacco etch virus (TEV) are interacting with host proteins. Other non-NBS-LRR receptors are also shown in this image.
3.1. Virus Receptor/Viral R Genes in Anti-Viral Immunity: NBS/LRR Genes
In bygone decades, several viral receptors have been characterized, which induce a multitude of defense responses against plant viruses based on their structure, protein nature, and ligand interaction. Of those, most belong to the NBS-LRR family, and few recline to the RLK family, which recognizes specific Avr proteins of viruses. NBS-LRR viral receptors are further classified into coiled-coil (CC)-NBS-LRR and Toll/Interleukin receptor (TIR)-NBS-LRR based on their N terminal region (Table 2). The majority of the viral R genes encode a CC-NBS-LRR protein, while few encode TIR-NBS-LRR proteins [77]. The first identified viral R gene to convene resistance against tobacco mosaic virus (TMV) was the Tobacco N gene (Table 2). Subsequently, several R genes imparting antiviral resistance in plants have been identified, such as Sw-5b for TSWV in tomatoes [78], Rx1 and Rx2 for Potato virus X (PVX) identified in potatoes [78], RTM1 and RTM2 for TEV, RCY1 for CMV in Arabidopsis [79], and the I locus for bean common mosaic virus (Table 2). Further, the mechanism of how plant viruses overcome the complete resistance offered by the R gene (zig-zag model) has also been discussed in subsequent sections.
3.1.1. N Gene
The Tobacco N gene [80] encodes TIR-NBS-LRR class proteins in the cytoplasm and nucleus. The N protein induces necrosis of plant cells upon indirect interaction with the 50 kDa helicase domain (p50 effector) of TMV replicase [81]. The effector p50 recruits chloroplast localized N-receptor-interacting protein 1 (NRIP1) to interact with N protein in the cytoplasm and nucleus to mediate complete and durable resistance for TMV. The host protein NRIP1 interacts with both the N protein TIR domain and the p50 effector. Consequently, the defense and necrosis of cells have been activated upon this indirect interaction of the TIR domain and p50 effectors (Table 2). Further, a study revealed that p50 effectors interact with the TIR domain, directly followed by the NB and LRR domain, which leads to a conformational change and oligomerization of the N protein [82]. Upon perception of p50 in the nucleus, N protein interacts with transcription factors to activate PR protein expression, for instance, the interaction of N protein and Squamosa Promoter-Binding Protein (SBP)-domain transcription factor SPL6, which activates the promoter for expression of PR protein [83].
3.1.2. Rx Gene
Rx gene of potato is another well-studied CC-NBS-LRR type of R protein that mediates extreme resistance against PVX and other potexviruses. In addition, by a single amino acid substitution in the LRR domain, it can recognize Carlavirus (Table 2). The CC domain undergoes intramolecular interaction with the NBS-LRR region of Rx and with the Rx cofactor RanGAP2 (Ran GTPase-activating protein 2), while the C terminus of LRR domain specifically recognizes the CP of PVX [84]. Perception of CP by Rx protein leads to suppression of virus accumulation in an early stage of infection rather than activating HR response at the site of infection [83,85]. The association of the chaperone complex and its signaling proteins with Rx protein modulates the immune responses and nucleocytoplasmic distribution of Rx protein [86]. In addition to the chaperone complex, Rx is activated upon recognition of the Ran GTPase-mediated interaction with the CP. Further, the physical interaction of RanGAP2 protein as a cytoplasmic retention factor with Rx mediates nucleocytoplasmic partitioning of Rx protein through relocation from the cytoplasm to the nucleus, which is crucial to elucidate complete resistance and effective immune signaling against PVX [87].
Further comparison of both N and Rx-mediated resistance concluded that, in both cases, the R proteins are activated in the cytoplasm; however, their complete functionality depends on their nucleocytoplasmic distribution inside the host. This R-signaling cascade complex in plant–virus interaction involves rapid activation of mitogen-activated protein kinases (MAPKs) and the contribution of molecular chaperone complexes towards controlling the stabilization and destabilization of R proteins [88].
3.1.3. Sw-5b and Tsw Genes
The most effective dominant resistant genes, namely Sw-5b and Tsw, offer durable and robust resistance against TSWV infection in tomatoes and peppers, respectively [89]. The genes of Sw-5b and Tsw were identified from Solanum peruvianum and Capsicum chinense, respectively (Table 2). The protein products of both genes belong to CC-NBS-LRR-type protein. The cognate Avr determinants for Sw-5b and Tsw gene proteins are Non-structural movement (NSm) and non-structural suppressor (NSs) proteins, respectively. The locations of avr proteins are also different; NSm protein is encoded by an M RNA fragment, while NSs protein is encoded by an S RNA fragment of TSWV. Specifically, Sw-5b encodes N terminal CC domain, central NB-ARC (Apaf-1 R protein, and CED-4) domain, and C-terminal LRR domain. Further, recently, a 21-conserved amino acid motif of Nsm protein has been proved to interact with Sw-5b protein and to induce hypersensitive responses (HR) at the site of virus entry and eventually lead to the abscission of leaves in resistant tomato [90]. The Sw-5b protein poses an additional Solanaceae-specific domain (SD) at the N terminal which helps to prevent the auto-inhibition and activation of resistant protein [91]. In the interaction of Sw-5b and NSm proteins, to enhance the specificity and sensitivity of NSm detection, Sw-5b undergoes two-step recognition of NSm by both SD and LRR domains [92]. Upon perception of avr (NSm), Sw-5b in association with the NRC protein family (NB-LRR proteins required for HR-associated cell death) induces HR in the host cells [93]. Like Sw-5b in tomatoes, Tsw has also been shown to confer resistance by HR to a vast variety of TSWV isolates.
Although ETI is robust and durable in plants having the R gene, synergistic interaction among the viruses and the emergence of several resistant-breaking strains upon repeated cultivation of resistant cultivars over the years results in the continuous arms race between the virus and the host. Mutation in the avr gene plays a key role in this process. For example, the TMV-Ob strain overcomes the resistance conferred by the N gene in tobacco due to a nucleotide change of a 126 kDa gene of TMV [94]. Similarly, multiple resistance-breaking strains were associated with TSWV in tomatoes and peppers. TSWV strains overcoming resistance against Sw-5b in tomato was reported from various regions at different time points [95,96]. The resistance-breaking phenotype was associated with amino acid substitutions in NSm, namely C118Y, T120N, and D122G [97,98,99]. Similarly, in pepper, TSWV-resistant breaking strains were reported from different regions [100,101]. Not much understanding has been established about the precise mechanism by which viruses overcome resistance; however, it can be safely assumed that due to mutation in the avr protein, the virus escapes from its interaction with the R protein, which has led the viruses to overcome the resistance offered by the R gene, which is the great evidence for the zig-zag model between plant and virus.
