Abstract
In an effort to provide an overview of the biophysical approaches used to study G-protein-coupled receptors, we chose to consider the adenosine A2A receptor as a model, as it is widely reported in the literature to explore the way GPCRs are studied nowadays. After a brief introduction of the receptor, we gathered descriptions of the various tools used to investigate the pharmacology and structure of the A2A receptor. We began by describing the key developments which have led to successful studies of GPCRs including the cloning, expression and purification of A2A, and the subsequent characterizations including quality control, binding and functional studies that have been necessary for the further understanding of the receptor. Then, we reviewed the reconstitution of A2A into nanodiscs as well as the use of this biological material in structural mass spectrometry, NMR, calorimetry and various other approaches to gain not only information about the structure and function of A2A, but also the dynamics of the receptor and the tools necessary to pursue such investigations. The body of techniques presented herein are applicable to all GPCRs amenable to purification.
1. Foreword: Biophysical Approaches to Answer Which Questions?
During the last two to three decades, as researchers in the GPCR area, we saw a profound evolution in the study of these membrane-bound proteins. It started with the description of whole genomes and the systematic comparison of sequences that revealed the remarkable conserved topology of this exceptionally large family of proteins. This in turn shed light on the necessity to explore the subtle molecular mechanisms and determinants that were responsible for recognizing such a variety of ligands, to understand how these receptors were selectively activated and how they are involved in so many cellular processes. Initially, the aim was to better understand the pharmacology of these proteins, and thus it became key to be able to study these entities in solution, or in reconstituted and controlled environments. The endpoint is to identify and develop directly interacting molecules that could become novel or “better” drugs. This includes molecules which provide safer pharmacological profiles, improved efficacy, in addition to greater precision, efficiency, and potency. Our scientific community therefore invested intensive efforts to produce and isolate these GPCRs and, hence, to assay and develop integrated analytical techniques to enable the characterization of GPCR structures, functions and dynamics, and further understand the way these receptors are modulated. To make such a characterization, a large diversity of biophysical principles have been exploited, including light, fluorescence, mass and NMR spectroscopies, surface plasmon resonance (SPR), X-ray diffraction and electron microscopy, among others. All of these techniques have been successfully applied to the biophysical dissection of the molecular functioning of GPCRs.
Like many other protein families in similar studies, a small number of prototypical receptors served the development of these GPCR-tailored methodologies, among which is the adenosine A2A receptor (A2A) which we chose to focus on in the present review. This receptor from the Class A GPCR family is indeed of continuing interest as a major therapeutic target in a panel of inflammatory, neurodegenerative and cancer disease types. More importantly, unlike a large proportion of GPCRs that are still recalcitrant to molecular investigations, this receptor rapidly appeared favorably throughout the literature to be produced and isolated in various experimental formats that prove compatible with a remarkable diversity.
The present review summarizes the main biophysical techniques that brought important information on GPCRs through the data obtained on the adenosine A2A receptor. In the first section, we will remind the reader of the main facts known about this receptor. The next sections extensively review the preparative conditions leading to the isolation of the receptor and the many biophysical approaches that contributed to our understanding of the structure and function of GPCRs in general.
2. Background and Introduction
Adenosine is a natural compound formed by an adenine attached to a ribose via a β-N9-glycosidic bond. It occurs widely in nature in the form of many diverse derivatives from multiple sources. All those molecules in which adenosine is a central chemical play important roles in living organisms. It is a part of one of the four building blocks of which the nucleic acids are made and its phosphorylated derivatives are key elements of the energetic system in all living systems. Finally, it exists also under a cyclic form, cAMP, which is a clear element of signaling pathways via the regulation of its key synthesis enzyme, adenylate cyclase. Adenosine is recognized essentially by four GPCRs: A1, A2A, A2B and A3. A full survey of these receptor characteristics can be found in Fredholm et al. [1]. The human A2A receptor is a 412-amino-acid-long protein encoded by the gene located at chromosome position 22q11.23. The A2A receptor was initially isolated and purified as early as 1974 [2]. Although membrane protein purification under an active form is not trivial, this receptor was widely used because its cloning and expression from human sources were reported early [3]. It was possible to express it in E. coli [4] and its characterization began from there.
A complete review of A2A pharmacology exists and is a reference for the matters concerning this receptor [5]. In brief, the A2A expression pattern concerns mainly the brain [6], peripheral T cells [7] and platelets as well as various organs such as the colon, heart and kidneys [5].
The main coupling routes of the receptors are in human platelets, cAMP [8], while coupling to ERK1/2 has also been reported on the human cloned receptor [9]. Early works clearly indicated that A2A is not linked to intracellular calcium modifications, at least upon reference to its agonist effects [10]. In 1995, interplays were suspected with other GPCRs, leading to the A2A-dependent reduction in dopamine receptor activation [10]. A very complex picture of the co-regulations of ion channels was drawn later (see, for example, Ikeuchi et al. [11] and Wang and Zhou [12]).
The key step for receptor studies is the binding assay on membranes, whether derived from native tissues or from hosts in which the receptor has been cloned. This is also dependent on the availability of specific radioligands, such as the agonists [3H]-CGS21680 [13] or [3H]-NECA [14], generally preferred over tritiated adenosine [15], and the antagonist [3H]-XAC (xanthine amine congener) [16], with pKd in the range of 8. New ligands were added, with pKd in the range of 9, such as [3H]-ZM241385 [17].
A complete survey of the clinical status of A2A receptor ligands as therapeutic drugs can be found in Ijzermann et al. [5]. More than 50 different drug trials were still active in 2022 for targeting A2A as treatments for cancer (Imaradenant), pain (Spongosine), Parkinson’s disease (BIIB014, Taminadenant) and sickle cell disease (Regadenoson), for which the infusion of the A2A agonist reduces the production of IFN-γ and enhances the production of IL-13 and CD39 [18].
3. Mass Production of A2A Receptors
3.1. Expression Systems
Owing to their finely regulated functions, GPCRs are naturally scarce in native biological sources. Accordingly, robust recombinant expression systems are needed to produce the milligram amounts required for their biophysical analyses. These systems should not only fit yield criteria but should also offer the most adapted environment for producing properly folded and functional GPCRs. Several systems have been developed with the aim of producing large amounts of the receptor properly folded with various degrees of sophistication and success rates. These include cell-free approaches [19,20], bacterial [21] or yeast [22,23,24] microorganisms, insect and animal cells using viral-infection or DNA-transfection strategies [25], photoreceptor cells of transgenic animals [26,27] and even whole organisms such as silkworm (Bombyx mori) larvae [28].
Among the many GPCRs investigated in these studies, A2A is remarkably one of the very few that has been successfully expressed at relatively high levels in nearly all recombinant systems. It was indeed produced in milligram amounts in E. coli, either unfolded in inclusion bodies [29], or as ligand-binding active receptors in the bacterial membranes [30,31,32]. A2A was also efficiently produced in the membranes of yeast cells, mainly Saccharomyces cerevisiae [33,34,35] and Pichia pastoris [22,36,37,38,39]. These hosts combine the advantages of handling the simplicity of microorganisms with the complex cellular machineries of eukaryotes. S. cerevisiae notably offers a large panel of molecular tools and engineering possibilities that were exploited for functional studies of many GPCRs [40] as well as for extensive screenings of stabilizing mutations and preparative conditions, including for A2A [41,42,43,44,45,46]. Concerning P. pastoris, its peculiar methylotrophic metabolism, the tightly regulatable expression conditions and the exceptional cell densities it can afford, make this yeast well suited for expressing membrane proteins for structural studies. Accordingly, it has not only allowed two of the high-resolution structures of A2A to be solved by X-ray crystallography [38,47], but it has also delivered various isotopically labelled samples for most of the NMR-based studies conducted on A2A [47,48,49,50].
Though technically more demanding and more time-consuming than microbes, animal cell systems offer a closer native-like environment and have therefore been extensively used as recombinant hosts for A2A production. Most notably, recombinant baculovirus infecting either Trichoplusia ni (Tni) or Spodoptera frugiperda (Sf9) insect cells became the reference systems for generating crystallography-grade GPCR samples over the years. For A2A in particular, the majority of high resolution structures referenced to date were obtained from various forms of the receptor produced with this insect cell expression system [51,52,53,54,55]. Concerning mammalian cells, low expression levels and cost issues initially precluded the direct use of the high-quality receptors they produce for further biophysical analyses. Until recently, CHO or HEK 293 cell lines were thus mainly used as orthogonal recombinant systems for evaluating and validating the pharmacological properties of engineered or mutated A2A receptors that were designed for structural studies with other systems [39,51,56,57,58,59]. With the development of HEK 293-derived cell lines adapted to suspension and high cell density culturing in optimized media, combined with their transient transfection with tetracycline-inducible plasmids, GPCRs produced in these quasi-homologous host cells, including A2A, have now become accessible to thorough biophysical and structural studies [60,61].
3.2. Molecular Constructs
The design of A2A sequences that were expressed in these different systems are representative of the strategies classically employed for the large-scale production and purification of membrane proteins. The A2A coding sequence, sometimes optimized to fit the codon preference of the host cell [38], has been N- or C-terminally fused to various tags (most commonly His6, His8, His10 and FLAG) to enable detection and purification schemes, or to reporter proteins (GFP, RFP) [33,41,62,63] to monitor expression and purification optimization. These supplementary sequences were often separated from the A2A coding sequence by a cleavage sequence (TEV or 3C protease sequence) [22,62,64,65]. Finally, signal peptide sequences adapted to the expression host (from influenza hemagglutinin HA in animal cells, or from α-Factor in yeast systems) were almost always added upstream of the fusion constructs to improve protein translocation in the ER and target the plasma membrane [22,33,51,66,67].
More specific modifications were further implemented in the A2A sequence when homogeneity and stability issues had to be addressed, notably for crystallography or cryo-electron microscopy (cryo-EM) structural studies. As further described in Section 8 below, besides a systematic truncation of the last C-terminal amino acids 317 to 412 [38,51,52], two complementary or combined sequence engineering strategies were employed to stabilize A2A. The first one consists of replacing flexible domains of the receptor with more structured and stable soluble proteins, the most common being the phage T4 lysozyme (T4L) [51,58,68] and the apo-cytochrome b562RIL (BRIL) [54,56,61,65,69,70]; however, other less-used proteins such as flavodoxin, xylanase and rubredoxin [71], as well as custom-made de-novo-designed α-helical proteins have been successfully employed [43]. The second approach relied on the introduction of thermostabilizing point mutations, identified via directed-evolution [31,45] or structure-guided [55] approaches, allowing crystallographic studies [52,72] and other biochemical analyses [42,59,62]. An additional point mutation aiming at suppressing a glycosylation site at position N154 to improve receptor homogeneity was often introduced [38,47,52,72]. However, similar constructs bearing no such mutation also led to homogenous and crystallography grade samples [51,56,68]. Finally, for some specific NMR studies, several amino acids exposed at the intracellular surface were replaced by cysteine residues for targeted labeling with 19F chemical probes [48,73,74,75].
4. Extraction and Purification
When the expression conditions are met, the next challenge before biophysical investigations of isolated membrane proteins consists of extracting and purifying the expected amounts of pure, homogenous, and correctly folded receptors. As for a large majority of GPCRs and other membrane proteins studied in solution [25,76], A2A was mainly solubilized using the mild detergent n-dodecyl-β-D-maltopyranoside (DDM), independently of the expression system used [30,38,62,63,67,77], or with the more recent lauryl maltose neopentyl glycol (LMNG) [47,48,50,61]. The physicochemical properties of these two surfactants are particularly suited for destabilizing biological membranes, while providing the receptors a reasonably stable hydrophobic environment to maintain their folding and function throughout their preparation and the planned analyses [78].
In the case of thermostabilized A2A sequences, the higher stability conferred by the selected point mutations allowed the solubilization of the receptors with n-decyl-β-D-maltopyranoside (DM), a harsher maltoside-based detergent. Compared to DDM, DM forms smaller micelles and has a higher critical micelle concentration, which was better adapted to further detergent exchanges and crystallographic studies using vapor diffusion crystallization conditions [52,69,72,79,80].
For the non-thermostabilized A2A constructs, the stability issues during solubilization and purification were addressed via the addition of specific ligands, locking the receptors into a limited number of conformations. Theophylline was the most frequently used [30,38,47,48,49,50,51,55,56,58,71,79,80,81,82], even if other ligands such as caffeine [69], DPCPX [77] and ZM241385 [61,63] appeared to also be useful. The stabilizing effect of these ligands [56,83,84,85] was very often combined with the supplementation of cholesteryl hemisuccinate (CHS), a cholesterol derivative that modulates the shape of detergent micelles in a pseudo-bicelle architecture. The addition of CHS into the purification buffers has been known to be beneficial for improving the stability of numerous solubilized GPCRs [86], including A2A [30,56,63,87]. Finally, other agents commonly applied to minimize the aggregation of membrane proteins in solution were also often added, such as glycerol which likely acts as an amphiphilic interface between hydrophobic domains and the polar solvent [88], and NaCl which contributes to maintaining proteins soluble in aqueous solutions through a “salting in” effect. Incidentally, sodium ions are also considered an allosteric modulator of A2A [89]. Iodoacetamide can also be used to prevent protein aggregation by intermolecular disulfide-bond formation.
The strategies used for purifying detergent-solubilized A2A were essentially based on affinity chromatography (AC) approaches. Since nearly all recombinant receptors were fused to a poly-histidine tag, an immobilized-metal affinity chromatography was most often performed as a first step, either on Co2+ [50,51,63,67,90,91] or Ni2+-grafted resins [30,34,52,77]. Some procedures also involved agarose resins covalently bound to XAC, a non-specific, high A2A affinity antagonist at adenosine receptors [92], which were implemented as ligand-affinity columns in a first [30] or second purification step [49,50,93,94]. Recently, AC procedures using anti-tag antibodies coupled to agarose resins, such as M1 anti-FLAG [61], or anti-Rho-1D4 [59] antibodies, were also reported.
Purification of A2A has often been finalized by a polishing step on size exclusion chromatography (SEC) Superdex 200 columns, allowing the recovery of homogenous populations of monomeric receptors, and the elimination of additional impurities and specific compounds eluted from the previous AC step(s).
5. Isolation in Lipid Nanoparticles
As detailed in the previous section, a variety of conditions has allowed the isolation of A2A in detergent, in a sufficiently stable and convenient environment for a panel of biochemical and biophysical studies. However, the hydrophobic environment brought by detergents is chemically and physically distinct from the complexity of biological membranes, which poses certain limitations and issues for a number of investigations. Examples of such issues include the interference of detergents with certain analytical methods, insufficient stability for prolonged periods of analyses, absence of critical lipid and/or protein interactants for a receptor’s structure and function [95]. To overcome these difficulties, several methods have been developed over the years to reinsert detergent-isolated GPCRs in a more stable and more native lipid environment, including lipid-detergent bicelles, lipid bilayer nanoparticles or unilamellar lipid vesicles (ULVs) [95,96,97].
Bicelles are discoidal particles made of a planar phospholipid bilayer surrounded by a belt of detergent. While the reconstitution of several purified GPCRs in bicelles proved useful for their structural and functional investigations [98,99,100], none of these approaches have been reported so far for the characterization of A2A.
Concerning the implementation of ULVs, or liposomes, the situation is just slightly better documented. In an NMR study on A2A purified in detergent, liposome reconstitution was used as a quality control material to evaluate the functionality of the highly deuterated receptor for its capacity to activate its heterotrimeric G protein [67]. In another study, an original liposome synthesis method called INSYRT (in situ lipid synthesis for protein reconstitution technology) was recently developed and validated with A2A [101]. The receptor was solubilized and purified with DDM thioester analogs that served as precursors for the synthesis of phospholipids via a native chemical ligation (NCL) reaction. The resulting proteoliposomes were further characterized to contain A2A, showing ligand-binding pharmacological properties similar to those described in cellular membranes, and to the receptor reconstituted in the so-called nanodiscs (NDs).
Nanodiscs are nanometric discoidal lipid bilayers stabilized by two copies of a membrane scaffold proteins (MSP) deriving from the Apo-A1 lipoprotein [102]. Since it has been demonstrated that likely every detergent-solubilized membrane protein can self-assemble in such particles and maintain its structure and activity in a stable environment [103], NDs have been the prevailing source of material for the biophysical investigation of numerous GPCRs reconstituted in a lipid bilayer in recent times [104]. This is particularly the case for A2A for which the use of ND particles generated led to the discovery of a number of findings on receptor–ligand interactions and the structural dynamics of the receptor. The different approaches used included surface plasmon resonance (SPR) [62,105], mass spectrometry (MS) [106], miniaturized weak affinity chromatography (nanoWAC) [77], single molecule fluorescence microscopy [75] and ligand- or receptor-oriented NMR techniques [49,50,74,75,107]. In these studies, detergent-purified A2A was successfully assembled with different versions of the engineered MSP, either MSP1ΔH5 [50,74], MSP1D1 [62,80,106,107], MSP1E3D1 [49,77] or the less widespread Apolipoprotein A1 from zebrafish, Zap1 [105], which essentially differ in their molecular size and in the disk size of the generated NDs (from approximately 9 to 12 nm in diameter [103]). Some of these MSP were genetically [62] or chemically [77] modified to allow the grafting of the resulting NDs via their MSP on different supports for SPR and WAC analyses. The lipid composition essentially consisted of POPC:POPG mixtures incubated with MSP and A2A at various molecular ratios, in some cases replaced by or supplemented with other components used to evaluate their effect on the ND assembly efficiency [80] and/or on the receptor function [49,50,74].
Amphiphilic polymers, mainly amphipols, have also been largely used for isolating GPCRs in a membrane-mimicking environment [108,109]. While these particles have proven to be highly stable and allowed many investigations in detergent-free solutions, they are essentially generated after the reconstitution of proteins already purified in detergents. Thus, they may lack some lipids important for the conformational dynamic and function of the receptors. To date, no publication has reported on A2A reconstitution in amphipols.
Alternative amphiphilic co-polymers made of styrene and maleic acid (SMAs) are being increasingly used for the characterization of isolated membrane proteins, notably GPCRs, in lipid bilayers [97,110]. Unlike the standard amphipols, which are acrylate-based polymers grafted with octylamine, SMA compounds hold the ability, under defined conditions, to destabilize biological membranes and spontaneously form SMA lipid disc particles (SMALPs). Those containing the protein of interest can then be directly purified. A2A has been successfully isolated in SMALPs generated from the membranes of Pichia pastoris and HEK recombinant cells, and was characterized for its ligand-binding capacity and its increased stability compared to the receptor isolated in detergent [39]. Further studies reported by the same group investigated A2A SMALPs from Pichia pastoris membranes to evaluate ligand-binding events via fluorescence correlation spectroscopy [111], as well as some ligand-induced conformational changes in A2A via fluorescence spectroscopy [112].
6. Quality Controls on Isolated Receptors
Once the protein material is isolated and purified, a panel of biochemical and/or biophysical tests are generally performed to assess the integrity of the receptor including the purity, homogeneity, stability and the activity of the isolated receptors. These are all important or even critical criteria to be met before initiating the functional and structural characterizations that are described in sections below.
Receptor integrity and purity are almost always assessed via classical SDS-PAGE analyses, sometimes complemented with A2A-specific immunodetection. The evaluation of sample homogeneity is often necessary to identify the presence of potential residual contaminant, but also to appreciate the oligomerization states of the receptor. This is essentially assessed through standard size-exclusion chromatography (SEC) analyses [35,51,71,94,106], or by discriminating the particle size either in the final sample via dynamic light scattering (DLS) [80] or during its purification via SEC-MALS [35,43,91,94]. Negative-staining EM imaging may also provide a useful qualitative complementary approach [61,77,91].
Native mass spectrometry (native MS) is often used as a first-line technique after the protein purification of soluble proteins to address oligomerization issues. This technique aims at transferring and detecting intact non-covalent interactions in the gas phase of a mass spectrometer. While native MS has been established for the analysis of soluble proteins and protein complexes, it is however challenging for membrane protein analyses [113,114], but has been successfully applied to A2A. Figure 1, panel B, represents a native mass spectrum of A2A solubilized in DDM, highlighting several species corresponding to different populations of receptors, including full-length, degraded or modified forms.
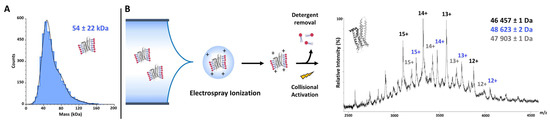
Figure 1.
A2A mass measurement using mass photometry and native MS. Mass Photometry (A) is an emerging technique for fast mass and oligomeric state determination of proteins. Analysis of A2A showed a good homogeneity and enabled the determination of a mass of 54 ± 22 kDa (molecular mass distribution histogram of A2A—the solid blue line represents major species fit with a Gaussian function). The quite large mass distribution obtained can be explained by the accuracy of the technique itself but also by the presence of potentially remaining micelles around the protein which may average out the overall mass obtained and thus increase the mass deviation. Native MS (B) is a powerful analytical tool for the study of membrane proteins embedded in detergent. The technique, preserving non-covalent interactions in the gas phase for mass analysis, enabled the visualization of the A2A protein with nano electrospray. Several populations (48.6 kDa, the expected mass of A2A; 46.4 and 47.9 kDa, truncated forms of the protein) have been identified after optimization of the instrumental parameters. These finely tuned and harsh conditions are essential for efficient detergent removal.
The implementation of mass photometry, a very recent label-free technique that quantifies molecular weights of biomolecules at the single-molecule level by interferometric detection of scattered light [115], provides an additional way for the characterization of membrane protein sample homogeneity [116]. In this context, we evaluated the technique for the analyses of A2A samples (Figure 1, panel A). The obtained results highlight the complementarity of mass photometry and native MS for the characterization of membrane proteins. Requiring low amounts of starting material (approximately 100 ng protein) and no additional buffer exchange steps during the preparation, mass photometry can be used to obtain information on the sample studied with less optimization than native MS, providing heterogeneity and oligomerization information very quickly but with considerably less accuracy.
As mentioned earlier, the stability of the isolated protein is often critical for a number of analyses, in particular for crystallography studies or when the analytical methods require extended periods of measurement. For A2A, this issue has been mostly addressed via methods assessing the thermal stability of the isolated protein. Those were mainly fluorescence-based techniques, either involving the thiol-reactive chemical probe N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide that covalently binds to internal cysteines and fluoresces upon thermal denaturation [55,56,71,83], consisting of a fluorescence size-exclusion chromatography (FSEC) evaluation that requires the presence of a fluorescent protein (i.e., GFP) fused to the receptor [62], or label-free tryptophan fluorescence monitoring upon unfolding [117]. A2A thermal stability was also assessed via the analyses of migration profiles of the protein on acrylamide gel after denaturation, either revealed via Western blotting [84] or via clear-native PAGE [63]. As an alternative to thermal denaturation, an isothermal chemical denaturation method using guanidium chloride has been successfully applied to assess the stability of A2A when isolated in the presence of various ligands [90].
Perhaps the most important indication of the quality of a protein sample relies on its capacity to fulfill its function. A variety of techniques have been applied to assess the activity of A2A via its ability either to bind specific ligands or to activate its associated heterotrimeric G-protein partner upon agonist stimulation. These methods include radioligand binding assays in filtration [36,77,91] or scintillation proximity assay (SPA) formats [62,105], GTPγS activity assays [67], fluorescence analyses [111], and a panel of SPR-, MS- or NMR-based approaches, which are described in more detail in the following sections.
7. Functionality of the Isolated A2A
7.1. Ligand Binding
7.1.1. Radioactive Ligand Binding
As far as specific radiolabeled compounds are available, for decades, this direct evaluation of receptor–ligand interactions has been the preferred technique to study and define the pharmacology of GPCRs. It is a highly sensitive approach based on the counting of the radiation emitted by the radioactive compounds bound to their receptors, from which are deduced precise affinity parameters either in saturation or competition modes. In the most classical methods, receptor-bound and free radioligands are separated via filter-based techniques that are particularly well suited to evaluating GPCRs in membrane fraction samples. When GPCRs are solubilized and isolated, standard filter-based assays are generally not compatible with the small size of the particles to be analyzed, so alternative separation methods were needed. For A2A, these mainly relied on the use of mini-spin gel filtration columns [30,31,32,36,39,49,118,119], on the filter-based separation of the receptor–ligand complexes either precipitated with polyethylene glycol (PEG) [77,91] or immobilized on agarose beads [34,83], or by immobilization on functionalized scintillating bead supports designed for SPA [47,62,77,79,105]. These studies were essentially conducted to characterize and validate the pharmacology of A2A isolated either in detergent micelles [30,31,36,39,49,91], in NDs [49,62,77,105], in SMALPs [39] or in liposomes [101]. Interestingly, one study conducted on different A2A samples generated from P. pastoris or HEK cells, either in membrane fractions or isolated in detergent or in SMA particles [39], determined comparable affinity values for several agonist and antagonist ligands. In addition, this work demonstrated the increased stability overtime of SMALP or ND samples over detergent micelles on the ligand-binding activity of the isolated receptor [39].
Radioactive ligand binding has also been the key method for evaluating the thermal stability of solubilized A2A, either for screening thermostabilizing mutations [31,52] or for evaluating the stabilizing effect of various detergents and additives [119].
7.1.2. Surface Plasmon Resonance
Several alternative biophysical methods, such as SPR, have emerged to determine binding affinities and kinetics, as they are essential for drug discovery and development [120]. Indeed, the kinetics of drug–receptor complex formation, and in particular the residence time of the ligand [121], is a crucial element that affects the functional properties of a ligand and its pharmacological profile [122,123,124].
SPR is a label-free technique that allows the quantitative analysis of molecular binding. One binding partner is immobilized on a biosensor surface and the other is introduced into a microfluidic system and flows in solution over the surface. The binding causes a small change in the refractive index at the biosensor surface due to the accumulated adsorbed mass. Continuous monitoring of this signal allows the determination of the kinetic parameters such as the association rate constant (kon), the dissociation rate constant (koff) and the affinity (Kd) of the interaction [125].
SPR has become the method of choice for studying ligand binding to various GPCRs [126,127,128,129]. This technique requires small amounts of membrane proteins in a native or similar environment. The challenges are to maintain high levels of binding activity of the immobilized receptor and to detect the binding of low-molecular-weight ligands, as this technology is based on mass changes [130,131,132]. This has been successfully applied to assess the ligand-binding affinities of a number of GPCRs. A variety of A2A/ligand interactions have been studied using SPR over the past decades. Findings from these studies illustrates how SPR technology supports structure-based ligand discovery and fragment screening and contributes to the better understanding of the molecular aspects of signaling. Detergent-solubilized A2A, either non modified or thermostabilized (A2A-StaR), has been widely investigated and generally extracted from insect cells membranes. Several A2A-StaR (purified or as crude solubilized extracts) were captured through their C-terminal 10His tag to a Ni-loaded NTA sensor surface (carboxymethylated dextran functionalized with nitrilotriacetic acid) [90,131,133,134,135]. Small antagonists (molecular weight ranging from 285 to 345 Da) were shown to interact with a Kd ranging from 0.3 to 160 nM in accordance with the classical competition radioligand binding assay [133]. Importantly, from the screening point of view, SPR is able to detect even weaker GPCR/compound complexes (Kd > 1 µM) [131]. The groundwork was laid for the cross-screening of a panel of antagonists with mutated A2A-StaR. The resulting matrix of binding kinetics data was then combined with molecular modelling and docking data from a refined map of the A2A binding site, thereby facilitating the structure-based drug design [135,136]. Affinity and kinetic rate constants measured at different temperatures allow the calculation of enthalpic and entropic energy components as well as the analysis of kinetics of the transition state, constituting the thermodynamic signature of a target-ligand complex [137,138,139]. Calculating the kinetic transition state components for six ligand–A2A–StaR complexes and combining them with supervised molecular dynamics (MD) and metadynamics approaches clarified the crucial role of receptor and antagonist solvation/desolvation in the transition states of entropic nature. Thermodynamics combined with kinetic analyses allows a better understanding of the details of the molecular interactions. They could also help rationalize the drug design since kinetics better correlates with EC50 than thermodynamic equilibrium affinity constant values for some GPCR–ligand complexes [138].
Solubilized GPCR micelles are unstable and tend to denature and/or aggregate over time. As detailed previously in Section 5, several strategies have been developed to reconstitute GPCRs in a lipid bilayer, such as proteoliposomes or high-density lipoproteins known as NDs, to provide more stable conditions for the receptor compared to detergents. The kinetic characterization, using SPR, of the binding of nine adenosine antagonists to a A2A-StaR reconstituted in NDs correlates well with those of the same receptor in a membrane environment obtained from the conventional radioligand binding assay. NDs likely correspond to a more native environment than detergent and may avoid the problems of ligand partitioning in micelles [105]. Bocquet et al. conducted different binding assays (SPA and SPR) with the A2A-StaR in membranes, in NDs and in detergent micelles [62]. Different set-ups were evaluated in the SPR assay, with NDs being immobilized via different tags fused either on the receptor or on the MSP of the NDs, without impacting the binding kinetics of small antagonists (337 to 428 Da). In addition, the receptor was shown to be more stable in NDs than in detergent micelles.
The SPR approach has also been validated for fragment screening investigations on A2A (see Section 9). The success of such experiments requires a high density of immobilized GPCRs with high binding capacity, as well as careful experimental design, to reduce false positive results since high concentrations of fragments are tested and tend to non-specifically bind to many targets (reviewed in Shepherd et al. [132] and Coyle and Walser [140]). This approach was first developed for a solubilized A2A-StaR with a panel of 70 fragments derived from xanthine, with molecular weights ranging from 136 to 194 Da. [134]. SPR was also used to validate putative ligands of a A2A-StaR issued from a mass spectrometry fragment screening [141]. In another study, the native, non-stabilized A2A receptor, as well as three other receptor subtypes from the same family (A1, A2B and A3), were investigated via SPR. [142].
As GPCRs interact with different intracellular partners of the downstream signaling cascade, SPR was also used to study some of these molecular interactions. It was notably shown that the binding of solubilized A2A to immobilized Gα subunit proteins was affected by GDP analogues and that the C-terminus of A2A was necessary for its high-affinity association to this G-protein subunit [143]. In another study on A2A in NDs bound to a full or a partial agonist, the binding characteristics for the heterotrimeric Gαβγ proteins were determined using SPR [50]. No differences in affinity and kinetics were observed, but complementary structural analyses using NMR suggested a model based on conformational changes in the receptor to explain their different functional output. Other aspects of A2A functioning have been explored via SPR, such as the impact of mutations in a cholesterol consensus motif on G-protein binding [59].
7.1.3. Calorimetry
Isothermal titration calorimetry (ITC) is a label-free biophysical technique that can measure kinetics and the thermodynamic parameters of binding, such as affinity, stoichiometry, enthalpy and entropy between macromolecules (e.g., between two proteins) or between a macromolecule and its small molecule ligands [144,145]. Applied to GPCRs, ITC allows for a deeper understanding of the binding interactions. Indeed, it reflects the subtle structural and dynamic changes between bound and unbound forms, identifying the most important regions of the binding interface [146,147] as well as receptor–ligand specificity, cell surface expression, endocytosis and recycling [148]. A2A is of particular interest because it possesses a long and flexible C-terminal region which favors interactions with several proteins, including G-protein-coupled receptor kinases, β-arrestins, α-actinins, calmodulin (CaM), ubiquitin-specific protease 4 (USP4), and neuronal calcium sensor protein 1 (NCS-1) [149,150,151,152,153,154,155,156]. The affinity and stoichiometry of A2A binding to two of its partners, CaM and α-actinin 1, have been recently studied together with their regulation by calcium using ITC [157,158]. In this study, human CaM and two A2A C-terminal regions of different lengths (A2A-ctL comprising 293-412 amino acids and A2A-ctS comprising 321-412 amino acids). CaM was titrated into the protein in the presence of calcium with or without the addition of EDTA. In the absence of EDTA, the binding between A2A and CaM displayed strong affinity (Kd = 97.9 nM) with a 1:1 stoichiometry. In the presence of calcium, the binding of CaM to A2A-ctL led to an exothermic reaction with a favorable enthalpy, but unfavorable entropy, possibly indicating conformational changes. No binding occurs in the presence of the calcium chelator, EDTA. The binding site of CaM on the A2A-ctL region was further identified using the A2A-ctS construct lacking the amino acids 293 to 320. CaM binding in the presence of calcium led to small exothermic peaks signalling a loss of affinity. These results, confirmed by NMR spectroscopy, identified the residues 293-310 as the binding epitope of CaM onto A2A [158].
Similarly, mutating arginine residues in this region abolished CaM binding. These data are in agreement with previously published results [151]. α-Actinin binding to A2A was suspected to be implicated in the internalization of the receptor after agonist activation. To characterize the domain responsible for the interaction between the C-terminal domains of A2A and α-actinin 1, α-actinin1 wild-type constructs limited to the calcium binding domain (CABD), the rod domain or the CABD-rod domains were used. In the absence of calcium, the titration of the rod-CABD domains led to exothermic peaks and to a saturable binding curve. The affinity of rod-CABD domains to α-actinin was found to be in the micromolar range (Kd = 39 µM), which was weaker than for A2A-ctL with CaM. Together, the results of the ITC experiments suggest that the A2A C-terminus binds to the distal part of α-actinin 1 C-terminal domain, either in the CABD or at the subsequent C-terminus, a region shown to interact with CaM, and in fact it has been demonstrated that these proteins compete when binding to A2A. It is worth highlighting that ITC is a classical approach to identify GPCR partners and should be systematically used to validate and characterize receptor partners at the molecular level.