3.1.4. RCY1 and HRT Locus
Two dominant locus, RCY1 and HRT, belong to the same family that encodes CC-NBS-LRR-type proteins and confer resistance against the yellow strain of CMV-Y and turnip crinkle virus (TCV), respectively, in different ecotypes of Arabidopsis (C24 for RCY1 and Dijon-17 for HRT). Upon perception of the coat protein, both genes activate HR through salicylic acid (SA), jasmonic acid (JA), and ethylene-mediated signaling responses (Table 2). An interaction of HRT protein and regulatory complex EDS/PAD4/SAG101 is required for SA-mediated resistance against TCV [102].
4. Conclusions
The successful infectious cycle of plant viruses is determined by the compatible host and the vector that transmits them. In other words, the virus–vector–host tripartite interaction is crucial for successful infection. The receptor molecules cement the bridge between the virus, host, and the vector in the tripartite interaction. The current review focused on the receptors for the viruses that exist in the vectors that help in transmission while the receptors we discussed in the plants play a role in sensing the virus and triggering defense against them. Understanding both aspects is important for devising efficient virus management strategies. The receptors in vectors for the viruses could be the potential targets. Our current review presents the abundance of viral receptors in major vectors, and, at the same time, stresses the need for characterizing them. Recent advances that are made in the area of molecular biology and biochemistry could be integrated into the field which could potentially address the functional characterization of insect receptors that interact with the viral factor. Since the species of plant viruses are specifically transmitted by specific vectors, targeting the vectors will benefit in controlling the virus. Therefore, shedding more light on insect receptors associated with viruses in vectors could be a part of the anti-viral strategy. It is more important to note that the strategy should have minimal off-target effects on other insects and the least toxic effects on humans and the environment which is essential for wide application and acceptance. Coming to the viral receptors in plants, the plant receptors we discussed here play an important role in plant defense against viruses. Since they are the source of resistance against viruses, identifying and understanding the resistance genes and their mechanisms present in plants is essential for developing resistant varieties of crops. Defense and counter-defense between the host and the viruses is a continuous and evolving process. With the incidences of resistance-breaking strains of viruses, it is becoming evident that there is a need to identify more resistance genes essential to identify resistance. Major studies only identify the resistance break, but much deeper characterization has not been carried out. Again, the application of recent advancements in biology could help in shedding light. It is also a known fact that cultivating a resistant variety along with proper vector management strategies are crucial parts of an integrated pest management system.
Author Contributions
Conceptualization, M.K. and R.D.; software, M.K. and B.S.; formal analysis, S.J. and S.C.; data curation, S.J., S.C. and B.S.; writing—original draft preparation, M.K., S.J., S.C., S.R.P. and B.S.; writing—review and editing, S.J., S.C., R.D. and M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Not applicable.
Acknowledgments
The authors appreciate anonymous reviewers for their constructive criticism and suggestions on the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Casasnovas, J.M. Virus-Receptor Interactions and Receptor-Mediated Virus Entry into Host Cells. Subcell. Biochem. 2013, 68, 441–466. [Google Scholar] [CrossRef] [PubMed]
- Hohn, T. Plant Virus Transmission from the Insect Point of View. Proc. Natl. Acad. Sci. USA 2007, 104, 17905. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.C.K.; Perry, K.L. Transmission of Plant Viruses by Aphid Vectors. Mol. Plant Pathol. 2004, 5, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Cox-Foster, D.; Gray, S.M.; Gildow, F. Vector Specificity of Barley Yellow Dwarf Virus (BYDV) Transmission: Identification of Potential Cellular Receptors Binding BYDV-MAV in the Aphid, Sitobion avenae. Virology 2001, 286, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, G. Identification of a Protein Associated with Circulative Transmission of Barley Yellow Dwarf Virus from Cereal Aphids, Schizaphis graminum and Sitobion avenae. Chin. Sci. Bull. 2003, 48, 2083–2087. [Google Scholar] [CrossRef]
- Seddas, P.; Boissinot, S.; Strub, J.M.; Van Dorsselaer, A.; Van Regenmortel, M.H.V.; Pattus, F. Rack-1, GAPDH3, and Actin: Proteins of Myzus persicae Potentially Involved in the Transcytosis of Beet Western Yellows Virus Particles in the Aphid. Virology 2004, 325, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Uzest, M.; Gargani, D.; Drucker, M.; Hébrard, E.; Garzo, E.; Candresse, T.; Fereres, A.; Blanc, S. A Protein Key to Plant Virus Transmission at the Tip of the Insect Vector Stylet. Proc. Natl. Acad. Sci. USA 2007, 104, 17959–17964. [Google Scholar] [CrossRef] [PubMed]
- Dombrovsky, A.; Gollop, N.; Chen, S.; Chejanovsky, N.; Raccah, B. In Vitro Association between the Helper Component-Proteinase of Zucchini Yellow Mosaic Virus and Cuticle Proteins of Myzus persicae. J. Gen. Virol. 2007, 88, 1602–1610. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Calvino, L.; Goytia, E.; López-Abella, D.; Giner, A.; Urizarna, M.; Vilaplana, L.; López-Moya, J.J. The Helper-Component Protease Transmission Factor of Tobacco Etch Potyvirus Binds Specifically to an Aphid Ribosomal Protein Homologous to the Laminin Receptor Precursor. J. Gen. Virol. 2010, 91, 2862–2873. [Google Scholar] [CrossRef]