7.1.4. Receptor Fluorescent Labeling
Many different techniques have been applied to illuminate the molecular bases of transmembrane signaling through GPCRs in general, and through A2A, in particular. Among them, fluorescence-based studies appear to be particularly adapted to investigate the functioning of these receptors, as they can be used in many different environments ranging from native and recombinant cell lines to isolated receptors in membrane-mimicking systems [159]. These techniques nevertheless require GPCRs to be specifically labelled with adapted fluorophores. For the last two decades, several approaches have been developed to label receptors for fluorescence-based applications. Besides the fluorescent ligands described later (Section 9), these include protein and peptide tags compatible with measurements in live cells (e.g., GFP, YFP, SnapTag and FlAsH-tetracysteine tag) [160]. Several of these labelling strategies have been applied to the A2A. For instance, the concomitant insertion of a Cyan Fluorescent Protein and the FlAsH-tetracysteine tag into human A2A was combined with fluorescence resonance energy transfer (FRET) measurements to provide a description of the activation process of this receptor in cellular model systems [161]. More recently, a covalent ligand was used to couple a fluorophore to A2A [162]. Although this latter approach was used in a cell-based system, it could be extended to isolated receptors as well. Besides cell-based studies, strategies to label purified receptors in detergent micelles or nanodiscs have also been developed. In many cases, the insertion of the fluorescent probe primarily involves the labelling of unique reactive cysteines with a fluorescent probe through a maleimide-based reaction. This strategy was applied to recombinant A2A to successfully introduce probes for NMR- and fluorescence-based applications [75]. For the latter, cyanine 3 was attached to the purified receptor expressed in P. pastoris and assembled into nanodiscs [163]. This opened the way to single-molecule fluorescence experiments aimed at describing the conformational transitions of A2A. In the same way, a purified mutant receptor containing single reactive cysteines was labeled with an environment-sensitive dye, BODIPY-FL, for fluorescence quenching experiments aimed at delineating the dynamics of the TM6 domain of A2A [164]. In addition, orthogonal labeling strategies were also described in this same study [164]. Specifically, a double-cysteine mutant with reactive Cys residues on TM4 and TM6 was used for double-labeling with a fluorescence donor (Alexa Fluor 488) and acceptor (Alexa Fluor 647), paving the way for single molecule-FRET analyses of receptor conformational fluctuations. In addition to these classical protein engineering approaches, alternative labeling strategies have been described for A2A. In particular, a modified aminoacyl-tRNA-synthetase and a suppressor tRNA were used in a cell-free system to incorporate an unnatural amino acid that was further coupled to a fluorescent dye [165]. Together, these studies provide evidence that the isolated adenosine A2A receptor in different membrane-mimicking environments can be efficiently labeled with fluorophores, providing a convenient system to describe its energy ensemble and the modulations by the environment using fluorescence-based techniques.
7.1.5. Ligand-Based NMR
Ligand-observed NMR experiments are widely used to measure and analyze small molecule binding to soluble and membrane proteins such as GPCRs [166]. No protein labeling is required, and experiments can be performed in micelles, membranes and nanodiscs. Ligand-observed NMR experiments are suitable for moderate- to low-affinity ligands (10 nM to mM affinities). This is of particular interest as far as screening approaches are concerned, as the net results of most of the screening campaigns are poor affinity binders. Nevertheless, the binding of higher-affinity ligands can be monitored through competition experiments, using the NMR binding signal of a moderate-affinity compound as a reference signal. Saturation transfer difference (STD), nuclear Overhauser effect spectroscopy (NOESY) experiments or 19F NMR on labeled ligands have been reported on A2A [74,91,107]). Experiments are typically compared to a control experiment where the receptor is either missing or blocked by a high-affinity ligand. The NMR experiments allow the identification of novel ligands and of their binding pockets when a reference compound at a known binding site is available. Furthermore, they may reveal other binding pockets on A2A. For example, the binding of adenosine in an allosteric pocket was observed through STD experiments recorded with A2A bound to ZM241358 [91].
Three-dimensional structures of A2A–ligand complexes can be calculated from NOESY experiments called INPHARMA (Interligand Noes for PHArmacophore Mapping) [107,167]. For example, the structure of the complex between A2A and 3-pyrrolidin-1-ylquinoxalin-2-amine (PQA) was obtained from the interligand NOESY observed between PQA and the antagonist compound ZM241358 [107].
Finally, ligand-observed NMR can be used to investigate allosteric mechanisms, as positive and negative allosteric modulators (PAMs and NAMs), respectively, increasing or decreasing the binding of a ligand and thus its NMR binding signal. Recently, Huang et al. [74] investigated the binding of cholesterol on A2A using 19F-labeled cholesterol analogs. They observed the broadening of the NMR resonances as well as a 0.5 ppm up-field shift for the 19F-labeled cholesterol bound to A2A in nanodiscs compared to empty nanodiscs. However, the NMR spectrum was not modified upon the addition of an inverse agonist, full agonist, or a G-protein mimetic, suggesting that the cholesterol interactions with A2A are transient and unspecific, and that the effect of cholesterol is due to changes in the membrane bilayer properties [74]. The investigation of the allosteric modulation of A2A by small molecules using ligand-observed NMR represents a rapid and robust approach that should be further used to explore the biology of A2A.
7.1.6. Native Mass Spectrometry
As described above (see Section 6), native MS is often used to check the quality of a purified protein and to address oligomerization issues. In addition, native MS is also a valuable method to evaluate molecular interactions between the protein and various partners. Yen et al. [168] used native MS to reveal that A2A in its apo state still binds to endogenous phosphatidylserines (PS) and phosphatidylinositol (PI). Native MS was next applied for the affinity ranking of such compounds, demonstrating that PI(4,5)P2 is a better binder compared to other phospholipids during binding assays [168]. Even more challenging is the native MS detection of a heterotrimeric G-protein complex involving the monomeric A2A, the Gα subunit and a nanobody to stabilize the entire assembly [168]. The lipids bound to this receptor complex were measured after quadrupole selection and collisional activation of the whole complex. To obtain such high-quality, high-resolution native MS data of GPCRs, all steps of the native MS workflow must be carefully optimized, from sample injection using nano-emitters, to MS methods. Furthermore, an optimization of collisional conditions is required to allow efficient desolvation and membrane-mimic removal while maintaining an intact membrane for protein analysis. Comparisons between ligand-binding native MS results obtained from classical NH4OAc buffer and NaCl-based buffers on A2A and glucagon receptors in the presence of different ligands demonstrated that ligand-binding interactions are better conserved on GPCRs using NaCl-based buffers than NH4OAc buffers [169]. For A2A, the authors could differentiate agonist from antagonist compounds based on the capacity of the receptor to retain sodium adducts from the storage buffer, thus paving the way for the use of native MS screenings on GPCRs for the detection and characterization of non-covalent interactions under more relevant physiological conditions.
7.2. Activity
7.2.1. G-Protein Coupling and Arrestin Recruitment
Advanced Resonance Energy Transfer (BRET/FRET) strategies have been developed during the last decade to monitor receptor–effector coupling and selectivity in signaling. These approaches have been extensively reviewed [170,171] and have been mostly used with recombinant cell systems expressing receptor and effectors labeled with fluorophores and/or fused to fluorescent/bioluminescent partners or fragments. In the case of isolated receptors, apart from the purification of stable signaling complexes for subsequent structural studies, in vitro assays monitoring the coupling to signaling partners are also currently used to assess the functional properties of the isolated GPCRs, in parallel to ligand-binding assays. Initially, these assays focused on the allosteric stabilization of the active state of the receptor and its impact on an agonist’s affinity and/or receptor stability. Such assays were used, for instance, to reveal the coupling of the isolated adenosine receptor to engineered mini-G proteins [53]. Specifically, mini-Gs were shown to increase the affinity of the purified A2A in detergents for NECA, and significantly increased the stability of the receptor in the presence of the agonist. Besides these assays, experimental designs have been implemented to monitor receptor-catalyzed G-protein activation through GTP turnover (Figure 2). This process can be visualized by monitoring the binding of a radiolabeled [35S]GTPγS [172] or the fluorescent analog of GTP (Bodipy-GTPγS) [173] to the Gα-subunit of the purified Gαβγ heterotrimer. [35S]GTPγS binding was used to decipher the G-protein coupling to A2A in Sf9 membranes [174]. Besides membrane fractions, labeled GTPγS binding assays can also be applied to GPCRs isolated in detergent micelles [175,176] or nanodiscs [177,178]. More recently, a GTP turnover assay has been devised that monitors the amount of free GTP after receptor-catalyzed G-protein activation. Initially described for other GPCRs such as the α2-adrenergic [179] or ghrelin [180] receptors, this assay has been recently used to reveal the allosteric modulation of the adenosine A2A receptor in nanodiscs by cholesterol [74].
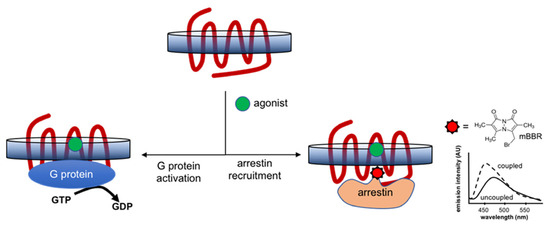
Figure 2.
Schematic representation of the in vitro assays for G-protein activation and arrestin recruitment. These assays monitor receptor-catalyzed G-protein activation through GDP-to-GTP exchange by measuring either the binding of a labeled GTP analog or the depletion of GTP in the solution. To study arrestin recruitment, the most popular assay relies on monitoring the changes in the emission profile of a fluorophore, monobromobimane (mBBR), attached to the finger loop of recombinant arrestin.
Regarding arrestin coupling, although some alternative assays have been reported that are based, for instance, on the use of radiolabeled arrestin [181], a popular in vitro assay with isolated GPCRs relies on the use of recombinant arrestin labeled on its finger loop with a specific fluorophore, monobromobimane (mBBR) (Figure 2). This fluorophore is attached to recombinant arrestin through a unique reactive cysteine [182]. The emission properties (intensity, maximum wavelength) of mBBR vary upon coupling to the receptor, and as such, allow for visualization of the GPCR–arrestin interaction. Initially developed with rhodopsin and visual arrestins [183], this assay has since been extended to other GPCRs such as the vasopressin, β2-adrenergic or ghrelin receptors in detergents, amphipols and nanodiscs [178,184,185,186]. To the best of our knowledge, this assay has not been used with purified A2A so far but could be directly applied to this receptor isolated in membrane-mimicking systems, allowing, for instance, screening for particular ligands that would selectively trigger arrestin recruitment.
It is clear from the last years in GPCR biology that our understanding of receptor signaling pathways has been completely changed by two concepts: (i) the description of the signaling bias of receptors [187,188,189] and (ii) the numerous proteins that have been found associated with receptors, such as the MT1 receptor [190]. For the first point, isolated systems are necessary to better understand how the activation of receptors are controlled. For the second point, an understanding of the activation routes of a receptor will be clearly defined from isolated systems for which potential coupling between purified receptors and their protein binding partners will be studied in the defined environment of nanodiscs.
7.2.2. Functionality of Isolated Receptors: Activity of Receptor Dimers and Oligomers
Even though a monomeric A2A receptor is sufficient to promote G-protein activation or β-arrestin recruitment, this receptor, like many other GPCRs, has been shown to form dimers and larger oligomers [191,192,193,194]. GPCR oligomers offer additional inter-receptor communications to potentially regulate their own functional properties. A2A receptor was shown to associate with itself [195], forming A2A homodimers, but also with other receptors to form heterodimers or larger oligomers. In particular, A2A could form heterodimers with the other adenosine receptors A1, A2B and A3 [196,197], as well as with the cannabinoid CB1 receptor [198], the dopamine D2 and D3 receptors [199,200,201,202] or the glutamate mGlu5 receptor [203]. Most of these studies were performed in heterologous systems, mainly using BRET or FRET approaches. Among the heterodimers, A2A/D2 has gained a lot of attention. First, its existence is well documented in vivo both in animal models [204] and in the post-mortem human brain [205]. Second, A2A/D2 heterodimers are located in the GABAergic striatopallidal neurons, a region critical in locomotor activity and central in the physiopathology of Parkinson’s disease [206]. Interestingly, A2A/D2 heteromers were shown to operate a reciprocal negative functional crosstalk and the interaction between the two receptors is reduced along the development of the pathology [206,207]. Therefore, A2A/D2 constitutes a major therapeutic target for the treatment of Parkinson’s disease.
While cellular and in vivo studies provide a general picture of the functional crosstalk within GPCR oligomers [208] (whether homodimers, heterodimers or heteromers) including A2A/D2, a precise characterization of the pharmacological, conformational or signaling specificity of each oligomer relative to the monomers or other oligomers is very difficult to reach in these complex systems where several oligomeric species are likely to co-exist. Therefore, isolated systems where a single receptor combination is present, either monomer or oligomer, bring invaluable information to elucidate the molecular, pharmacological and signaling signatures. Additionally, in isolated systems, specific mutations (e.g., non-binding or non-coupling mutations) can easily be introduced to a given protomer. They can analyze the overall effect of the absence of the oligomer function. To date, a single publication reported A2A oligomers in isolated systems [94]. However, other GPCR oligomers have been analyzed in such systems illustrating the power of the method but also highlighting the technical challenges to overcome [197,209].
As indicated above, purified receptors can be obtained either in detergent micelles, or reconstituted in proteoliposomes or in lipid nanodiscs. Depending on the protein concentration, isolation procedures and purification steps, samples containing the desired receptor composition are obtained. For example, the neurotensin NTS1 receptor is mostly monomeric in detergent at low protein concentrations and becomes dimeric at higher concentrations [210]. Leukotriene BLT2 receptor has been shown to be present in a mixture of monomers and dimers with each species successfully isolated after HPLC fractionation [176]. For nanodiscs, the incorporation of monomeric or dimeric GPCRs can be achieved by acting on the receptor-to-scaffold protein ratio as reported for rhodopsin [211]. Alternatively, the reconstitution of the purified glutamate mGlu2 receptor into nanodiscs has been shown to lead to a mixture of discs containing monomers or dimers that can be isolated by HPLC fractionation [212]. Upon reconstitution, the dimer can assemble in parallel or anti-parallel orientations, with only the former having a physiological relevance. A general assessment of the relative orientation can be conducted using fluorescently labeled protomers either at the N- or C-terminus. Indeed, FRET would occur only at a short distance, i.e., when both fluorophores are located at the same side of the receptor relative to the lipids. Thus, if FRET is detected between probes inserted at the N- and C-terminus, anti-parallel dimers are present [176]. To overcome this phenomenon and to purify only the parallel dimers from a mixture, an original strategy has been developed using the stable CaM binding simultaneously to two similar sequences derived from the CaM binding domain from Petunia glutamate decarboxylase (PGD). Accordingly, after the fusion of the 26-residue PGD-tag to the N-terminus of the receptor of interest, only parallel dimers can be purified using a CaM binding step followed by SEC. This method was efficiently used on lipid nanodiscs containing LTB1 receptor dimers or ghrelin GHSR receptor dimers [213]. When considering heterodimers, their formation in detergents or in nanodiscs gives a mixture of several species such as monomers, homodimers and heterodimers. In order to select the heterodimers from this mixture, double tag purification strategies have been developed. Indeed, the fusion of a different tag to each receptor of interest, followed by two successive purification steps, each using one of the tags, lead to the purification of only dimers presenting one of each tag [214]. This method has been used to prepare BLT1 receptor dimers in detergent where one of the subunits only is mutated in its binding site for coupling with a G protein [214]. It is worth noting that some receptors assemble in complexes larger than dimers. Therefore, nanodisc size is not compatible with the insertions of these large complexes. To overcome this limitation, proteoliposomes are an alternative that have been used to reconstitute a heterotetramer composed of two GHSR and two D2 receptors [215]. The heterotetramers were further isolated from the proteoliposomes using SMALPs.
Following the isolation of receptor complexes, functional and conformational experiments can be performed similarity to those carried out on isolated monomeric receptors, highlighting the specific properties of the different complexes. For example, in a rhodopsin dimer, only one of the protomers is in a high-affinity state upon recruitment of G protein while the dimers have a lower affinity for β-arrestin 1 than the monomer [211,216]. The analysis of the GHSR/D2 heteromer highlights that heteromerization directly modulated D2-mediated Gi protein activation. More specifically, such heterocomplexes act directly on the conformation of the Gαi subunit [215]. Applying similar strategies to the analysis of the A2A homodimer or A2A/D2 heteromer analysis would bring important information on the precise mechanism underlying the function of the complexes and would lead to new hypotheses of how to more precisely regulate these pathophysiologically relevant heteromers.
8. Structure of A2A
The early evidence for a necessary transfer of information from structure to function has been reviewed by Piirainen et al. [217] and clearly showed an early path for the rationalization of GPCR structure determination to gain an understanding of the function of the A2A receptor and GPCRs in general. The central role of biophysical techniques in the determination of the structure and function of GPCRs has been nicely reviewed by Langelaan et al. [218]. Furthermore, artificial intelligence approaches, such as with AlphaFold, are powerful tools to predict three-dimensional protein structures and protein–protein interactions. This technology can be accessed for the prediction of the human A2A three-dimensional structure, in addition to almost all other GPCRs [219,220].
8.1. Structural Mass Spectrometry Approaches
Among the large panel of biophysical methods available, structural MS techniques have become a valuable tool for either ligand screening or for more detailed structural characterization of purified GPCRs, including the A2A receptor. The structural MS toolbox encompasses a series of techniques, such as native MS for stoichiometry and oligomerization determination, ion mobility MS for the assessment of global conformation, and labeling methods such as hydrogen/deuterium exchange MS (HDX-MS) and chemical cross-linking to investigate interacting regions and/or conformational changes [221,222].
While native MS and mass photometry have great capacity for the characterization of oligomeric state or binding events (see Section 6 and Section 7 above), both techniques are not adapted to provide either amino acid resolution information on binding regions, or the dynamics of assembly or disassembly [113,114]. In recent years, HDX-MS has become a very powerful labeling method for membrane protein analysis, adapted to tackle conformational issues related to ligand binding and even allostery [223]. A typical HDX-MS experiment includes the incubation of a protein of interest in two or more states (apo- or holo-states for example) in deuterated buffer at different time points to allow hydrogens from backbone amides to exchange with deuterium. The exchange reaction is then quenched by switching to acidic pH and low temperature followed by online digestion with acid-functional proteases. The generated peptides are then separated on a reversed phase-HPLC system and detected using MS. Dynamics and binding-site mapping are then extracted by comparing apo- and holo-states. While HDX-MS is becoming a well-established technique for soluble proteins with standardized workflows and protocols [224], the study of membrane proteins remains technically challenging. Thus, extensive work on the optimization of the experimental conditions (composition of buffers, choice of digestion protease, etc.) must be carried out systematically. For instance, nepenthesin protease seems to be particularly suited for membrane protein studies compared to the standard pepsin one. Similarly, the addition of urea as denaturant in quench buffers seems to be beneficial when working with membrane proteins [225]. Other dedicated workflows and methodological developments have been set up to prevent the early deterioration of the instruments caused by the large amounts of detergents and phospholipids in the samples, including an additional off-line cleaning of the LC columns, or the use of zirconium beads for lipid depletion, etc. [226,227].
Figure 3 presents a typical HDX-MS workflow used to monitor the binding of the antagonist ZM241385 on A2A and the induced conformational changes. Online digestion of the protein allowed to cover 70.8% of the sequence with 79 identified peptides and a redundancy of 2.7, due to the use of nepenthesin II protease, was reported as more efficient on GPCRs. Experiments carried out on apo A2A showed a low deuterium incorporation on transmembrane domains, reflecting their known low accessibility to the solvent. Among these domains, some peptides (20-31; 58-62; 86-93; 134-141; 234-241; 245-253; 271-280) exhibited a dramatically low deuterium incorporation typically less than 10%, which is consistent with the known structure of the protein. Upon ZM241385 binding, a significant part of the protein was protected meaning that A2A incorporates less deuterium in the presence of the ligand. Several peptides exhibited statistically significant differences in deuterium incorporation (p-value of 0.01). Peptides 168-175, 245-250 and 245-253 in the known ligand binding region [38] (F168, N253 and I274 residues) presented a H/D protection higher than 5% confirming the ZM241385 binding site. The significant protection of additional peptides 59-65, 276-288, 277-282 and 277-288 supports the known ligand fixation, as involved in the following residues (A63, H278), forming a cleft to accommodate the phenol group of ZM241385, as well as for the end of the transmembrane domain 7 (TM7) represented by residues 281 to 298 [72]. Other regions showing differences in incorporation (TM3, TM4 and the loop in between) may reflect the induced-structural rearrangement of the receptor upon ligand binding. This example highlights the ability of HDX-MS experiments to measure membrane protein conformational dynamics in their native state.
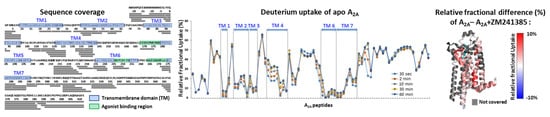
Figure 3.
Structural characterization of A2A upon ligand ZM241385 binding with HDX-MS. HDX-MS identified peptides shown on the A2A sequence; transmembrane domains are highlighted in blue and the agonist ligand-binding region in green (left). The relative deuterium uptake of A2A of each identified peptide at each deuteration times are plotted, with peptides in the transmembrane regions framed in blue (middle). Relative fractional uptake differences between A2A and A2A with the ligand ZM241385 are represented on the protein structure (PDB code: 4EIY [56]) at a 30 min deuteration time; regions not covered are colored in grey (right).
In addition to continuous labeling studies (deuteration time points from a few seconds to hours), HDX-MS can also be used in a time-resolved manner to monitor sequential conformational changes in the protein of interest over time, using very short “pulsed” deuteration time [223] either in the apo state to report on folding and aggregation [228] or in an holo state to report on protein–protein interaction. To our knowledge, this pulsed HDX-MS approach has been reported only once on A2A by Du et al. [229], in order to better understand why GPCR/G-protein complexes are stabilized in a GDP free-state, how this nucleotide is released and to lay the structural basis of coupling specificity between G-protein subtypes and GPCRs. Interestingly, the authors used hydroxyl radical protein footprinting with mass spectrometry [230,231], which aims at looking at protein conformational changes after the irreversible labeling of amino acid side chains by hydroxyl radicals (generated by radiolysis or photolysis reactions), as an orthogonal technique to detect very early events (from a few milliseconds to seconds).
Among the emerging structural MS methods, chemical cross-linking followed by MS analysis is still rarely used for in vitro membrane protein analysis. The first step consists of the covalent binding of a cross-linker agent to the side chain of amino acids, followed by the quenching of the reaction and the reduction, alkylation and digestion step. Then, peptides including cross-linked dipeptides are separated and analyzed using LC-MS/MS and finally identified via different software due to the mass increment due to the cross-linking reagent [232,233]. The A2A receptor was used as a model to assess which chemical cross-linkers are suitable for the characterization of GPCRs by cross-linking mass spectrometry (XL-MS) [234]. Similar to HDX-MS, all steps of the XL-MS workflow have to be adapted to membrane protein analyses, especially the choice of the cross-linker reagent and the cross-linking conditions, enzymatic digestion and sample clean up, along with adapted LC-MS methods and data treatment. By comparing the MS-identified cross-links to the structurally compatible cross-links from available GPCR structures, Jones et al. concluded arginine-arginine cross-linker and lysine-arginine are best adapted for XL-MS analyses of GPCRs and are highly complementary to the existing classical cross-linkers present in the toolbox of specialists [235]. Altogether, structural MS methods have great potential to complement more classical high-resolution structural biology techniques, and are becoming more and more popular.
8.2. X-ray Crystallography of GPCRs: Uncovering Conserved Activation Mechanisms Using A2A as a Model
GPCRs are highly dynamic and become particularly unstable during the purification process once extracted from the cell membrane. As a consequence, crystallization of GPCRs remains a challenging task. To overcome such difficulties, protein engineering strategies have been developed to reduce the flexibility and to increase the stability of these membrane receptors. A2A was one of the first receptors for which the high-resolution structure was solved using X-ray crystallography. The T4 Lysozyme (T4L) fusion partner, initially established for resolving the high-resolution crystal structure of the beta-2 adrenergic receptor (β2AR) [236], was later applied to solve the first structure of A2A [51]. The T4L increases the hydrophilic surface of detergent-solubilized GPCRs to promote crystal contacts, and reduces the flexibility associated with the TM6 helix [236]. Subsequent developments led to the application of the BRIL fusion partner into intracellular loop 3 to solve the A2A crystal structure at 1.8 Å [56]. Another key engineering strategy to overcome the instability of GPCRs in detergents is by introducing point mutations in the 7TM domain to select thermostabilized ligand-bound conformations. The development of conformational thermostabilization of the A2A [31,237] has been successfully applied for a range of co-crystallized agonist- and antagonist-bound structures [52,72].
Ciancetta pointed out in 2019 that [238] “A2A represents to date (2019) one of the rare Class A GPCR that has been solved in the inactive (R) [79], active-intermediate (R*) [68], and fully active state coupled to an engineered Gs protein α subunit (R * G) [53,64]”. Most of these structures have been obtained using X-ray crystallography. During the last 3 years, more structures were solved in similar configurations for other Class A GPCRs.
The structural determination of multiple A2A conformational states has contributed greatly to the understanding of Class A GPCR activation mechanisms [38,51,52,53,68,72,239]. The antagonist-binding pocket has been thoroughly characterized by the co-crystallization of A2A bound to ZM241385 (Figure 4A) and to various xanthine-containing compounds including caffeine and XAC, using either thermostabilization or fusion protein strategies [72]. Later developments combining the antagonist-thermostabilized point mutations with the BRIL fusion partner provided the highly utilized construct known as A2A-StaR2-bRIL [69,79,240]. Multiple structures of antagonist-bound states also revealed the sodium allosteric modulation of A2A [56,58]. The allosteric sodium ion-binding site is nested in a set of highly conserved residues, located below the orthosteric binding pocket, and bound to the highly conserved Asp2.50 [56,58].
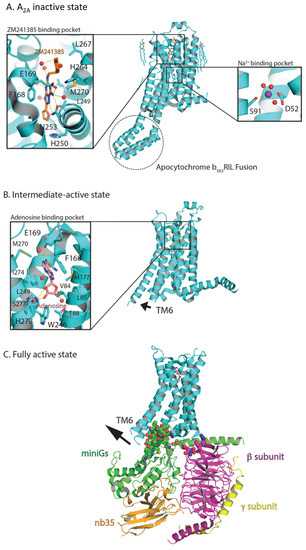
Figure 4.
Three-dimensional structures of the human adenosine A2A receptor. The A2A receptor is bound to the antagonist ZM241385 (A; PDB: 4EIY [56]), agonist adenosine (B; PDB: 2YDO [52]), NECA and mini-Gs protein (C; PDB: 6GDG [241]). These figures represent the inactive, intermediate-active, and fully active states, respectively. Polar contacts are represented as yellow dashed lines, with the sodium ion depicted as a purple sphere and water molecules as red spheres.
Agonist-binding modes of UK-432097 [68], NECA and adenosine (Figure 4B) to the thermostabilized A2A highlight the molecular interaction of the ribose moiety of all agonists with Ser277 and His278 and contraction of the receptor binding site [52]. Agonist binding also initiates a slight outward movement of TM6 that defines an intermediate-active conformation [52,57,58,68,242]. The crystal structure of the A2A bound to an engineered Gs protein (known as mini-Gs) illustrated the large outward movement of TM6 that is required for the mini-Gs to bind, in addition to slight movements of TM5 and TM7 and rotamer shifts of several amino acids near the G-protein binding site (Figure 4C) [53].
This was further validated by solving the cryo-EM structure of A2A bound to the heterotrimeric mini-Gs (Figure 4C) [241]. However, comparisons between the NECA binding pocket in the intermediate [52] and fully activated conformation [53] show identical binding modes. Together, these studies provided an overview of the conserved activation mechanisms of Class A GPCRs [243].
High-resolution crystal structures of A2A have also led to the advancement of structural-based drug design (SBDD) [70,135]. Recent reports have outlined chemical modifications of an adenosine scaffold, commonly attributed to A2A agonist activity, which led to the alteration of the compound from an agonist to an antagonist [244]. Although the ribose moiety is accepted to be important for A2A agonist activity, recent structural and functional data have demonstrated the binding mode of a non-riboside partial agonist [242]. Although allosteric regulation of the human A2A remains relatively unexplored, differences between antagonist-bound conformations have suggested the possibility of an allosteric binding site within the 7TM binding pocket [245]. Taken together, these studies highlight future avenues for the design of selective orthosteric A2A modulators and the possibility of development of allosteric ligands.
Technical advances have been made in macromolecular crystallography for speeding up the workflow for screening, data collection and processing, proving to be essential for the progress of GPCR structural biology. The infrastructure developments of synchrotron microfocus beamlines have also been key for solving A2A structures and GPCRs in general. Moreover, the use X-ray free-electron lasers (XFELs) and lipid cubic phase (LCP) injectors for sample delivery has allowed the structural determination of A2A at room temperature, opening new possibilities for investigating the highly dynamic nature of GPCRs [78,81,246,247,248,249,250,251]. Currently, X-ray crystallography has become one of the more accessible approaches for the structural determination of GPCRs. Understanding the dynamic activation mechanism of A2A remains of strong interest. Accordingly, the use of integrated approaches, such as cryo-EM, will uncover more details of A2A in complex with its signaling partners in the future.
8.3. Cryo-Electron Microscopy
Cryo-EM is a powerful technique for solving structures of challenging and dynamic proteins that are difficult to obtain using X-ray crystallography. While there is a debate on the respective virtues of cryo-EM versus X-ray crystallography for the determination of protein structures, particularly those of membrane-bound and integral membrane proteins [252,253,254], it is clear to us that the complementarity between those approaches opens avenues for a better description of membrane-bound proteins in the future, as reviewed by others [255,256].
A key advantage of cryo-EM is that it does not require the preparation of protein crystals, rather it allows for the imaging of single isolated particles in isomorphous ice prepared directly from purified samples. Conversely, the application of the cryo-EM technology for proteins or complexes smaller than 60 kDa remains challenging due to the inherent low signal-to-noise ratio (or low contrast) that limits the accurate alignment of projections during image analyses. Accordingly, the relatively small size of a ligand-bound Class A GPCR such as A2A (approximately 35–45 kDa) is typically not adapted to cryo-EM studies but is suitable for X-ray crystallography which remains the preferred technique for the determination of GPCR structure in the inactive state [257].
This size limitation has been recently circumvented by increasing the molecular weight of GPCRs isolated in protein complexes. Most remarkably, advances of the near-atomic resolution of activated GPCRs in complex with G-protein binding partners have been achievable with the use of cryo-EM [258]. Cryo-EM has since been successfully employed to solve the structure of an engineered A2A thioredoxin fusion protein (TrxA-A2A) in complex with the mini-Gs protein [241]. A recent study made use of an anti-BRIL Fab in complex with the A2A-BRIL to increase the molecular weight of A2A and to facilitate cryo-EM data processing [61].
One method recently developed, known as microcrystal electron diffraction (MicroED), combines crystallization in LCP with the acquisition of cryo-EM for structural determination. Here, a beam of electrons is transmitted through protein microcrystals grown in LCP, rather than X-rays by conventional crystallography. A2A is currently the only GPCR for which this method has been successfully employed [259]. Future work ought to apply the use of cryo-EM to investigate the structures of the purified A2A in complex with signaling partners such as the full-length Gs heterotrimer and other intracellular signaling partners.
8.4. NMR Approaches
At a molecular level, GPCR signaling can be described through the concept of free energy landscape [260] where orthosteric ligands, intracellular effectors and allosteric modulators influence both the population and lifetime of various inactive, intermediate and active states. High-resolution NMR spectroscopy is a powerful tool to investigate these complex energy landscapes. While NMR can be used to obtain de novo structures of proteins, in the case of large complexes such as GPCRs associated with various surfactants or lipids, most of the NMR studies that are conducted in solution concern conformational landscapes or interactions, with considerations sometimes relating to chemical exchanges. They rely advantageously on the plethora of atomic scale structures obtained from X-ray crystallography and cryo-EM to highlight the coexistence of several inactive and active sub-states, including lowly populated states, but also, to a much lesser extent, the detection of some chemical exchanges. Along with the β2AR, A2A is one of the GPCRs most studied with NMR. These studies are based on different isotopic labeling strategies and/or expression systems.
An appreciable amount of NMR studies of A2A is based on fluorine (19F) NMR [48,50,73,93]. This nucleus can be easily incorporated in proteins via biosynthetic pathways or chemical reactions and has the advantage of displaying a large chemical shift dispersion compared to other nuclei that are traditionally detected to study biomolecules [261,262]. This partly counterbalances the fast 19F transversal relaxation due to the large chemical shift anisotropy, which becomes the major source of linewidth in high magnetic fields. As GPCRs are usually expressed in eukaryotic expression systems, 19F labeling via chemical reactions is generally preferred. However, in this case, only solvent-exposed residues can be labeled due to the accessibility for the chemical reaction to occur and, consequently, all these studies are based on single extra-membrane 19F probes. Following such a strategy, the conformational landscape of the receptor was observed using one-dimensional (1D) 19F NMR at the three following positions that are all located at the extracellular tip of TM helices: V229C6.31 (TM helix VI, TM6) [48,93], L225C6.27 (TM6) and A289C7.54 (TM7) [73]. 1D 19F spectra in these locations display a pattern characteristic of a complex conformational ensemble, composed of several inactive and active sub-states in equilibrium. Their population strongly depends on the presence or absence of ligands and their associated pharmacological properties, as well as the presence or absence of effectors. This illustrates that signal transduction by A2A relies on a dynamic equilibrium of several conformations that can be detected through a fluorine probe. Interestingly, several coexisting active states, whose populations are related to specific ligands were observed, fitting with the concept of partial agonism, i.e., when a GPCR is activated to a sub-maximal level.