- Tamborindeguy, C.; Bereman, M.S.; DeBlasio, S.; Igwe, D.; Smith, D.M.; White, F.; MacCoss, M.J.; Gray, S.M.; Cilia, M. Genomic and Proteomic Analysis of Schizaphis graminum Reveals Cyclophilin Proteins Are Involved in the Transmission of Cereal Yellow Dwarf Virus. PLoS ONE 2013, 8, e71620. [Google Scholar] [CrossRef]
- Linz, L.B.; Liu, S.; Chougule, N.P.; Bonning, B.C. In Vitro Evidence Supports Membrane Alanyl Aminopeptidase N as a Receptor for a Plant Virus in the Pea Aphid Vector. J. Virol. 2015, 89, 11203. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Gao, X.W. The Cuticle Protein Gene MPCP4 of Myzus persicae (Homoptera: Aphididae) Plays a Critical Role in Cucumber Mosaic Virus Acquisition. J. Econ. Entomol. 2017, 110, 848–853. [Google Scholar] [CrossRef]
- Webster, C.G.; Pichon, E.; van Munster, M.; Monsion, B.; Deshoux, M.; Gargani, D.; Calevro, F.; Jimenez, J.; Moreno, A.; Krenz, B.; et al. Identification of Plant Virus Receptor Candidates in the Stylets of Their Aphid Vectors. J. Virol. 2018, 92, e00432-18. [Google Scholar] [CrossRef]
- Mulot, M.; Monsion, B.; Boissinot, S.; Rastegar, M.; Meyer, S.; Bochet, N.; Brault, V. Transmission of Turnip Yellows Virus by Myzus persicae Is Reduced by Feeding Aphids on Double-Stranded RNA Targeting the Ephrin Receptor Protein. Front. Microbiol. 2018, 9, 457. [Google Scholar] [CrossRef]
- Bahrami Kamangar, S.; Christiaens, O.; Taning, C.N.T.; De Jonghe, K.; Smagghe, G. The Cuticle Protein MPCP2 Is Involved in Potato Virus Y Transmission in the Green Peach Aphid Myzus persicae. J. Plant Dis. Prot. 2019, 126, 351–357. [Google Scholar] [CrossRef]
- Deblasio, S.L.; Wilson, J.R.; Tamborindeguy, C.; Johnson, R.S.; Pinheiro, P.V.; Maccoss, M.J.; Gray, S.M.; Heck, M. Affinity Purification-Mass Spectrometry Identifies a Novel Interaction between a Polerovirus and a Conserved Innate Immunity Aphid Protein That Regulates Transmission Efficiency. J. Proteome Res. 2021, 20, 3365–3387. [Google Scholar] [CrossRef]
- Liang, Y.; Ma, K.S.; Liang, P.Z.; Yang, L.W.; Zhang, L.; Gao, X.W. Combined Transcriptomic and Proteomic Analysis of Myzus persicae, the Green Peach Aphid, Infected with Cucumber Mosaic Virus. Insects 2021, 12, 372. [Google Scholar] [CrossRef]
- He, M.J.; Zuo, D.P.; Zhang, Z.Y.; Wang, Y.; Han, C.G. Transcriptomic and Proteomic Analyses of Myzus persicae Carrying Brassica Yellows Virus. Biology 2023, 12, 908. [Google Scholar] [CrossRef]
- Guoying, Z.; Xiongbin, L.; Huijuan, L.; Juanli, L.; Shengxiang, C.; Zuxun, G. Rice Ragged Stunt Oryzavirus: Role of the Viral Spike Protein in Transmission by the Insect Vector. Ann. Appl. Biol. 1999, 135, 573–578. [Google Scholar] [CrossRef]
- Li, S.; Xiong, R.; Wang, X.; Zhou, Y. Five Proteins of Laodelphax striatellus Are Potentially Involved in the Interactions between Rice Stripe Virus and Vector. PLoS ONE 2011, 6, e26585. [Google Scholar] [CrossRef]
- Huo, Y.; Liu, W.; Zhang, F.; Chen, X.; Li, L.; Liu, Q.; Zhou, Y.; Wei, T.; Fang, R.; Wang, X. Transovarial Transmission of a Plant Virus Is Mediated by Vitellogenin of Its Insect Vector. PLoS Pathog. 2014, 10, e1003949. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, W.; Li, J.; Luo, L.; Kang, L.; Cui, F. The C-Jun N-Terminal Kinase Pathway of a Vector Insect Is Activated by Virus Capsid Protein and Promotes Viral Replication. eLife 2017, 6, e26591. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Gray, S.; Huo, Y.; Li, L.; Wei, T.; Wang, X. Proteomic Analysis of Interaction between a Plant Virus and Its Vector Insect Reveals New Functions of Hemipteran Cuticular Protein. Mol. Cell. Proteom. 2015, 14, 2229–2242. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, H.; Ren, T.; Xie, L.; Wei, T. Interaction between Non-Structural Protein Pns10 of Rice Dwarf Virus and Cytoplasmic Actin of Leafhoppers Is Correlated with Insect Vector Specificity. J. Gen. Virol. 2015, 96, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.J.; Liu, C.W.; Zhou, X.; Zhang, C.X.; Bao, Y.Y. A Mitochondrial Membrane Protein Is a Target for Rice Ragged Stunt Virus in Its Insect Vector. Virus Res. 2017, 229, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, X.; Zhou, Y. Ribosomal Protein L18 Is an Essential Factor That Promote Rice Stripe Virus Accumulation in Small Brown Planthopper. Virus Res. 2018, 247, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Liu, W.; Wu, N.; Zhang, L.; Zhang, Z.; Zhou, X.; Wang, X. Invasion of Midgut Epithelial Cells by a Persistently Transmitted Virus Is Mediated by Sugar Transporter 6 in Its Insect Vector. PLoS Pathog. 2018, 14, e1007201. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, D.; Hu, J.; Zhang, K.; Kang, L.; Chen, Y.; Huang, L.; Zhang, L.; Xiang, Y.; Song, Q.; et al. The α-Tubulin of Laodelphax striatellus Mediates the Passage of Rice Stripe Virus (RSV) and Enhances Horizontal Transmission. PLoS Pathog. 2020, 16, e1008710. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Qiao, L.; Lu, H.; Chen, X.; Wang, X.; Yu, J.; Zhu, J.; Xiao, Y.; Ma, Y.; Wu, Y.; et al. Flotillin 2 Facilitates the Infection of a Plant Virus in the Gut of Insect Vector. J. Virol. 2022, 96, e0214021. [Google Scholar] [CrossRef]
- Alviar, K.B.; Rotenberg, D.; Martin, K.M.; Whitfield, A.E. The Physical Interactome between Peregrinus maidis Proteins and the Maize Mosaic Virus Glycoprotein Provides Insights into the Cellular Biology of a Rhabdovirus in the Insect Vector. Virology 2022, 577, 163–173. [Google Scholar] [CrossRef]