By investigating these three positions of the 19F probe, different distributions of populations in the conformational ensemble could be identified in the apo or holo (e.g., in the presence of the full agonist NECA) states. Between V229C6.31 [48,93], or L225C6.27 [73], and A289C7.54 [73], this may reflect a different response to drug efficacy at the intracellular ends of TM6 and TM7 [73]. In contrast, it is most puzzling that the distribution of populations is not the same between V229C6.31 and L225C6.27, as these two residues are localized very closely with each other at the extracellular tip of TM6 [48,73]. Moreover, these two studies were performed in the same detergent solution. This discrepancy could be associated with different possible effects of aromatic ring current fields in the vicinity of these two 19F NMR probes that could partially respond differently to conformational changes [263].
Interestingly, looking at the conformational ensemble of V229C6.31 in the apo state or in the presence of NECA of A2A embedded in a lipid nanodiscs, the 1D 19F NMR spectra are very different from what has been previously observed in detergent [50] (Figure 5), highlighting the importance of studying the energy landscape of membrane proteins in a native-like lipid bilayer environment, even if using very small nanodiscs where the receptor is surrounded by just one layer of lipid (lipoprotein ΔH5 [264]), such as those used in Huang et al. [50]. Furthermore, this study is also remarkable as they could additionally depict the conformational ensemble of V229C6.31 in the presence of a heterotrimeric G protein, revealing a G-protein-bound intermediate and distinct nucleotide-free state in the presence of either a partial or a full agonist. This study further suggests a scaffolding role for the Gβγ-associated subunits, facilitating the allosteric transmission through the transmembrane part of the receptor from the orthosteric ligand site to the intra-cellular side in a ligand-dependent manner [50].
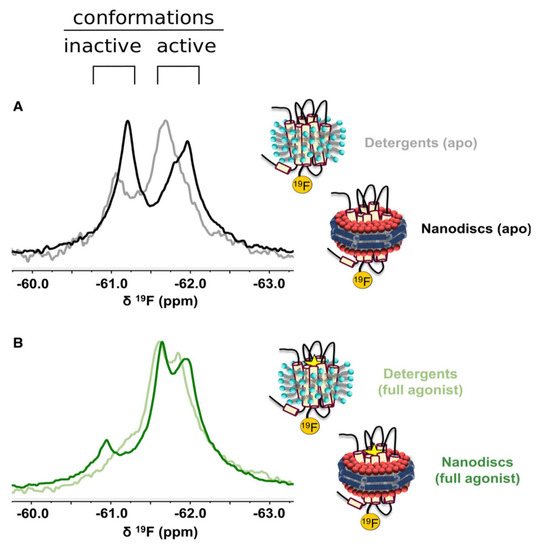
Figure 5.
A2A exhibits a different distribution of states and slower exchange dynamics in a lipid bilayer environment than in detergent micelles (A,B). Comparison of the 19F NMR spectra of apo (A) or agonist-bound (B) A2A reconstituted in either LMNG micelles or phospholipid nanodiscs (legend and figure adapted from Huang et al. [50] with automatic permission from Elsevier).
A common feature identified in all of these studies is that most of the sub-states sampled by the 19F probe suggest a slow chemical exchange at the 19F chemical shift timescale (millisecond timescale), under the assumption that some chemical exchanges are present between the conformations revealed by the fluorine probe. A strong indication that such chemical exchanges are indeed present is given by the observed variations in either chemical shifts and/or signal linewidths in association with measurements of the transversal relaxation time (T2) upon the addition of various ligands and/or a heterotrimeric G protein or a Gα mimetic peptide. 1D 19F NMR can also be expanded with a second 19F frequency axe to directly visualized conformational exchanges by 2D [19F-19F] exchange spectroscopy (EXSY) spectra [73] (Figure 6).
Figure 6.
Conformational exchange in the A2A complex with the full agonist NECA observed using 2D exchange spectroscopy. A contour plot is shown of a 2D [19F, 19F]-EXSY spectrum collected at 280 °K with a mixing time of 100 ms. The diagonal peak positions of the active-like sub-states P1, P2, and P4 are labeled, and a dashed box indicates cross-peaks observed between conformations P1 and P2. This spectrum indicates the existence of a low-energy barrier for interconversion between sub-states P1 and P2 (legend and figure adapted from Susac et al. [73] with automatic permission from PNAS).
Among the various allosteric modulators that have been identified in the activation of GPCRs, lipids play an important role by either binding to specific sites, e.g., cholesterol or phosphatidylinositol molecules acting as specific cofactors [180], or by modulating the physical properties of the lipid bilayer [265], thereby impacting the activation of these receptors. Docosahexaenoic acid (DHA) phospholipid chains are well represented in the membrane of cells where A2A is highly expressed. With the help of the introduction of a 13C-labeled and protonated methyl group in methionine residues, Mizumura et al. investigated the effect of these acyl chains on the activation of A2A [49]. Thanks to deuteration, this study could be performed on quite large nanodiscs stabilized by the lipoprotein MSP1E3 [103]. Based on 2D [1H,13C] NMR spectra, they deduced that in the presence of a partial agonist, A2A is in an equilibrium between inactive and active states, while a full agonist shifts the equilibrium towards the active state only. Interestingly, the addition of DHA enhances the G-protein activation by A2A, a positive allosteric effect that has also been observed, for instance, with phosphatidylinositol (4,5)-bisphosphate and an analog of cholesterol in the activation of ghrelin [180] and leukotriene receptors [266], respectively. Based on these observations, they make the assumption that DHA acyl chains modify the conformational equilibrium centered at the NPxxY motif that would induce a large clockwise rotation of TM6, which is based on the lipid-dependent chemical shift variations of methionine residues located at the cytoplasmic ends of TM3 and TM6 (M106 and M232, respectively).
Another important class of allosteric modulators of GPCRs are cations. Deciphering A2A conformation landscape in various situations, Ye et al. confirmed the negative allosteric modulation of A2A activity by sodium ions, as already observed in pharmacological and structural studies and confirmed as being common to many Class A GPCRs [56]. Interestingly, they observed an opposite trend upon the addition of divalent cations, e.g., Ca2+ and Mg2+, which had positive allosteric effects that were boosted by the presence of an agonist and a G-protein-derived peptide [93]. With the aid of molecular dynamic (MD) simulations, they propose that divalent cations would establish bridges between extra-cellular acidic residues belonging to TM5 and TM6, which would allosterically contribute to the opening of the G-protein binding cavity on the intracellular side of the receptor.
19F NMR can be useful to study GPCR conformational ensembles thanks to a large chemical shift dispersion, a high sensitivity (a gyromagnetic ratio close to the proton) and the fact that there are no background signals. However, if using just a single incorporated probe, in particular, localized in an extra-membrane region, it is very difficult to obtain a global and coherent view of the molecular mechanism governing activation. To overcome these limitations, Eddy et al. used uniform-labeling of the receptor with 15N and 2H isotopes to investigate the allosteric coupling between the drug binding event and intracellular signaling [47]. The authors took advantage of the 1H,15N correlation peak chemical shifts of exchangeable Gly amide (1HN) and Trp indole protons, as their 1H/15N cross-peaks resonate in well-isolated parts in typical 2D [1H,15N] correlation spectra. Thanks to an additional partial deuteration (~70%), this strategy enabled them to identify well resolved NMR cross-peaks that could be assigned to six individual Trp indole protons and eight Gly amide protons by single mutagenesis. As discussed in this study, multiplying the number of sampling points in the receptor is only interesting if most of the assigned residues are located in the vicinity of “ring current fields” to increase the sensitivity of these nuclei to subtle changes in the conformational polymorphism of the receptor. Hence, significant 1H and/or 15N chemical shift variations for 11 of the 14 assigned residues were observed between A2A bound to an agonist and to an antagonist. Incidentally, no data were reported on the apo state in this work. The synthesis of all their observations leads to a model where there is a strong interplay between the very-well-conserved residue of A522.50 and W2566.48; the latter, which is located just below the ligand orthosteric pocket, has previously been hypothesized to change its rotameric state upon activation (“toggle switch”) [267]. The authors also extended the use of indole protons of tryptophan to introduce extrinsic tryptophan residues in A2A to sample the energy landscape of the receptor at additional locations, confirming that the receptor responds differently to ligands with variable efficacy and that the intracellular end of TM6 samples wide conformational variations upon activation [268]. More recently, they applied this same strategy to compare the conformational ensembles of A2A associated with two different partial agonists (LUF5834 and Regadenoson) and the full agonist NECA [269]. Their study suggests a different signal propagation pathway based on variations in populations of simultaneously coexisting conformations at equilibrium [269].
Solution-state NMR undoubtedly represents a key ally to classical structural approaches using crystallography or electron microscopy to further explore the energy landscape of very dynamic proteins, such as GPCRs. Numerous studies performed at or nearly at physiological temperatures with no or just a few chemical modifications depict a complex conformational landscape often associated with high kinetic barriers. A2A is no exception, and beyond representing an important target for several human diseases, it also portrays an interesting model to study the fundamental properties of GPCRs, such as biased or partial agonism and the impact of allosteric modulators in the activation process. From the studies performed so far, it appears to be better to sample several locations at the same time within the receptor, whenever possible. In order to connect different parts of the receptor in the molecular activation mechanism(s), it will also be important to sample novel positions beyond the water-exposed regions. This should also limit the discrepancies between different approaches relying on nuclei with different physical properties and/or sample conditions. Moreover, as a membrane protein in solution is highly sensitive to its swimming belt, it seems to be essential to try to study the receptor in a membrane-like environment. Furthermore, in consideration of the importance of the constitutive activity in GPCR signaling, it will be crucial to include the apo state in all subsequent studies, as this has not been conducted systematically in the studies mentioned here. One major objective for NMR remains the determination of the kinetic barriers separating the different sub-states observed, which, in addition to the determination of populations, will finally provide fundamental kinetic and thermodynamic data to better understand the signaling process through these integral membrane receptors such as A2A. One possible path to help to increase the amount of information gained from an NMR experiment would be to improve the deuteration level of the receptor in conjunction with the use of appropriate isotope-labeling schemes, high magnetic fields and other suitable methodologies [270]. This would allow not only base NMR-derived activation models on chemical shift variations or intensities of NMR signals, but would also be of particular advantage to investigate the dynamics in GPCRs using the relaxation phenomena of nuclear spins [271].
8.5. Future Trends for GPCR Structure Studies
8.5.1. Cryo-Electron Tomography
Cryo-electron tomography (cryo-ET) data can be collected with the same transmission electron microscopes as single-particle analysis data. Cryo-ET is label-free and provides three-dimensional snapshots of organelles and proteins at nanometer resolutions within their functional cellular environments, allowing users to visualize and understand how they, and other molecules, work together to carry out major processes in a cell.
The majority of Class A GPCRs are relatively small and thus very difficult to identify in vivo in a cellular environment. However, one can cite the very interesting example on visual rhodopsin, a member of the GPCR family [272]. In this work, the authors identified the highly ordered arrangement of rhodopsin molecules in the disk membrane of intact photoreceptors with molecular resolution. Due to the small size of rhodopsin (40 kDa), the resulting molecular map was not amenable to fitting with the X-ray structure of rhodopsin. However, the resolution was sufficient to identify that rhodopsin was forming dimers as building blocks of a hierarchical supramolecular architecture.
This structure is characterized by aligned rows of dimers, and pairs of rows. The authors proposed that GPCR dimerization and oligomerization offer possibilities for the allosteric regulation of GPCR activity. These results support previously published work, which show for the first time that rhodopsin molecules assemble in dimers and form paracrystals in native rod outer segment membranes using atomic force microscopy (AFM) images [273].
8.5.2. Structural Flexibility of GPCR Investigated Using Single Molecule Fluorescence and Atomic Force Microscopy
Evidence has emerged that, rather than being activated by ligands in an on/off manner switching from an inactive to an active state, GPCRs exhibit high structural flexibility in the absence and even in the presence of ligands. The physiological as well as pharmacological impact of this structural flexibility remains puzzling. Single molecule fluorescence offers the advantage of monitoring the structural dynamics of biomolecules in real-time, with minimal structural invasiveness and in the context of complex biological environments; in particular, single-molecule Förster resonance energy transfer is a key method to measure actual distance changes between two probes, and highlight conformational changes occurring at timescales relevant for protein conformational movements [159].
AFM-based high-resolution imaging and force spectroscopy have been also successfully used to better understand the molecular mechanism details of how GPCR conformations are modulated by ligand-binding or GPCR-transducer complex formation [274].
9. Pharmacology of the Isolated Protein A2A
9.1. Background
While finding inhibitors or ligands for soluble proteins is relatively easy, and basically concerns the number of molecules one can screen, for membrane-associated proteins, such as GPCRs, it is far more complicated. A key to understand GPCR function still resides in the availability of molecules with chemically diverse structures—in other words, structurally diverse ligands (agonists as well as antagonists) are needed. Although this is more important for some GPCRs (melatonin, dopamine) than for others (histamine, serotonin), it is crucial that efforts are made in screening large libraries of molecules with the hope of finding new ligands. The availability of new approaches, particularly in silico, will assist in the discovery of new molecules, although many obstacles remain on these avenues.
9.2. Background on Ligand Chemistry
9.2.1. A2A Receptor Agonists
Not surprisingly, most adenosine receptor agonists are derivatives of adenosine, while only a few examples of non-nucleoside agonists have been found, as shown in Figure 7 [5,275]. Due to the well-known fact that adenine is the “address portion” of the adenosine scaffold, while ribose encodes the “message portion”, most modifications of the ribose part usually resulted in a decrease in efficacy. The only well-tolerated modification identified so far was the exchange of the primary hydroxyl with an ethyl amide. Besides that, a thioether analogue, GS9667, and a molecule containing a rigid pseudo-ribose moiety, MRS-5151, also proved to be successful agonists [276]. In contrast, adenines lacking the entire ribose entity most often act as antagonists. Modifications at the C-2 and C-6 positions on the adenine ring were well explored and tolerated, and led to the discovery of the first A2A selective agonist, CGS 21,680 [277]. There are two main families of non-nucleoside agonists, based on other heterocyclic scaffolds such as the dicyanopiridines [278].
Figure 7.
Structures of the main A2A receptors non-nucleoside based agonists.
Most of these compounds have pKi ranging from 9.4 (Apadenoson) to 5 (MRS-3558) and display some specificity over the other adenosine receptor subtypes. However, the specificity profile of these compounds remains poorly explored outside the adenosine receptor family. Activities of agonists at A2A and other receptors have generally been poorly reported, and most of them came from industrial origins (e.g., Pfizer, AstraZeneca, Ciba-Geigy). Comprehensive molecular details of available A2A receptor ligands, including functional motif, binding sites, EC50/IC50, pKi, pA2, and related references can be found either in the IUPHAR data base [1] or in IJzermann et al. [5].
9.2.2. A2A Receptor Antagonists
For the discovery of antagonists, theophylline and adenine initially served as a nature-derived starting points (shown in Figure 8 and Figure 9), from which compounds with diverse modifications were produced. Nevertheless, alternative scaffolds including flavonoids or antimalarial quinolines were also used to design antagonists of adenosine receptors (Figure 10). Alkylxanthines, such as CSC or Rolophylline, are the prototypical adenosine receptor antagonists and hundreds of analogues have been reported, leading to selective compounds for each adenosine receptor subtype [279]. The best A2A receptor antagonists were developed with the incorporation of 8-styryl groups on the xanthine scaffold, resulting in the first approved A2A antagonist drug, Istradefylline (Figure 8). A large number of adenine derivatives without the ribose moiety have been reported as adenosine receptor antagonists, starting with the A1-selective N-0840 first reported in 1987 [280]. It was followed by many alternative but closely related heterocycles, including fused five- and six-membered rings. One of the tricyclic series has progressed to Preladenant [281], which reached the clinical phase (Figure 9). It is worth noting that the most powerful antagonist is SCH442416 [282,283], although the actual antagonistic property seemed not to be experimentally determined, but was based on the fact that this compound displaced the potent antagonist, SCH58261 [284]. This compound and its reported analogues were also structurally related to adenine. The development of this molecule led to a positron emission tomography (PET) imaging compound applied to visualize the A2A expression in vivo [283,285].
Figure 8.
Structures of the main A2A receptors agonists derived from theophylline.
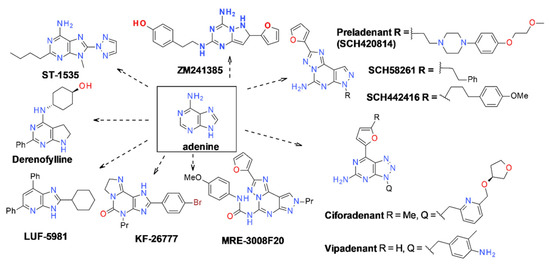
Figure 9.
Structures of the main A2A receptors agonists derived from adenine.
Figure 10.
Structures of the main A2A receptors antagonists.
Molecular pharmacology shows that the A2A agonist and antagonist families are well-populated. They form a large ensemble of molecules with a large panel of properties, although their chemical diversity is somewhat restricted, as is often the case for small molecule GPCRs, such as melatonin [286] or serotonin [287] receptors.
9.3. Allosteric Modulators
Among the new descriptions of GPCR ligands, besides the classical antagonists, agonists and inverse agonists, are also the allosteric modulators of GPCR signaling which have previously been reviewed in depth [288,289,290]. In brief, such compounds bind to the receptor at a site different to the orthosteric site, leading to subtle modifications of the area, and hence to changes in either the affinity for the agonist or of the activity of orthosteric ligands. Two major classes of allosteric ligands have been described, i.e., negative allosteric modulators (NAMs) or positive allosteric modulators (PAMs). NAMs inhibit the activation of the receptor and the downstream signal transduction. On the other hand, PAMs increase the activation of the receptor. Furthermore, ions are also considered as allosteric modulators for multiple GPCRs, such as Na+ for the adenosine A2A receptor [89,291] or Zn2+ for the β2-adrenergic receptor [292] for which they have been shown to bind at allosteric sites which differ from the orthosteric site as reviewed by van der Westhuizen et al. [290]. While they offer new ways to modulate and fine-tune GPCR activity, they are quite difficult to screen for, as they might be dependent on the nature of the agonist they work in conjunction with. This poses a great challenge for the screening format, especially when functional assays are performed on living cells. Furthermore, to characterize this novel, more peculiar type of binding sites of NAMs and PAMs, a large panel of biophysical techniques is required, including molecular dynamics (MD) simulations, ligand structure–activity relationship studies, and in-solution NMR analyses, to name a few [293].
The main example of an A2A PAM is perhaps DFIABA (3,4-difluoro-2-((2-fluoro-4-iodophenyl)amino)benzoic acid), a compound that enhances adenosine signaling at the A2A receptor, and thereby induces slow wave sleep without affecting body temperature in mice [294]. This remarkable feature exemplifies the role of these types of molecules, promising precise modulations of signaling processes with reduced side effects. Other molecules with similar functions include, AEA061 [295], or inosine. Inosine is an endogenous molecule, that could open avenues in the concept of natural PAMs or NAMs, adding to the arsenal of molecules modulating GPCRs functions [296]. The interplay between several receptors, in pathological models illustrates the complexity of various pharmacological ligands, as soon as they interfere with several profoundly different GPCRs, e.g., M4 and A2A [297], mGlu4 and A2A [298] or (TLR)4 and A2A [299]. Adenosine receptors have also been screened for NAMs, as adenosine receptor subtype-specific negative allosteric modulators would offer access to new potential treatments for analgesia, possibly with minimum negative off-target effects, an area where immense treatment progress remains to be done [300].
9.4. Fluorescent Ligands
Adenosine receptors are of therapeutic interest and, as such, selective ligands attract particular attention. Indeed, a plethora of ligands with well-defined pharmacological profiles have been developed during the last years for A2A. Due to the accumulation of the three-dimensional structures of A2A in different activation states, the design of ligands is now largely structure-based. Different compounds with particular pharmacological profiles have been identified and reviewed exhaustively [5], as well as their modified counterparts such as radiolabeled, covalent or bivalent ligands [301]. We will focus here on a particular class of labeled compounds, fluorescent ligands. Indeed, such compounds are of major interest when developing ligand-binding assays with isolated receptors [302]. A full series of fluorescent A2A-selective ligands have been devised to be used in native membranes or cell-based systems [303]. These include agonists such as the initial compound FITC-APEC that was derived from the conjugation of the A2A agonist APEC to fluorescein [304]. An alternative version of FITC-APEC, with Alexa Fluor 488 as the fluorophore instead of fluorescein, has been more recently used to study the kinetics of A2A redistribution in living cells [305]. Besides agonists, fluorescent antagonists have also been designed, for instance by coupling the selective A2A antagonist preladenant to Alexa Fluor 647 [306], or using either of the two pharmacophores, 1,3-dipropyl-8-phenylxanthine and triazolo [1,5-c]quinazolin-5-yl)amine [307]. Regarding isolated receptors, the number of studies with fluorescent ligands is much more limited, and these compounds have been mostly used in the context of ligand-binding experiments. A fluorescent antagonist, CA600245, was recently used to assess the ligand-binding ability of A2A encapsulated into SMALPs generated by solubilizing membranes of P. pastoris expressing A2A with the SMA polymer. In this case, the fluorescent ligand comprised the antagonist xanthine amine moiety linked to a red BODIPY630/650 fluorophore (Figure 11A). The resulting study demonstrated that A2A in SMALPS is a powerful model system to characterize ligand-receptor complexes when used in combination with fluorescence spectroscopy [111]. More recently, the fluorescent compound FITC-APEC described above (Figure 11B) was used to provide an experimental and theoretical framework for monitoring ligand-binding kinetics to the A2A in detergent micelles under ligand depletion conditions using fluorescence anisotropy measurements [308]. Taken together, the availability of different fluorescent probes from distinct pharmacological classes now provides a toolbox of labeled ligands to develop radioactive-free ligand-binding assays with purified receptors, as well as to probe the pharmacological behavior of A2A isolated in detergent micelles and nanodiscs.
Figure 11.
Chemical structures of the fluorescent ligands used for in vitro assays with purified A2A. (A) CA600245 was used to assess the functional properties of the isolated receptor in SMALPS [111]. (B) FITC-APEC was used to monitor the ligand-binding kinetics to the A2A in detergent micelles [308].
9.5. Structure-Based Ligand Discovery for A2A
Structure-based methods in drug discovery comprise a variety of different computer-assisted strategies, including either ligand- or structure-based virtual screening approaches, as well as pharmacophore-based, fragment-based and comparative molecular field analysis [309,310,311,312]. These methods are widely used to identify novel ligands exerting either an agonistic or an antagonistic effect on a specific protein target and they also have had considerable impact on the drug-discovery-related pharmacology of A2A. There are several examples in the literature that illustrate the practical application of structural information for the discovery of A2A antagonists which have been reviewed by Jazayeri et al. [313]. Furthermore, molecular docking has also been employed for the design of either A2A agonists or PDE10A inhibitors, and has been a recent focus in the search of novel antiproliferative agents [314]. In addition, MD simulations have been used extensively to investigate the conformational dynamics at the A2A or PDE10A [315,316,317,318,319,320,321]. In a recent paper, Kalash et al. studied a novel structure-based method for identifying ligands that activate A2A, while simultaneously inhibiting the PDE10A [322]. The focus of this approach was on the key interacting residues that are reported in the literature to discriminate between agonist and antagonist activity of A2A ligands [52,57,64,68]. It has been postulated that the motion of the residue Val84 in theTM3, upon A2A ligand binding, might discriminate between agonist and antagonist activity, which had not been studied previously by any of the MD simulations of A2A [64,323]. Kalash et al. show that the motion of this residue can be used as a conformational descriptor for the characterization of receptor activation by A2A ligands [322]. Furthermore, it is suggested that a selective interaction of ligands with His250 accounts for A2A sub-type selectivity. This interaction has only been detected in co-crystal structures with selective A2A agonists, such as CGS21680 [57] and UK432097 [58]. Moreover, MD simulation analysis demonstrated that this residue undergoes conformational changes only when selective A2A agonists are bound and not when non-selective agonists bind to A2A [322]. Additionally, the analysis of the MD trajectories highlighted the motion of Val84 in TM3 as an essential requirement for A2A activation, which is a strong indicator for the potential application of structure-based methods in the discovery of novel A2A ligands.
9.6. Fragment-Based Ligand Discovery for A2A
Fragment-based ligand design is a well-established approach for the generation of novel protein inhibitors or modulators [324]. In a first step, fragment hits are identified from the screening of a fragment library against the protein target. In a second step, selected fragments are iteratively optimized using three-dimensional structures of the protein–ligand complex until a high-affinity compound is obtained. Fragment libraries typically regroup thousands of highly soluble small molecules with low molecular weight (<300 g/mol). Fragment screening using biophysical methods has been achieved for A2A using target immobilized NMR screening [325], SPR [134], STD-NMR [91] and Affinity Mass Spectrometry [293] with A2A samples prepared in detergents, or weak-affinity chromatography with A2A prepared in nanodiscs [77]. Whereas main studies are conducted with the wild-type receptor, a thermostabilized receptor was used in the study reported by Congreve et al. [134]. All techniques, except for STD-NMR, require the immobilization of A2A onto a solid support. To avoid the identification of false positive hits, the binding signal observed in the presence of A2A is compared to the one observed in the absence of A2A or in the presence of the receptor blocked with a reference ligand. In addition to experimental biophysical screening, fragment-based computer-aided design was reported by several groups [293,311,326,327,328,329]. To probe the conformational changes in the receptor upon fragment-based ligand binding, serial crystallography could play a key role in the future as a method for obtaining high-resolution conformational dynamics of protein–inhibitor complexes.
9.7. Mass Spectrometry-Based Ligand Screening
Among the available modern screening methods described in this Section, affinity selection-mass spectrometry (AS-MS) is emerging as a technique of choice for the identification and confirmation of binders from large compound libraries, due to its high throughput capacity. AS-MS has been applied by the group of Shui to find new A2A receptor binders [106]. As a proof of concept, they first developed AS-MS using a small model library consisting of a fifteen-compound mixture including eight known binders (such as adenosine, NECA, caffeine or ZM241385) and A2A reconstituted in nanodiscs. All the known compounds were identified as positive binders from the AS-MS screen, validating the set-up for hit discovery on GPCRs. They next highlighted the potential of AS-MS to screen large compound libraries (approximately 20,000 compounds) on A2A in one pool, leading to the discovery of three new antagonist compounds [141]. Interestingly, affinity MS experiments were successfully performed on a purified A2A immobilized on nickel agarose beads as well as on the receptor embedded in cell-membranes. More recently [293], the same authors adapted their methodology to screen a library of 1100 fragments with the purpose of finding new NAMs of A2A. Among the 28 initial hits, 17 have been confirmed to be specific binders at A2A, and 9 as strong binders by SPR. These nine binders were then analyzed using affinity mass spectrometry in the presence of A2A, free or bound to ZM241385. Thanks to an extensive optimization of immobilization and incubation conditions (see scheme in Figure 12), their method was initially validated as it allowed the detection of three known weak antagonists with binding affinity above 20 μM (theophylline, NG-52 and LH-846). The six fragments that retained 50% binding in the presence of ZM241385 were further tested in competition with HMA (5-(N,N-hexamethylene)amiloride), one of the most potent NAMs of A2A. Two new HMA-competitive binders were discovered using this approach and confirmed as NAMs using a competitive radioligand binding assay, molecular docking and 19F-labeled protein NMR experiments [293].
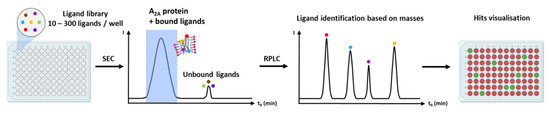
Figure 12.
Schematic workflow of modern affinity selection mass spectrometry (AS-MS) used for A2A hit identification. Additionally, called automated ligand identification system (ALIS), this technique is performed to identify small molecules that engage a specific target, such as A2A. The protein of interest is incubated with a mixture of compounds, each with unique molar mass. The protein–ligand complexes are separated from unbound compounds using SEC. The protein peak (depicted in blue on the figure) is then directed to a reverse-phase liquid chromatography (RPLC) column for dissociation, desalting, and elution of any ligands into an ESI-MS system for identification. Alternatively to this 2D-LC set-up, an ultracentrifugation or an offline gel filtration step can be performed prior to LC-MS. This high-throughput screening technique enables hit identification for large collections of ligands.
10. Concluding Remarks and Perspectives
As the isolation of A2A and its reconstitution in an active form were achieved relatively early compared to most other GPCRs, its handling became more streamlined compared to other GPCRs. Accordingly, A2A has become a highly amenable model for the application and development of novel innovative approaches for the biophysical characterization of GPCRs. Moreover, A2A is also a very important drug target in several human disease areas, and it goes without saying that the accumulation of knowledge of the structure and function of A2A was also driven by the desire to discover novel and more specific pharmacological compounds. Indeed, the progress made in many of the cellular biology and cellular chemistry domains, together with the in silico approach of modelling the receptor structures and the three-dimensional structure of ligands, lead to the discovery of a series of novel ligands including compounds containing novel chemical scaffolds. Nonetheless, it remains to be seen if this knowledge will be transferable to the studies of other, more challenging to obtain GPCRs, especially from other families.
Many other areas could be further explored for A2A that might be more advanced in the context of other receptors. For example, the structural effects of the variety of agonists existing for this receptor should be further investigated by cryo-EM, possibly also in complex with different G proteins, to investigate the possibility of biased agonism at this receptor and how it can be explained at the structural level. Similarly, the structural effects on signal transduction of allosteric modulators are still largely unknown for the A2A receptor. To further understand these mechanisms, as well as the effects of subtle variations of the agonists on the structure of the G-protein binding site, dynamics kinetics need to further be studied. To this end, single molecule studies, molecular dynamics and also ensemble NMR studies will be invaluable.
While studies of the isolated receptor are crucial for understanding the mechanisms of signal transduction, it is of the upmost importance to also study receptors in their cellular context. Indeed, the progress recently made in the chemical alkylation of proteins in situ (for instance by click chemistry) might eventually allow high resolution studies of the receptor in a “native” milieu, surrounded by its “natural” neighbors in A2A-richly expressing membranes isolated from human tissues. However, the prerequisite of such experimental endeavors will be to introduce a handle at the receptor that can be recognized specifically by the chemical reaction. Combining the use of stem cells with the expression of such handles in a neutral genomic area of the transgene could provide a closer model of the physiological situation.
Finally, A2A has been proven to be an excellent model in many cases of biophysical, structural and even pharmacological characterizations of a GPCR. This is probably due to the relative easiness with which the receptor has been cloned, expressed and isolated, together with a natural stability that helped keep it active once purified.
Funding
The following funding sources are acknowledged. LJC: ANR UnderPressure (ANR-22-CE29-0020-03); ANR DYNALAND; Laboratoire d’Excellence (LabEx) DYNAMO (ANR-11-LABX-0011); Equipements d’Excellence (EQUIPEX) CACSICE (ANR-11-EQPX-0008); SC: CNRS, the University of Strasbourg, the Agence Nationale de la Recherche, and the French Proteomic Infrastructure (ProFI; ANR-10-INBS-08-03); GL: Switch-On (ANR-20-CE11-0019); IK, CD & RW: ANR-21-CE29-0012-02; CVB: AFM-Téléthon #23207.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
A2A, adenosine A2A receptor; AC, affinity chromatography; AFM, atomic force microscopy; AS-MS, affinity selection-mass spectrometry; BRIL, apo-cytochrome b562RIL; CABD, calcium binding domain; CaM, calmodulin; cryo-EM, electron cryo-microscopy; cryo-ET, cryo-electron tomography; DDM, n-dodecyl-β-D-maltopyranoside; DHA, docosahexaenoic acid; EXSY, exchange spectroscopy; FRET, fluorescence resonance energy transfer; GFP, green fluorescent protein; HDX, H/D exchange mass spectrometry; ITC, isothermal titration calorimetry; LCP, lipid cubic phase; LMNG, lauryl maltose neopentyl glycol; mBBR, monobromobimane; MS, mass spectrometry; MSP, membrane scaffold proteins; NAM, negative allosteric modulators; NDs, nanodiscs; NOESY, nuclear Overhauser effect spectroscopy; PAM, positive allosteric modulators; PGD, petunia glutamate decarboxylase; PI, Phosphatidylnositol; PQA, 3-pyrrolidin-1-ylquinoxalin-2-amine; PS, Phosphatidylserines; SEC, size-exclusion chromatography; Sf9, Spodoptera frugiperda; SMALPs, SMA lipid disc particles; SPA, Scintillation Proximity Assay; SPR, surface plasmon resonance; StaR, thermostabilized; STD, saturation transfer difference; T4L, T4 Lysozyme; TM, GPCR transmembrane domain; ULVs, unilamellar lipid vesicles; XAC, xanthine amine congener; XFEL, X-ray free-electron lasers; XL-MS, Cross-linking mass spectrometry.