- Zhang, L.; Li, L.; Huang, L.; Li, X.; Xu, C.; Hu, W.; Sun, Y.; Liu, F.; Li, Y. Voltage-Dependent Anion Channel 2 (VDAC2) Facilitates the Accumulation of Rice Stripe Virus in the Vector Laodelphax striatellus. Virus Res. 2023, 324, 199019. [Google Scholar] [CrossRef] [PubMed]
- Castrosanto, M.A.; Clemente, A.J.N.; Whitfield, A.E.; Alviar, K.B. In Silico Analysis of the Predicted Protein-Protein Interaction of Syntaxin-18, a Putative Receptor of Peregrinus maidis Ashmead (Hemiptera: Delphacidae) with Maize Mosaic Virus Glycoprotein. J. Biomol. Struct. Dyn. 2023, 41, 3956–3963. [Google Scholar] [CrossRef]
- Ghosh, A.; Jangra, S.; Dietzgen, R.G.; Yeh, W. Bin Frontiers Approaches to the Diagnosis of Thrips (Thysanoptera): How Effective Are the Molecular and Electronic Detection Platforms? Insects 2021, 12, 920. [Google Scholar] [CrossRef] [PubMed]
- Jangra, S.; Ghosh, A.; Mukherjee, S.; Baranwal, V.K.; Dietzgen, R.G. Development of a Polymerase Spiral Reaction-Based Isothermal Assay for Rapid Identification of Thrips Palmi. Front. Mol. Biosci. 2022, 9, 853339. [Google Scholar] [CrossRef]
- Ghosh, A.; Basavaraj, Y.B.; Jangra, S.; Das, A. Exposure to Watermelon Bud Necrosis Virus and Groundnut Bud Necrosis Virus Alters the Life History Traits of Their Vector, Thrips palmi (Thysanoptera: Thripidae). Arch. Virol. 2019, 164, 2799–2804. [Google Scholar] [CrossRef] [PubMed]
- Jangra, S.; Mittal, A.; Dhall, H.; Jain, R.K.; Ghosh, A. A Multiplex PCR Assay for Rapid Identification of Major Tospovirus Vectors Reported in India. BMC Genom. 2020, 21, 170. [Google Scholar] [CrossRef] [PubMed]
- Mahanta, D.K.; Jangra, S.; Priti, S.; Ghosh, A.; Sharma, P.K.; Iquebal, M.A.; Jaiswal, S.; Baranwal, V.K.; Kalia, V.K.; Chander, S. Groundnut Bud Necrosis Virus Modulates the Expression of Innate Immune, Endocytosis, and Cuticle Development-Associated Genes to Circulate and Propagate in Its Vector, Thrips Palmi. Front. Microbiol. 2022, 13, 773238. [Google Scholar] [CrossRef] [PubMed]
- Bandla, M.D.; Campbell, L.R.; Ullman, D.E.; Sherwood, J.L. Interaction of Tomato Spotted Wilt Tospovirus (TSWV) Glycoproteins with a Thrips Midgut Protein, a Potential Cellular Receptor for TSWV. Phytopathology 1998, 88, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Kikkert, M.; Meurs, C.; Van De Wetering, F.; Dorfmüller, S.; Peters, D.; Kormelink, R.; Goldbach, R. Binding of Tomato Spotted Wilt Virus to a 94-KDa Thrips Protein. Phytopathology 1998, 88, 63–69. [Google Scholar] [CrossRef]
- Jagdale, S.S.; Ghosh, A. In Silico Analyses of Molecular Interactions between Groundnut Bud Necrosis Virus and Its Vector, Thrips Palmi. Virusdisease 2019, 30, 245–251. [Google Scholar] [CrossRef]
- Badillo-Vargas, I.E.; Chen, Y.; Martin, K.M.; Rotenberg, D.; Whitfield, A.E. Discovery of Novel Thrips Vector Proteins That Bind to the Viral Attachment Protein of the Plant Bunyavirus Tomato Spotted Wilt Virus. J. Virol. 2019, 93, e00699-19. [Google Scholar] [CrossRef]
- Zheng, X.-B.; Wan, Y.-R.; Zhang, Y.-J.; Wu, Q.-J. Identifying Proteins in Western Flower Thrips That Interact with Tomato Spotted Wilt Orthotospovirus GN. Chin. J. Appl. Entomol. 2020, 57, 640–657. [Google Scholar]
- Shi, J.; Zhou, J.; Jiang, F.; Li, Z.; Zhu, S. The Effects of the E3 Ubiquitin-Protein Ligase UBR7 of Frankliniella occidentalis on the Ability of Insects to Acquire and Transmit TSWV. PeerJ 2023, 11, e15385. [Google Scholar] [CrossRef]
- Khan, F.; Stanley, D.; Kim, Y. Two Alimentary Canal Proteins, Fo-GN and Fo-Cyp1, Act in Western Flower Thrips, Frankliniella occidentalis TSWV Infection. Insects 2023, 14, 154. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Wan, Y.; Tao, M.; Yuan, J.; Zhang, K.; Wang, J.; Zhang, Y.; Liang, P.; Wu, Q. Obstructor, a Frankliniella occidentalis Protein, Promotes Transmission of Tomato Spotted Wilt Orthotospovirus. Insect Sci. 2023, 30, 741–757. [Google Scholar] [CrossRef]
- Czosnek, H.; Hariton-Shalev, A.; Sobol, I.; Gorovits, R.; Ghanim, M. The Incredible Journey of Begomoviruses in Their Whitefly Vector. Viruses 2017, 9, 273. [Google Scholar] [CrossRef] [PubMed]
- Devendran, R.; Kumar, M.; Ghosh, D.; Yogindran, S.; Karim, M.J.; Chakraborty, S. Capsicum-Infecting Begomoviruses as Global Pathogens: Host-Virus Interplay, Pathogenesis, and Management. Trends Microbiol. 2022, 30, 170–184. [Google Scholar] [CrossRef] [PubMed]
- Ohnesorge, S.; Bejarano, E.R. Begomovirus Coat Protein Interacts with a Small Heat-Shock Protein of Its Transmission Vector (Bemisia tabaci). Insect Mol. Biol. 2009, 18, 693–703. [Google Scholar] [CrossRef]
- Götz, M.; Popovski, S.; Kollenberg, M.; Gorovits, R.; Brown, J.K.; Cicero, J.M.; Czosnek, H.; Winter, S.; Ghanim, M. Implication of Bemisia tabaci Heat Shock Protein 70 in Begomovirus-Whitefly Interactions. J. Virol. 2012, 86, 13241–13252. [Google Scholar] [CrossRef]
- Wang, Z.Z.; Shi, M.; Huang, Y.C.; Wang, X.W.; Stanley, D.; Chen, X.X. A Peptidoglycan Recognition Protein Acts in Whitefly (Bemisia tabaci) Immunity and Involves in Begomovirus Acquisition. Sci. Rep. 2016, 6, 37806. [Google Scholar] [CrossRef]
- Kanakala, S.; Ghanim, M. Implication of the Whitefly Bemisia tabaci Cyclophilin B Protein in the Transmission of Tomato Yellow Leaf Curl Virus. Front. Plant Sci. 2016, 7, 1702. [Google Scholar] [CrossRef] [PubMed]
- Rana, V.S.; Popli, S.; Saurav, G.K.; Raina, H.S.; Chaubey, R.; Ramamurthy, V.V.; Rajagopal, R. A Bemisia tabaci Midgut Protein Interacts with Begomoviruses and Plays a Role in Virus Transmission. Cell Microbiol. 2016, 18, 663–678. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; He, Y.Z.; Guo, Q.; Guo, T.; Liu, Y.Q.; Zhou, X.P.; Liu, S.S.; Wang, X.W. Vector Development and Vitellogenin Determine the Transovarial Transmission of Begomoviruses. Proc. Natl. Acad. Sci. USA 2017, 114, 6746–6751. [Google Scholar] [CrossRef] [PubMed]
- Thesnim, P.; Jangra, S.; Kumar, M.; Ghosh, A. Effect of Silencing Bemisia tabaci TLR3 and TOB1 on Fitness and Begomovirus Transmission. Front. Plant Sci. 2023, 14, 1136262. [Google Scholar] [CrossRef] [PubMed]
- Rana, V.S.; Popli, S.; Saurav, G.K.; Raina, H.S.; Jamwal, R.; Chaubey, R.; Ramamurthy, V.V.; Natarajan, K.; Rajagopal, R. Implication of the Whitefly, Bemisia tabaci, Collagen Protein in Begomoviruses Acquisition and Transmission. Phytopathology 2019, 109, 1481–1493. [Google Scholar] [CrossRef] [PubMed]
- Saurav, G.K.; Rana, V.S.; Popli, S.; Daimei, G.; Rajagopal, R. A Thioredoxin-like Protein of Bemisia tabaci Interacts with Coat Protein of Begomoviruses. Virus Genes 2019, 55, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chi, Y.; Zhang, X.J.; Wang, X.W.; Liu, S.S. Implication of Whitefly Vesicle Associated Membrane Protein-Associated Protein B in the Transmission of Tomato Yellow Leaf Curl Virus. Virology 2019, 535, 210–217. [Google Scholar] [CrossRef] [PubMed]
- He, Y.Z.; Wang, Y.M.; Yin, T.Y.; Fiallo-Olivé, E.; Liu, Y.Q.; Hanley-Bowdoin, L.; Wang, X.W. A Plant DNA Virus Replicates in the Salivary Glands of Its Insect Vector via Recruitment of Host DNA Synthesis Machinery. Proc. Natl. Acad. Sci. USA 2020, 117, 16928–16937. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Lei, T.; Zhang, X.J.; Yin, T.Y.; Wang, X.W.; Liu, S.S. A Vector Whitefly Endocytic Receptor Facilitates the Entry of Begomoviruses into Its Midgut Cells via Binding to Virion Capsid Proteins. PLoS Pathog. 2020, 16, e1009053. [Google Scholar] [CrossRef]
- Zhao, J.; Guo, T.; Lei, T.; Zhu, J.C.; Wang, F.; Wang, X.W.; Liu, S.S. Proteomic Analyses of Whitefly-Begomovirus Interactions Reveal the Inhibitory Role of Tumorous Imaginal Discs in Viral Retention. Front. Immunol. 2020, 11, 1596. [Google Scholar] [CrossRef]
- Fan, Y.Y.; Zhong, Y.W.; Zhao, J.; Chi, Y.; Bouvaine, S.; Liu, S.S.; Seal, S.E.; Wang, X.W. Bemisia tabaci Vesicle-Associated Membrane Protein 2 Interacts with Begomoviruses and Plays a Role in Virus Acquisition. Cells 2021, 10, 1700. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Pan, L.L.; Liu, S.S.; Mansoor, S.; Wang, X.W. Implication of the Whitefly Protein Vps Twenty Associated 1 (Vta1) in the Transmission of Cotton Leaf Curl Multan Virus. Microorganisms 2021, 9, 304. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Bello, V.H.; Ghanim, M. Transmission Parameters of Pepper Whitefly-Borne Vein Yellows Virus (PeWBVYV) by Bemisia tabaci and Identification of an Insect Protein with a Putative Role in Polerovirus Transmission. Virology 2021, 560, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Prasannakumar, N.R.; Maruthi, M.N. Identification of Whitefly (Bemisia tabaci) Proteins Interacting with Tomato Leaf Curl Bangalore Virus Coat Protein Gene Using Y2H System. Int. J. Trop. Insect Sci. 2022, 42, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Guo, H.; Zhu-Salzman, K.; Ge, F.; Sun, Y. PEBP Balances Apoptosis and Autophagy in Whitefly upon Arbovirus Infection. Nat. Commun. 2022, 13, 846. [Google Scholar] [CrossRef] [PubMed]
- He, H.F.; Zhao, C.C.; Zhu, C.Q.; Yan, W.L.; Yan, M.H.; Zhang, Z.L.; Liu, J.L.; Shi, B.Z.; Bai, R.E.; Li, J.J.; et al. Discovery of Novel Whitefly Vector Proteins That Interact with a Virus Capsid Component Mediating Virion Retention and Transmission. Int. J. Biol. Macromol. 2023, 226, 1154–1165. [Google Scholar] [CrossRef]
- Ghosh, S.; Srinivasan, R.; Ghanim, M. A C2H2 Zinc Finger Transcription Factor of the Whitefly Bemisia tabaci Interacts with the Capsid Proteins of Begomoviruses and Inhibits Virus Retention. Insect Mol. Biol. 2023, 32, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Gao, Y.; Shi, X.; Yang, C.; Xie, G.; Tang, T.; Wang, D.; Zheng, L.; Liu, Y.; Zhang, D. Zinc Finger Protein 330 Regulates Ramie Mosaic Virus Infection in the Whitefly Bemisia tabaci MED. Pest Manag. Sci. 2023, 79, 1750–1759. [Google Scholar] [CrossRef]
- Wang, Y.M.; He, Y.Z.; Ye, X.T.; He, W.Z.; Liu, S.S.; Wang, X.W. Whitefly HES1 Binds to the Intergenic Region of Tomato Yellow Leaf Curl China Virus and Promotes Viral Gene Transcription. Virology 2020, 542, 54–62. [Google Scholar] [CrossRef]
- Macho, A.P.; Zipfel, C. Plant PRRs and the Activation of Innate Immune Signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef]
- Fritz-Laylin, L.K.; Krishnamurthy, N.; Tör, M.; Sjölander, K.V.; Jones, J.D.G. Phylogenomic Analysis of the Receptor-Like Proteins of Rice and Arabidopsis. Plant Physiol. 2005, 138, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Katagiri, F. Comparing Signaling Mechanisms Engaged in Pattern-Triggered and Effector-Triggered Immunity. Curr. Opin. Plant Biol. 2010, 13, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling Mechanisms in Pattern-Triggered Immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef] [PubMed]
- Hogenhout, S.A.; Van Der Hoorn, R.A.L.; Terauchi, R.; Kamoun, S. Emerging Concepts in Effector Biology of Plant-Associated Organisms. Mol. Plant Microbe Interact. 2009, 22, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The Plant Immune System. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Gouveia, B.C.; Calil, I.P.; Machado, J.P.B.; Santos, A.A.; Fontes, E.P.B. Immune Receptors and Co-Receptors in Antiviral Innate Immunity in Plants. Front. Microbiol. 2016, 7, 2139. [Google Scholar] [CrossRef]