References
- Fredholm, B.B.; Frenguelli, B.G.; Hills, R.; IJzerman, A.P.; Jacobson, K.A.; Klotz, K.-N.; Linden, J.; Müller, C.E.; Schwabe, U.; Stiles, G.L. Adenosine receptors in GtoPdb v.2021.2. IUPHAR/BPS Guide Pharmacol. CITE 2021, 2021. [Google Scholar] [CrossRef]
- Blackmore, R.V.; Linn, S. Partial purification and characterization of four endodeoxyribonuclease activities from Escherichia coli K-12. Nucleic Acids Res. 1974, 1, 1–17. [Google Scholar] [CrossRef]
- Pierce, K.D.; Furlong, T.J.; Selbie, L.A.; Shine, J. Molecular cloning and expression of an adenosine A2b receptor from human brain. Biochem. Biophys. Res. Commun. 1992, 187, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Michalke, K.; Huyghe, C.; Lichière, J.; Gravière, M.-E.; Siponen, M.; Sciara, G.; Lepaul, I.; Wagner, R.; Magg, C.; Rudolph, R.; et al. Mammalian G protein-coupled receptor expression in Escherichia coli: II. Refolding and biophysical characterization of mouse cannabinoid receptor 1 and human parathyroid hormone receptor 1. Anal. Biochem. 2010, 401, 74–80. [Google Scholar] [CrossRef] [PubMed]
- IJzerman, A.P.; Jacobson, K.A.; Müller, C.E.; Cronstein, B.N.; Cunha, R.A. International Union of Basic and Clinical Pharmacology. CXII: Adenosine Receptors: A Further Update. Pharmacol. Rev. 2022, 74, 340–372. [Google Scholar] [CrossRef]
- Yu, L.; Frith, M.C.; Suzuki, Y.; Peterfreund, R.A.; Gearan, T.; Sugano, S.; Schwarzschild, M.A.; Weng, Z.; Fink, J.S.; Chen, J.-F. Characterization of genomic organization of the adenosine A2A receptor gene by molecular and bioinformatics analyses. Brain Res. 2004, 1000, 156–173. [Google Scholar] [CrossRef] [PubMed]
- Koshiba, M.; Rosin, D.L.; Hayashi, N.; Linden, J.; Sitkovsky, M.V. Patterns of A2A extracellular adenosine receptor expression in different functional subsets of human peripheral T cells. Flow cytometry studies with anti-A2A receptor monoclonal antibodies. Mol. Pharmacol. 1999, 55, 614–624. [Google Scholar]
- Cooper, J.A.; Hill, S.J.; Alexander, S.P.; Rubin, P.C.; Horn, E.H. Adenosine receptor-induced cyclic AMP generation and inhibition of 5-hydroxytryptamine release in human platelets. Br. J. Clin. Pharmacol. 1995, 40, 43–50. [Google Scholar] [CrossRef]
- Schulte, G.; Fredholm, B.B. Human adenosine A1, A2A, A2B, and A3 receptors expressed in Chinese hamster ovary cells all mediate the phosphorylation of extracellular-regulated kinase ½. Mol. Pharmacol. 2000, 58, 477–482. [Google Scholar] [CrossRef]
- Yang, S.N.; Dasgupta, S.; Lledo, P.M.; Vincent, J.D.; Fuxe, K. Reduction of dopamine D2 receptor transduction by activation of adenosine A2a receptors in stably A2a/D2 (long-form) receptor co-transfected mouse fibroblast cell lines: Studies on intracellular calcium levels. Neuroscience 1995, 68, 729–736. [Google Scholar] [CrossRef]
- Ikeuchi, Y.; Nishizaki, T.; Mori, M.; Okada, Y. Adenosine activates the K+ channel and enhances cytosolic Ca2+ release via a P2Y purinoceptor in hippocampal neurons. Eur. J. Pharmacol. 1996, 304, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhou, F.M. cAMP-producing chemogenetic and adenosine A2a receptor activation inhibits the inwardly rectifying potassium current in striatal projection neurons. Neuropharmacology 2019, 148, 229–243. [Google Scholar] [CrossRef]
- Varani, K.; Gessi, S.; Dalpiaz, A.; Borea, P.A. Pharmacological and biochemical characterization of purified A2a adenosine receptors in human platelet membranes by [3H]-CGS 21680 binding. Br. J. Pharmacol. 1996, 117, 1693–1701. [Google Scholar] [CrossRef] [PubMed]
- Buckley, N.; Burnstock, G. Autoradiographic demonstration of peripheral adenosine binding sites using [3H]NECA. Brain Res. 1983, 269, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Schütz, W.; Brugger, G. Characterization of [3H]-adenosine binding to media membranes of hog carotid arteries. Pharmacology 1982, 24, 26–34. [Google Scholar] [CrossRef]
- Ji, X.D.; Stiles, G.L.; van Galen, P.J.; Jacobson, K.A. Characterization of human striatal A2-adenosine receptors using radioligand binding and photoaffinity labeling. J. Recept. Res. 1992, 12, 149–169. [Google Scholar] [CrossRef]
- Alexander, S.P.; Millns, P.J. [3H]ZM241385—An antagonist radioligand for adenosine A2A receptors in rat brain. Eur. J. Pharmacol. 2001, 411, 205–210. [Google Scholar] [CrossRef]
- Yu, J.C.; Lin, G.; Field, J.J.; Linden, J. Induction of antiinflammatory purinergic signaling in activated human iNKT cells. JCI Insight 2018, 3, e91954. [Google Scholar] [CrossRef]
- Klammt, C.; Schwarz, D.; Eifler, N.; Engel, A.; Piehler, J.; Haase, W.; Hahn, S.; Dötsch, V.; Bernhard, F. Cell-free production of G protein-coupled receptors for functional and structural studies. J. Struct. Biol. 2007, 158, 482–493. [Google Scholar] [CrossRef]
- Kögler, L.M.; Stichel, J.; Beck-Sickinger, A.G. Structural investigations of cell-free expressed G protein-coupled receptors. Biol. Chem. 2019, 401, 97–116. [Google Scholar] [CrossRef]
- Banères, J.-L.; Popot, J.-L.; Mouillac, B. New advances in production and functional folding of G-protein-coupled receptors. Trends Biotechnol. 2011, 29, 314–322. [Google Scholar] [CrossRef]
- André, N.; Cherouati, N.; Prual, C.; Steffan, T.; Zeder-Lutz, G.; Magnin, T.; Pattus, F.; Michel, H.; Wagner, R.; Reinhart, C. Enhancing functional production of G protein-coupled receptors in Pichia pastoris to levels required for structural studies via a single expression screen. Protein Sci. 2006, 15, 1115–1126. [Google Scholar] [CrossRef]
- Krettler, C.; Reinhart, C.; Bevans, C.G. Expression of GPCRs in Pichia pastoris for structural studies. Methods Enzymol. 2013, 520, 1–29. [Google Scholar] [CrossRef]
- Blocker, K.M.; Britton, Z.T.; Naranjo, A.N.; McNeely, P.M.; Young, C.L.; Robinson, A.S. Recombinant G protein-coupled receptor expression in Saccharomyces cerevisiae for protein characterization. Methods Enzymol. 2015, 556, 165–183. [Google Scholar] [CrossRef]
- Errey, J.C.; Fiez-Vandal, C. Production of membrane proteins in industry: The example of GPCRs. Protein Expr. Purif. 2020, 169, 105569. [Google Scholar] [CrossRef] [PubMed]
- Panneels, V.; Kock, I.; Krijnse-Locker, J.; Rezgaoui, M.; Sinning, I. Drosophila photoreceptor cells exploited for the production of eukaryotic membrane proteins: Receptors, transporters and channels. PLoS ONE 2011, 6, e18478. [Google Scholar] [CrossRef]
- Zhang, L.; Salom, D.; He, J.; Okun, A.; Ballesteros, J.; Palczewski, K.; Li, N. Expression of functional G protein-coupled receptors in photoreceptors of transgenic Xenopus laevis. Biochemistry 2005, 44, 14509–14518. [Google Scholar] [CrossRef] [PubMed]
- Kajikawa, M.; Sasaki, K.; Wakimoto, Y.; Toyooka, M.; Motohashi, T.; Shimojima, T.; Takeda, S.; Park, E.Y.; Maenaka, K. Efficient silkworm expression of human GPCR (nociceptin receptor) by a Bombyx mori bacmid DNA system. Biochem. Biophys. Res. Commun. 2009, 385, 375–379. [Google Scholar] [CrossRef]
- Michalke, K.; Gravière, M.-E.; Huyghe, C.; Vincentelli, R.; Wagner, R.; Pattus, F.; Schroeder, K.; Oschmann, J.; Rudolph, R.; Cambillau, C.; et al. Mammalian G-protein-coupled receptor expression in Escherichia coli: I. High-throughput large-scale production as inclusion bodies. Anal. Biochem. 2009, 386, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Weiss, H.M.; Grisshammer, R. Purification and characterization of the human adenosine A2a receptor functionally expressed in Escherichia coli. Eur. J. Biochem. 2002, 269, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Magnani, F.; Shibata, Y.; Serrano-Vega, M.J.; Tate, C.G. Co-evolving stability and conformational homogeneity of the human adenosine A2a receptor. Proc. Natl. Acad. Sci. USA 2008, 105, 10744–10749. [Google Scholar] [CrossRef]
- Lebon, G.; Bennett, K.; Jazayeri, A.; Tate, C.G. Thermostabilisation of an Agonist-Bound Conformation of the Human Adenosine A2A Receptor. J. Mol. Biol. 2011, 409, 298–310. [Google Scholar] [CrossRef]
- Niebauer, R.T.; Robinson, A.S. Exceptional total and functional yields of the human adenosine (A2a) receptor expressed in the yeast Saccharomyces cerevisiae. Protein Expr. Purif. 2006, 46, 204–211. [Google Scholar] [CrossRef]
- O’Malley, M.A.; Lazarova, T.; Britton, Z.T.; Robinson, A.S. High-level expression in Saccharomyces cerevisiae enables isolation and spectroscopic characterization of functional human adenosine A2a receptor. J. Struct. Biol. 2007, 159, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Schonenbach, N.S.; Rieth, M.D.; Han, S.; O’Malley, M.A. Adenosine A2a receptors form distinct oligomers in protein detergent complexes. FEBS Lett. 2016, 590, 3295–3306. [Google Scholar] [CrossRef] [PubMed]
- Fraser, N.J. Expression and functional purification of a glycosylation deficient version of the human adenosine 2a receptor for structural studies. Protein Expr. Purif. 2006, 49, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Gras, A.; Fiez-Vandal, C.; Ruprecht, J.; Rana, R.; Martinez, M.; Strange, P.G.; Wagner, R.; Byrne, B. Large-scale functional expression of WT and truncated human adenosine A2A receptor in Pichia pastoris bioreactor cultures. Microb. Cell Fact. 2008, 7, 28. [Google Scholar] [CrossRef]
- Hino, T.; Arakawa, T.; Iwanari, H.; Yurugi-Kobayashi, T.; Ikeda-Suno, C.; Nakada-Nakura, Y.; Kusano-Arai, O.; Weyand, S.; Shimamura, T.; Nomura, N.; et al. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature 2012, 482, 237–240. [Google Scholar] [CrossRef]
- Jamshad, M.; Charlton, J.; Lin, Y.-P.; Routledge, S.J.; Bawa, Z.; Knowles, T.J.; Overduin, M.; Dekker, N.; Dafforn, T.R.; Bill, R.M.; et al. G-protein coupled receptor solubilization and purification for biophysical analysis and functional studies, in the total absence of detergent. Biosci. Rep. 2015, 35, e00188. [Google Scholar] [CrossRef]
- Lengger, B.; Jensen, M.K. Engineering G protein-coupled receptor signalling in yeast for biotechnological and medical purposes. FEMS Yeast Res. 2020, 20, foz087. [Google Scholar] [CrossRef]
- Shiroishi, M.; Tsujimoto, H.; Makyio, H.; Asada, H.; Yurugi-Kobayashi, T.; Shimamura, T.; Murata, T.; Nomura, N.; Haga, T.; Iwata, S.; et al. Platform for the rapid construction and evaluation of GPCRs for crystallography in Saccharomyces cerevisiae. Microb. Cell Fact. 2012, 11, 78. [Google Scholar] [CrossRef]
- Bertheleme, N.; Strege, A.; Bunting, S.E.; Dowell, S.J.; Byrne, B. Arginine 199 and leucine 208 have key roles in the control of adenosine A2A receptor signalling function. PLoS ONE 2014, 9, e89613. [Google Scholar] [CrossRef]
- Mitsumoto, M.; Sugaya, K.; Kazama, K.; Nakano, R.; Kosugi, T.; Murata, T.; Koga, N. State-Targeting Stabilization of Adenosine A2A Receptor by Fusing a Custom-Made de Novo Designed α-Helical Protein. Int. J. Mol. Sci. 2021, 22, 12906. [Google Scholar] [CrossRef]
- Wang, X.; van Westen, G.J.P.; Heitman, L.H.; IJzerman, A.P. G protein-coupled receptors expressed and studied in yeast. The adenosine receptor as a prime example. Biochem. Pharmacol. 2021, 187, 114370. [Google Scholar] [CrossRef]
- Meltzer, M.; Zvagelsky, T.; Hadad, U.; Papo, N.; Engel, S. Yeast-based directed-evolution for high-throughput structural stabilization of G protein-coupled receptors (GPCRs). Sci. Rep. 2022, 12, 8657. [Google Scholar] [CrossRef]
- Yamamoto, T.; Yasuda, S.; Kasai, R.S.; Nakano, R.; Hikiri, S.; Sugaya, K.; Hayashi, T.; Ogasawara, S.; Shiroishi, M.; Fujiwara, T.K.; et al. A methodology for creating mutants of G-protein coupled receptors stabilized in active state by combining statistical thermodynamics and evolutionary molecular engineering. Protein Sci. 2022, 31, e4425. [Google Scholar] [CrossRef] [PubMed]
- Eddy, M.T.; Lee, M.-Y.; Gao, Z.-G.; White, K.L.; Didenko, T.; Horst, R.; Audet, M.; Stanczak, P.; McClary, K.M.; Han, G.W.; et al. Allosteric Coupling of Drug Binding and Intracellular Signaling in the A2A Adenosine Receptor. Cell 2018, 172, 68–80.e12. [Google Scholar] [CrossRef]
- Ye, L.; van Eps, N.; Zimmer, M.; Ernst, O.P.; Prosser, R.S. Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature 2016, 533, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Mizumura, T.; Kondo, K.; Kurita, M.; Kofuku, Y.; Natsume, M.; Imai, S.; Shiraishi, Y.; Ueda, T.; Shimada, I. Activation of adenosine A2A receptor by lipids from docosahexaenoic acid revealed by NMR. Sci. Adv. 2020, 6, eaay8544. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.K.; Pandey, A.; Tran, D.P.; Villanueva, N.L.; Kitao, A.; Sunahara, R.K.; Sljoka, A.; Prosser, R.S. Delineating the conformational landscape of the adenosine A2A receptor during G protein coupling. Cell 2021, 184, 1884–1894.e14. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, V.-P.; Griffith, M.T.; Hanson, M.A.; Cherezov, V.; Chien, E.Y.T.; Lane, J.R.; IJzerman, A.P.; Stevens, R.C. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 2008, 322, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Lebon, G.; Warne, T.; Edwards, P.C.; Bennett, K.; Langmead, C.J.; Leslie, A.G.W.; Tate, C.G. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature 2011, 474, 521–525. [Google Scholar] [CrossRef]
- Carpenter, B.; Nehmé, R.; Warne, T.; Leslie, A.G.W.; Tate, C.G. Structure of the adenosine A2A receptor bound to an engineered G protein. Nature 2016, 536, 104–107. [Google Scholar] [CrossRef]
- Miyagi, H.; Asada, H.; Suzuki, M.; Takahashi, Y.; Yasunaga, M.; Suno, C.; Iwata, S.; Saito, J.-I. The discovery of a new antibody for BRIL-fused GPCR structure determination. Sci. Rep. 2020, 10, 11669. [Google Scholar] [CrossRef]
- Claff, T.; Klapschinski, T.A.; Tiruttani Subhramanyam, U.K.; Vaaßen, V.J.; Schlegel, J.G.; Vielmuth, C.; Voß, J.H.; Labahn, J.; Müller, C.E. Single Stabilizing Point Mutation Enables High-Resolution Co-Crystal Structures of the Adenosine A2A Receptor with Preladenant Conjugates. Angew. Chem. Int. Ed Engl. 2022, 61, e202115545. [Google Scholar] [CrossRef]
- Liu, W.; Chun, E.; Thompson, A.A.; Chubukov, P.; Xu, F.; Katritch, V.; Han, G.W.; Roth, C.B.; Heitman, L.H.; IJzerman, A.P.; et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science 2012, 337, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Lebon, G.; Edwards, P.C.; Leslie, A.G.W.; Tate, C.G. Molecular Determinants of CGS21680 Binding to the Human Adenosine A2A Receptor. Mol. Pharmacol. 2015, 87, 907–915. [Google Scholar] [CrossRef] [PubMed]