- Kumar, M.; Devendran, R.; Kumar, R.V. Geminivirus Infections Co-Opt Posttranslational Modification of Proteins during Viral Pathogenesis. In Geminivirus: Detection, Diagnosis and Management; Gaur, R.K., Sharma, P., Czosnek, H., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 443–453. [Google Scholar] [CrossRef]
- Brommonschenkel, S.H.; Frary, A.; Frary, A.; Tanksley, S.D. The Broad-Spectrum Tospovirus Resistance Gene Sw-5 of Tomato Is a Homolog of the Root-Knot Nematode Resistance Gene Mi. Mol. Plant Microbe Interact. 2000, 13, 1130–1138. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Suzuki, M.; Natsuaki, K.; Shigyo, T.; Hino, K.; Teraoka, T.; Hosokawa, D.; Ehara, Y. Mapping the Virus and Host Genes Involved in the Resistance Response in Cucumber Mosaic Virus-Infected Arabidopsis thaliana. Plant Cell Physiol. 2001, 42, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Whitham, S.; Dinesh-Kumar, S.P.; Choi, D.; Hehl, R.; Corr, C.; Baker, B. The Product of the Tobacco Mosaic Virus Resistance Gene N: Similarity to Toll and the Interleukin-1 Receptor. Cell 1994, 78, 1101–1115. [Google Scholar] [CrossRef]
- Ueda, H.; Yamaguchi, Y.; Sano, H. Direct Interaction between the Tobacco Mosaic Virus Helicase Domain and the ATP-Bound Resistance Protein, N Factor during the Hypersensitive Response in Tobacco Plants. Plant Mol. Biol. 2006, 61, 31–45. [Google Scholar] [CrossRef]
- Mestre, P.; Baulcombe, D.C. Elicitor-Mediated Oligomerization of the Tobacco N Disease Resistance Protein. Plant Cell 2006, 18, 491–501. [Google Scholar] [CrossRef]
- Padmanabhan, M.S.; Ma, S.; Burch-Smith, T.M.; Czymmek, K.; Huijser, P.; Dinesh-Kumar, S.P. Novel Positive Regulatory Role for the SPL6 Transcription Factor in the N TIR-NB-LRR Receptor-Mediated Plant Innate Immunity. PLoS Pathog. 2013, 9, e1003235. [Google Scholar] [CrossRef] [PubMed]
- Candresse, T.; Marais, A.; Faure, C.; Dubrana, M.P.; Gombert, J.; Bendahmane, A. Multiple Coat Protein Mutations Abolish Recognition of Pepino mosaic potexvirus (PepMV) by the Potato Rx Resistance Gene in Transgenic Tomatoes. Mol. Plant Microbe Interact. 2010, 23, 376–383. [Google Scholar] [CrossRef]
- Bendahmane, A.; Kanyuka, K.; Baulcombe, D.C. The Rx Gene from Potato Controls Separate Virus Resistance and Cell Death Responses. Plant Cell 1999, 11, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Hoser, R.; Zurczak, M.; Lichocka, M.; Zuzga, S.; Dadlez, M.; Samuel, M.A.; Ellis, B.E.; Stuttmann, J.; Parker, J.E.; Hennig, J.; et al. Nucleocytoplasmic Partitioning of Tobacco N Receptor Is Modulated by SGT1. New Phytol. 2013, 200, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Tameling, W.I.L.; Nooijen, C.; Ludwig, N.; Boter, M.; Slootweg, E.; Goverse, A.; Shirasu, K.; Joosten, M.H.A.J. RanGAP2 Mediates Nucleocytoplasmic Partitioning of the NB-LRR Immune Receptor Rx in the Solanaceae, Thereby Dictating Rx Function. Plant Cell 2012, 22, 4176–4194. [Google Scholar] [CrossRef]
- Kadota, Y.; Shirasu, K. The HSP90 Complex of Plants. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2012, 1823, 689–697. [Google Scholar] [CrossRef]
- Chinnaiah, S.; Gautam, S.; Workneh, F.; Crosby, K.; Rush, C.; Gadhave, K.R. First Report of Sw-5 Resistance-Breaking Strain of Tomato Spotted Wilt Orthotospovirus Infecting Tomato in Texas. Plant Dis. 2023, 107, 2569. [Google Scholar] [CrossRef]
- Zhu, M.; Jiang, L.; Bai, B.; Zhao, W.; Chen, X.; Li, J.; Liu, Y.; Chen, Z.; Wang, B.; Wang, C.; et al. The Intracellular Immune Receptor Sw-5b Confers Broad-Spectrum Resistance to Tospoviruses through Recognition of a Conserved 21-Amino Acid Viral Effector Epitope. Plant Cell 2017, 29, 2214–2232. [Google Scholar] [CrossRef]
- Chen, X.; Zhu, M.; Jiang, L.; Zhao, W.; Li, J.; Wu, J.; Li, C.; Bai, B.; Lu, G.; Chen, H.; et al. A Multilayered Regulatory Mechanism for the Autoinhibition and Activation of a Plant CC-NB-LRR Resistance Protein with an Extra N-Terminal Domain. New Phytol. 2016, 212, 161–175. [Google Scholar] [CrossRef]
- Li, J.; Huang, H.; Zhu, M.; Huang, S.; Zhang, W.; Dinesh-Kumar, S.P.; Tao, X. A Plant Immune Receptor Adopts a Two-Step Recognition Mechanism to Enhance Viral Effector Perception. Mol. Plant 2019, 12, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Qian, X.; Chen, X.; Yang, T.; Feng, M.; Chen, J.; Cheng, R.; Hong, H.; Zheng, Y.; Mei, Y.; et al. Cytoplasmic and Nuclear Sw-5b NLR Act Both Independently and Synergistically to Confer Full Host Defense against Tospovirus Infection. New Phytol. 2021, 231, 2262–2281. [Google Scholar] [CrossRef] [PubMed]
- Padgett, H.S.; Beachy, R.N. Analysis of a Tobacco Mosaic Virus Strain Capable of Overcoming N Gene-Mediated Resistance. Plant Cell 1993, 5, 577–586. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ciuffo, M.; Finetti-Sialer, M.M.; Gallitelli, D.; Turina, M. First Report in Italy of a Resistance-Breaking Strain of Tomato spotted wilt virus Infecting Tomato Cultivars Carrying the Sw5 Resistance Gene. Plant Pathol. 2005, 54, 564. [Google Scholar] [CrossRef]
- Di Rienzo, V.; Bubici, G.; Montemurro, C.; Cillo, F. Rapid Identification of Tomato Sw-5 Resistance-Breaking Isolates of Tomato Spotted Wilt Virus Using High Resolution Melting and TaqMan SNP Genotyping Assays as Allelic Discrimination Techniques. PLoS ONE 2018, 13, e0196738. [Google Scholar] [CrossRef] [PubMed]
- López, C.; Aramburu, J.; Galipienso, L.; Soler, S.; Nuez, F.; Rubio, L. Evolutionary Analysis of Tomato Sw-5 Resistance-Breaking Isolates of Tomato Spotted Wilt Virus. J. Gen. Virol. 2011, 92, 210–215. [Google Scholar] [CrossRef]
- Huang, H.; Zuo, C.; Zhao, Y.; Huang, S.; Wang, T.; Zhu, M.; Li, J.; Tao, X. Determination of Key Residues in Tospoviral NSm Required for Sw-5b Recognition, Their Potential Ability to Overcome Resistance, and the Effective Resistance Provided by Improved Sw-5b Mutants. Mol. Plant Pathol. 2022, 23, 622–633. [Google Scholar] [CrossRef]
- Almási, A.; Pinczés, D.; Tímár, Z.; Sáray, R.; Palotás, G.; Salánki, K. Identification of a New Type of Resistance Breaking Strain of Tomato Spotted Wilt Virus on Tomato Bearing the Sw-5b Resistance Gene. Eur. J. Plant Pathol. 2023, 166, 219–225. [Google Scholar] [CrossRef]
- Macedo, M.A.; Rojas, M.R.; Gilbertson, R.L. First Report of a Resistance-Breaking Strain of Tomato Spotted Wilt Orthotospovirus Infecting Sweet Pepper with the Tsw Resistance Gene in California, U.S.A. Plant Dis. 2019, 103, 1048. [Google Scholar] [CrossRef]
- Jiang, L.; Huang, Y.; Sun, L.; Wang, B.; Zhu, M.; Li, J.; Huang, C.; Liu, Y.; Li, F.; Liu, Y.; et al. Occurrence and Diversity of Tomato Spotted Wilt Virus Isolates Breaking the Tsw Resistance Gene of Capsicum chinense in Yunnan, Southwest China. Plant Pathol. 2017, 66, 980–989. [Google Scholar] [CrossRef]
- Zhu, S.; Jeong, R.D.; Venugopal, S.C.; Lapchyk, L.; Navarre, D.R.; Kachroo, A.; Kachroo, P. SAG101 Forms a Ternary Complex with EDS1 and PAD4 and Is Required for Resistance Signaling against Turnip Crinkle Virus. PLoS Pathog. 2011, 7, e1002318. [Google Scholar] [CrossRef]
- Cosson, P.; Schurdi-Levraud, V.; Le, Q.H.; Sicard, O.; Caballero, M.; Roux, F.; Le Gall, O.; Candresse, T.; Revers, F. The RTM Resistance to Potyviruses in Arabidopsis thaliana: Natural Variation of the RTM Genes and Evidence for the Implication of Additional Genes. PLoS ONE 2012, 7, e39169. [Google Scholar] [CrossRef]
- Whitham, S.A.; Anderberg, R.J.; Chisholm, S.T.; Carrington, J.C. Arabidopsis RTM2 Gene Is Necessary for Specific Restriction of Tobacco Etch Virus and Encodes an Unusual Small Heat Shock-like Protein. Plant Cell 2000, 12, 569–582. [Google Scholar] [CrossRef]
- Chisholm, S.T.; Mahajan, S.K.; Whitham, S.A.; Yamamoto, M.L.; Carrington, J.C. Cloning of the Arabidopsis RTM1 Gene, Which Controls Restriction of Long-Distance Movement of Tobacco Etch Virus. Proc. Natl. Acad. Sci. USA 2000, 97, 489–494. [Google Scholar] [CrossRef]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.F.; Scott, J.W.; Edwards, J.D.; Bai, Y. The Tomato Yellow Leaf Curl Virus Resistance Genes Ty-1 and Ty-3 Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef] [PubMed]
- Butterbach, P.; Verlaan, M.G.; Dullemans, A.; Lohuis, D.; Visser, R.G.F.; Bai, Y.; Kormelink, R. Tomato Yellow Leaf Curl Virus Resistance by Ty-1 Involves Increased Cytosine Methylation of Viral Genomes and Is Compromised by Cucumber Mosaic Virus Infection. Proc. Natl. Acad. Sci. USA 2014, 111, 12942–12947. [Google Scholar] [CrossRef]
- Carr, J.P.; Lewsey, M.G.; Palukaitis, P. Signaling in Induced Resistance. Adv. Virus Res. 2010, 76, 57–121. [Google Scholar] [CrossRef] [PubMed]
- Calil, I.P.; Fontes, E.P.B. Plant Immunity against Viruses: Antiviral Immune Receptors in Focus. Ann. Bot. 2017, 119, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yuan, Y.R.; Pei, Y.; Lin, S.S.; Tuschl, T.; Patel, D.J.; Chua, N.H. Cucumber Mosaic Virus-Encoded 2b Suppressor Inhibits Arabidopsis Argonaute1 Cleavage Activity to Counter Plant Defense. Genes. Dev. 2006, 20, 3255–3268. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, J.; Wang, J.; Chen, L.; Fan, S.L.; Wu, J.W.; Wang, X.; Wang, Z.X. Structural Insights into the Negative Regulation of BRI1 Signaling by BRI1-Interacting Protein BKI1. Cell Res. 2014, 24, 1328–1341. [Google Scholar] [CrossRef]
- Kovalev, N.; Pogany, J.; Nagy, P.D. Template Role of Double-Stranded RNA in Tombusvirus Replication. J. Virol. 2014, 88, 5638. [Google Scholar] [CrossRef] [PubMed]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-Stranded RNAs Induce a Pattern-Triggered Immune Signaling Pathway in Plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Padgett, H.S.; Watanabe, Y.; Beachy, R.N. Identification of the TMV Replicase Sequence That Activates the N Gene-Mediated Hypersensitive Response. Mol. Plant-Microbe Interact 2007, 10, 709–715. [Google Scholar] [CrossRef]
- Bendahmane, A.; Querci, M.; Kanyuka, K.; Baulcombe, D.C. Agrobacterium Transient Expression System as a Tool for the Isolation of Disease Resistance Genes: Application to the Rx2 Locus in Potato. Plant J. 2000, 21, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Spassova, M.I.; Prins, T.W.; Folkertsma, R.T.; Klein-Lankhorst, R.M.; Hille, J.; Goldbach, R.W.; Prins, M. The Tomato Gene Sw5 Is a Member of the Coiled Coil, Nucleotide Binding, Leucine-Rich Repeat Class of Plant Resistance Genes and Confers Resistance to TSWV in Tobacco. Mol. Breed. 2001, 7, 151–161. [Google Scholar] [CrossRef]
- Hallwass, M.; de Oliveira, A.S.; de Campos Dianese, E.; Lohuis, D.; Boiteux, L.S.; Inoue-Nagata, A.K.; Resende, R.O.; Kormelink, R. The Tomato Spotted Wilt Virus Cell-to-Cell Movement Protein (NSM) Triggers a Hypersensitive Response in Sw-5-Containing Resistant Tomato Lines and in Nicotiana benthamiana Transformed with the Functional Sw-5b Resistance Gene Copy. Mol. Plant Pathol. 2014, 15, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Margaria, P.; Ciuffo, M.; Pacifico, D.; Turina, M. Evidence That the Nonstructural Protein of Tomato Spotted Wilt Virus Is the Avirulence Determinant in the Interaction with Resistant Pepper Carrying the TSW Gene. Mol. Plant Microbe Interact. 2007, 20, 547–558. [Google Scholar] [CrossRef] [PubMed]
- de Ronde, D.; Pasquier, A.; Ying, S.; Butterbach, P.; Lohuis, D.; Kormelink, R. Analysis of Tomato Spotted Wilt Virus NSs Protein Indicates the Importance of the N-Terminal Domain for Avirulence and RNA Silencing Suppression. Mol. Plant Pathol. 2014, 15, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Qu, F.; Morris, T.J. HRT Gene Function Requires Interaction between a NAC Protein and Viral Capsid Protein to Confer Resistance to Turnip Crinkle Virus. Plant Cell 2000, 12, 1917–1925. [Google Scholar] [CrossRef]
- Cooley, M.B.; Pathirana, S.; Wu, H.J.; Kachroo, P.; Klessig, D.F. Members of the Arabidopsis HRT/RPP8 Family of Resistance Genes Confer Resistance to Both Viral and Oomycete Pathogens. Plant Cell 2000, 12, 663–676. [Google Scholar] [CrossRef]
- Lanfermeijer, F.C.; Dijkhuis, J.; Sturre, M.J.G.; de Haan, P.; Hille, J. Cloning and Characterization of the Durable Tomato Mosaic Virus Resistance Gene Tm-22 from Lycopersicon esculentum. Plant Mol. Biol. 2003, 52, 1039–1051. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hayes, A.J.; Jeong, S.C.; Gore, M.A.; Yu, Y.G.; Buss, G.R.; Tolin, S.A.; Saghai Maroof, M.A. Recombination within a Nucleotide-Binding-Site/Leucine-Rich-Repeat Gene Cluster Produces New Variants Conditioning Resistance to Soybean Mosaic Virus in Soybeans. Genetics 2004, 166, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.N.; Ye, X.R.; Molina, J.; Roose, M.L.; Mirkov, T.E. Sequence Analysis of a 282-Kilobase Region Surrounding the Citrus Tristeza Virus Resistance Gene (Ctv) Locus in Poncirus trifoliata L. Raf. Plant Physiol. 2003, 131, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Maiti, S.; Paul, S.; Pal, A. Isolation, Characterization, and Structure Analysis of a Non-TIR-NBS-LRR Encoding Candidate Gene from MYMIV-Resistant Vigna Mungo. Mol. Biotechnol. 2012, 52, 217–233. [Google Scholar] [CrossRef]
- Moury, B.; Verdin, E. Viruses of Pepper Crops in the Mediterranean Basin: A Remarkable Stasis. Adv. Virus Res. 2012, 84, 127–162. [Google Scholar] [CrossRef] [PubMed]
- Tomita, R.; Sekine, K.T.; Mizumoto, H.; Sakamoto, M.; Murai, J.; Kiba, A.; Hikichi, Y.; Suzuki, K.; Kobayashi, K. Genetic Basis for the Hierarchical Interaction between Tobamovirus Spp. and L Resistance Gene Alleles from Different Pepper Species. Mol. Plant Microbe Interact. 2011, 24, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Tomita, R.; Murai, J.; Miura, Y.; Ishihara, H.; Liu, S.; Kubotera, Y.; Honda, A.; Hatta, R.; Kuroda, T.; Hamada, H.; et al. Fine Mapping and DNA Fiber FISH Analysis Locates the Tobamovirus Resistance Gene L3 of Capsicum chinense in a 400-Kb Region of R-like Genes Cluster Embedded in Highly Repetitive Sequences. Theor. Appl. Genet. 2008, 117, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Sawada, H.; Takeuchi, S.; Hamada, H.; Kiba, A.; Matsumoto, M.; Hikichi, Y. A New Tobamovirus-Resistance Gene, L1a, of Sweet Pepper (Capsicum annuum L.). J. Jpn. Soc. Hortic. Sci. 2004, 73, 552–557. [Google Scholar] [CrossRef]
- Anagnostou, K.; Jahn, M.; Perl-Treves, R. Inheritance and Linkage Analysis of Resistance to Zucchini Yellow Mosaic Virus, Watermelon Mosaic Virus, Papaya Ringspot Virus and Powdery Mildew in Melon. Euphytica 2000, 116, 265–270. [Google Scholar] [CrossRef]
- Vidal, S.; Cabrera, H.; Andersson, R.A.; Fredriksson, A.; Valkonen, J.P.T. Potato Gene Y-1 Is an N Gene Homolog That Confers Cell Death upon Infection with Potato Virus Y. Mol. Plant Microbe Interact. 2002, 15, 717–727. [Google Scholar] [CrossRef]
- Ma, J.; Hou, X.; Xiao, D.; Qi, L.; Wang, F.; Sun, F.; Wang, Q. Cloning and Characterization of the BcTuR3 Gene Related to Resistance to Turnip Mosaic Virus (TuMV) from Non-Heading Chinese Cabbage. Plant Mol. Biol. Rep. 2010, 28, 588–596. [Google Scholar] [CrossRef]
- Bijaisoradat, M.; Kuhn, C.W. Nature of Resistance in Soybean to Cowpea Chlorotic Mottle Virus. Phytopathology 1985, 75, 351. [Google Scholar] [CrossRef]
- Ishibashi, K.; Masuda, K.; Naito, S.; Meshi, T.; Ishikawa, M. An Inhibitor of Viral RNA Replication Is Encoded by a Plant Resistance Gene. Proc. Natl. Acad. Sci. USA 2007, 104, 13833–13838. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Kezuka, Y.; Kobayashi, C.; Kato, M.; Inoue, T.; Nonaka, T.; Ishikawa, M.; Matsumura, H.; Katoh, E. Structural Basis for the Recognition-Evasion Arms Race between Tomato Mosaic Virus and the Resistance Gene Tm-1. Proc. Natl. Acad. Sci. USA 2014, 111, E3486–E3495. [Google Scholar] [CrossRef] [PubMed]
- Grube, R.C.; Wintermantel, W.M.; Hand, P.; Aburomia, R.; Pink, D.A.C.; Ryder, E.J. Genetic Analysis and Mapping of Resistance to Lettuce Dieback: A Soilborne Disease Caused by Tombusviruses. Theor. Appl. Genet. 2005, 110, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Gou, X.; He, K.; Xi, D.; Du, J.; Lin, H.; Li, J. BAK1 and BKK1 in Arabidopsis thaliana Confer Reduced Susceptibility to Turnip Crinkle Virus. Eur. J. Plant Pathol. 2010, 127, 149–156. [Google Scholar] [CrossRef]
- Julie KØrner, C.; Klauser, D.; Niehl, A.; Domínguez-Ferreras, A.; Chinchilla, D.; Boller, T.; Heinlein, M.; Hann, D.R. The Immunity Regulator BAK1 Contributes to Resistance against Diverse RNA Viruses. Mol. Plant Microbe Interact. 2013, 26, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.M.; Santos, A.A.; Pires, S.R.; Rocha, C.S.; Saraiva, D.I.; Machado, J.P.B.; Mattos, E.C.; Fietto, L.G.; Fontes, E.P.B. Regulated Nuclear Trafficking of RpL10A Mediated by NIK1 Represents a Defense Strategy of Plant Cells against Virus. PLoS Pathog. 2008, 4, e1000247. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).