- White, K.L.; Eddy, M.T.; Gao, Z.-G.; Han, G.W.; Lian, T.; Deary, A.; Patel, N.; Jacobson, K.A.; Katritch, V.; Stevens, R.C. Structural Connection between Activation Microswitch and Allosteric Sodium Site in GPCR Signaling. Structure 2018, 26, 259–269.e5. [Google Scholar] [CrossRef]
- McGraw, C.; Koretz, K.S.; Oseid, D.; Lyman, E.; Robinson, A.S. Cholesterol Dependent Activity of the Adenosine A2A Receptor Is Modulated via the Cholesterol Consensus Motif. Molecules 2022, 27, 3529. [Google Scholar] [CrossRef]
- Yeliseev, A.; van den Berg, A.; Zoubak, L.; Hines, K.; Stepnowski, S.; Williston, K.; Yan, W.; Gawrisch, K.; Zmuda, J. Thermostability of a recombinant G protein-coupled receptor expressed at high level in mammalian cell culture. Sci. Rep. 2020, 10, 16805. [Google Scholar] [CrossRef]
- Zhang, K.; Wu, H.; Hoppe, N.; Manglik, A.; Cheng, Y. Fusion protein strategies for cryo-EM study of G protein-coupled receptors. Nat. Commun. 2022, 13, 4366. [Google Scholar] [CrossRef]
- Bocquet, N.; Kohler, J.; Hug, M.N.; Kusznir, E.A.; Rufer, A.C.; Dawson, R.J.; Hennig, M.; Ruf, A.; Huber, W.; Huber, S. Real-time monitoring of binding events on a thermostabilized human A2A receptor embedded in a lipid bilayer by surface plasmon resonance. Biochim. Biophys. Acta 2015, 1848, 1224–1233. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Takamuku, Y.; Asakawa, T.; Inai, M.; Hino, T.; Iwata, S.; Kan, T.; Murata, T. An efficient screening method for purifying and crystallizing membrane proteins using modified clear-native PAGE. Anal. Biochem. 2018, 548, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, B.; Lebon, G. Human Adenosine A2A Receptor: Molecular Mechanism of Ligand Binding and Activation. Front. Pharmacol. 2017, 8, 898. [Google Scholar] [CrossRef]
- Cui, M.; Zhou, Q.; Xu, Y.; Weng, Y.; Yao, D.; Zhao, S.; Song, G. Crystal structure of a constitutive active mutant of adenosine A2A receptor. IUCrJ 2022, 9, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Lundstrom, K.; Wagner, R.; Reinhart, C.; Desmyter, A.; Cherouati, N.; Magnin, T.; Zeder-Lutz, G.; Courtot, M.; Prual, C.; André, N.; et al. Structural genomics on membrane proteins: Comparison of more than 100 GPCRs in 3 expression systems. J. Struct. Funct. Genom. 2006, 7, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.D.; Dikiy, I.; Chapman, K.; Rödström, K.E.; Aramini, J.; LeVine, M.V.; Khelashvili, G.; Rasmussen, S.G.; Gardner, K.H.; Rosenbaum, D.M. Ligand modulation of sidechain dynamics in a wild-type human GPCR. eLife 2017, 6, e28505. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wu, H.; Katritch, V.; Han, G.W.; Jacobson, K.A.; Gao, Z.-G.; Cherezov, V.; Stevens, R.C. Structure of an agonist-bound human A2A adenosine receptor. Science 2011, 332, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.K.Y.; Segala, E.; Robertson, N.; Deflorian, F.; Doré, A.S.; Errey, J.C.; Fiez-Vandal, C.; Marshall, F.H.; Cooke, R.M. Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity. Structure 2017, 25, 1275–1285.e4. [Google Scholar] [CrossRef]
- Jespers, W.; Verdon, G.; Azuaje, J.; Majellaro, M.; Keränen, H.; García-Mera, X.; Congreve, M.; Deflorian, F.; de Graaf, C.; Zhukov, A.; et al. X-ray Crystallography and Free Energy Calculations Reveal the Binding Mechanism of A2A Adenosine Receptor Antagonists. Angew. Chem. Int. Ed Engl. 2020, 59, 16536–16543. [Google Scholar] [CrossRef] [PubMed]
- Chun, E.; Thompson, A.A.; Liu, W.; Roth, C.B.; Griffith, M.T.; Katritch, V.; Kunken, J.; Xu, F.; Cherezov, V.; Hanson, M.A.; et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure 2012, 20, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Doré, A.S.; Robertson, N.; Errey, J.C.; Ng, I.; Hollenstein, K.; Tehan, B.; Hurrell, E.; Bennett, K.; Congreve, M.; Magnani, F.; et al. Structure of the adenosine A2A receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure 2011, 19, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Sušac, L.; Eddy, M.T.; Didenko, T.; Stevens, R.C.; Wüthrich, K. A2A adenosine receptor functional states characterized by 19F-NMR. Proc. Natl. Acad. Sci. USA 2018, 115, 12733–12738. [Google Scholar] [CrossRef]
- Huang, S.K.; Almurad, O.; Pejana, R.J.; Morrison, Z.A.; Pandey, A.; Picard, L.-P.; Nitz, M.; Sljoka, A.; Prosser, R.S. Allosteric modulation of the adenosine A2A receptor by cholesterol. eLife 2022, 11, e73901. [Google Scholar] [CrossRef]
- Thakur, N.; Wei, S.; Ray, A.P.; Lamichhane, R.; Eddy, M.T. Production of human A2AAR in lipid nanodiscs for 19F-NMR and single-molecule fluorescence spectroscopy. STAR Protoc. 2022, 3, 101535. [Google Scholar] [CrossRef] [PubMed]
- Choy, B.C.; Cater, R.J.; Mancia, F.; Pryor, E.E. A 10-year meta-analysis of membrane protein structural biology: Detergents, membrane mimetics, and structure determination techniques. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183533. [Google Scholar] [CrossRef]
- Lecas, L.; Hartmann, L.; Caro, L.; Mohamed-Bouteben, S.; Raingeval, C.; Krimm, I.; Wagner, R.; Dugas, V.; Demesmay, C. Miniaturized weak affinity chromatography for ligand identification of nanodiscs-embedded G-protein coupled receptors. Anal. Chim. Acta 2020, 1113, 26–35. [Google Scholar] [CrossRef]
- Lee, M.-Y.; Geiger, J.; Ishchenko, A.; Han, G.W.; Barty, A.; White, T.A.; Gati, C.; Batyuk, A.; Hunter, M.S.; Aquila, A.; et al. Harnessing the power of an X-ray laser for serial crystallography of membrane proteins crystallized in lipidic cubic phase. IUCrJ 2020, 7, 976–984. [Google Scholar] [CrossRef]
- Segala, E.; Guo, D.; Cheng, R.K.Y.; Bortolato, A.; Deflorian, F.; Doré, A.S.; Errey, J.C.; Heitman, L.H.; IJzerman, A.P.; Marshall, F.H.; et al. Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength. J. Med. Chem. 2016, 59, 6470–6479. [Google Scholar] [CrossRef]
- Reis, R.I.; Moraes, I. Probing Membrane Protein Assembly into Nanodiscs by In Situ Dynamic Light Scattering: A2A Receptor as a Case Study. Biology 2020, 9, 400. [Google Scholar] [CrossRef]
- Batyuk, A.; Galli, L.; Ishchenko, A.; Han, G.W.; Gati, C.; Popov, P.A.; Lee, M.-Y.; Stauch, B.; White, T.A.; Barty, A.; et al. Native phasing of x-ray free-electron laser data for a G protein-coupled receptor. Sci. Adv. 2016, 2, e1600292. [Google Scholar] [CrossRef] [PubMed]
- Dikiy, I.; Clark, L.D.; Gardner, K.H.; Rosenbaum, D.M. Isotopic Labeling of Eukaryotic Membrane Proteins for NMR Studies of Interactions and Dynamics. Methods Enzymol. 2019, 614, 37–65. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, M.A.; Naranjo, A.N.; Lazarova, T.; Robinson, A.S. Analysis of Adenosine A 2 a Receptor Stability: Effects of Ligands and Disulfide Bonds. Biochemistry 2010, 49, 9181–9189. [Google Scholar] [CrossRef] [PubMed]
- Ashok, Y.; Nanekar, R.; Jaakola, V.-P. Defining thermostability of membrane proteins by western blotting. Protein Eng. Des. Sel. 2015, 28, 539–542. [Google Scholar] [CrossRef]
- Zhang, X.; Stevens, R.C.; Xu, F. The importance of ligands for G protein-coupled receptor stability. Trends Biochem. Sci. 2015, 40, 79–87. [Google Scholar] [CrossRef]
- Thompson, A.A.; Liu, J.J.; Chun, E.; Wacker, D.; Wu, H.; Cherezov, V.; Stevens, R.C. GPCR stabilization using the bicelle-like architecture of mixed sterol-detergent micelles. Methods 2011, 55, 310–317. [Google Scholar] [CrossRef]
- O’Malley, M.A.; Helgeson, M.E.; Wagner, N.J.; Robinson, A.S. The morphology and composition of cholesterol-rich micellar nanostructures determine transmembrane protein (GPCR) activity. Biophys. J. 2011, 100, L11–L13. [Google Scholar] [CrossRef]
- Vagenende, V.; Yap, M.G.S.; Trout, B.L. Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. Biochemistry 2009, 48, 11084–11096. [Google Scholar] [CrossRef]
- Wittmann, H.-J.; Seifert, R.; Strasser, A. Mathematical analysis of the sodium sensitivity of the human histamine H3 receptor. Silico Pharmacol. 2014, 2, 1. [Google Scholar] [CrossRef]
- Ross, P.; Weihofen, W.; Siu, F.; Xie, A.; Katakia, H.; Wright, S.K.; Hunt, I.; Brown, R.K.; Freire, E. Isothermal chemical denaturation to determine binding affinity of small molecules to G-protein coupled receptors. Anal. Biochem. 2015, 473, 41–45. [Google Scholar] [CrossRef]
- Igonet, S.; Raingeval, C.; Cecon, E.; Pučić-Baković, M.; Lauc, G.; Cala, O.; Baranowski, M.; Perez, J.; Jockers, R.; Krimm, I.; et al. Enabling STD-NMR fragment screening using stabilized native GPCR: A case study of adenosine receptor. Sci. Rep. 2018, 8, 8142. [Google Scholar] [CrossRef]
- Ukena, D.; Jacobson, K.A.; Kirk, K.L.; Daly, J.W. A [3H]amine congener of 1,3-dipropyl-8-phenylxanthine. FEBS Lett. 1986, 199, 269–274. [Google Scholar] [CrossRef]
- Ye, L.; Neale, C.; Sljoka, A.; Lyda, B.; Pichugin, D.; Tsuchimura, N.; Larda, S.T.; Pomès, R.; García, A.E.; Ernst, O.P.; et al. Mechanistic insights into allosteric regulation of the A2A adenosine G protein-coupled receptor by physiological cations. Nat. Commun. 2018, 9, 1372. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.D.Q.; Vigers, M.; Sefah, E.; Seppälä, S.; Hoover, J.P.; Schonenbach, N.S.; Mertz, B.; O’Malley, M.A.; Han, S. Homo-oligomerization of the human adenosine A2A receptor is driven by the intrinsically disordered C-terminus. eLife 2021, 10, e66662. [Google Scholar] [CrossRef]
- Jones, A.J.Y.; Gabriel, F.; Tandale, A.; Nietlispach, D. Structure and Dynamics of GPCRs in Lipid Membranes: Physical Principles and Experimental Approaches. Molecules 2020, 25, 4729. [Google Scholar] [CrossRef]
- Serebryany, E.; Zhu, G.A.; Yan, E.C.Y. Artificial membrane-like environments for in vitro studies of purified G-protein coupled receptors. Biochim. Biophys. Acta 2012, 1818, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Overduin, M.; Trieber, C.; Prosser, R.S.; Picard, L.-P.; Sheff, J.G. Structures and Dynamics of Native-State Transmembrane Protein Targets and Bound Lipids. Membranes 2021, 11, 451. [Google Scholar] [CrossRef]
- Park, S.H.; Das, B.B.; Casagrande, F.; Tian, Y.; Nothnagel, H.J.; Chu, M.; Kiefer, H.; Maier, K.; de Angelis, A.A.; Marassi, F.M.; et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature 2012, 491, 779–783. [Google Scholar] [CrossRef] [PubMed]
- Duc, N.M.; Du, Y.; Zhang, C.; Lee, S.Y.; Thorsen, T.S.; Kobilka, B.K.; Chung, K.Y. Effective application of bicelles for conformational analysis of G protein-coupled receptors by hydrogen/deuterium exchange mass spectrometry. J. Am. Soc. Mass Spectrom. 2015, 26, 808–817. [Google Scholar] [CrossRef]
- Schmidt, P.; Bender, B.J.; Kaiser, A.; Gulati, K.; Scheidt, H.A.; Hamm, H.E.; Meiler, J.; Beck-Sickinger, A.G.; Huster, D. Improved in Vitro Folding of the Y2 G Protein-Coupled Receptor into Bicelles. Front. Mol. Biosci. 2017, 4, 100. [Google Scholar] [CrossRef]
- Brea, R.J.; Cole, C.M.; Lyda, B.R.; Ye, L.; Prosser, R.S.; Sunahara, R.K.; Devaraj, N.K. In Situ Reconstitution of the Adenosine A2A Receptor in Spontaneously Formed Synthetic Liposomes. J. Am. Chem. Soc. 2017, 139, 3607–3610. [Google Scholar] [CrossRef]
- Bayburt, T.H.; Grinkova, Y.V.; Sligar, S.G. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Lett. 2002, 2, 853–856. [Google Scholar] [CrossRef]
- Ritchie, T.K.; Grinkova, Y.V.; Bayburt, T.H.; Denisov, I.G.; Zolnerciks, J.K.; Atkins, W.M.; Sligar, S.G. Chapter 11—Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009, 464, 211–231. [Google Scholar] [CrossRef]
- Lavington, S.; Watts, A. Lipid nanoparticle technologies for the study of G protein-coupled receptors in lipid environments. Biophys. Rev. 2020, 12, 1287–1302. [Google Scholar] [CrossRef]
- Segala, E.; Errey, J.C.; Fiez-Vandal, C.; Zhukov, A.; Cooke, R.M. Biosensor-based affinities and binding kinetics of small molecule antagonists to the adenosine A2A receptor reconstituted in HDL like particles. FEBS Lett. 2015, 589, 1399–1405. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Lu, Y.; Wu, D.; Peng, Y.; Loa-Kum-Cheung, W.; Peng, C.; Quinn, R.J.; Shui, W.; Liu, Z.-J. Ligand identification of the adenosine A2A receptor in self-assembled nanodiscs by affinity mass spectrometry. Anal. Methods 2017, 9, 5851–5858. [Google Scholar] [CrossRef]
- Fredriksson, K.; Lottmann, P.; Hinz, S.; Onila, I.; Shymanets, A.; Harteneck, C.; Müller, C.E.; Griesinger, C.; Exner, T.E. Nanodiscs for INPHARMA NMR Characterization of GPCRs: Ligand Binding to the Human A2A Adenosine Receptor. Angew. Chem. Int. Ed Engl. 2017, 56, 5750–5754. [Google Scholar] [CrossRef]
- Mary, S.; Damian, M.; Rahmeh, R.; Mouillac, B.; Marie, J.; Granier, S.; Banères, J.-L. Amphipols in G protein-coupled receptor pharmacology: What are they good for? J. Membr. Biol. 2014, 247, 853–860. [Google Scholar] [CrossRef]
- Zoonens, M.; Popot, J.-L. Amphipols for each season. J. Membr. Biol. 2014, 247, 759–796. [Google Scholar] [CrossRef] [PubMed]
- Overduin, M.; Klumperman, B. Advancing membrane biology with poly(styrene-co-maleic acid)-based native nanodiscs. Eur. Polym. J. 2019, 110, 63–68. [Google Scholar] [CrossRef]
- Grime, R.L.; Goulding, J.; Uddin, R.; Stoddart, L.A.; Hill, S.J.; Poyner, D.R.; Briddon, S.J.; Wheatley, M. Single molecule binding of a ligand to a G-protein-coupled receptor in real time using fluorescence correlation spectroscopy, rendered possible by nano-encapsulation in styrene maleic acid lipid particles. Nanoscale 2020, 12, 11518–11525. [Google Scholar] [CrossRef]
- Routledge, S.J.; Jamshad, M.; Little, H.A.; Lin, Y.-P.; Simms, J.; Thakker, A.; Spickett, C.M.; Bill, R.M.; Dafforn, T.R.; Poyner, D.R.; et al. Ligand-induced conformational changes in a SMALP-encapsulated GPCR. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183235. [Google Scholar] [CrossRef] [PubMed]
- Marty, M.T.; Hoi, K.K.; Robinson, C.V. Interfacing Membrane Mimetics with Mass Spectrometry. Acc. Chem. Res. 2016, 49, 2459–2467. [Google Scholar] [CrossRef] [PubMed]
- Keener, J.E.; Zhang, G.; Marty, M.T. Native Mass Spectrometry of Membrane Proteins. Anal. Chem. 2021, 93, 583–597. [Google Scholar] [CrossRef]
- Young, G.; Hundt, N.; Cole, D.; Fineberg, A.; Andrecka, J.; Tyler, A.; Olerinyova, A.; Ansari, A.; Marklund, E.G.; Collier, M.P.; et al. Quantitative mass imaging of single biological macromolecules. Science 2018, 360, 423–427. [Google Scholar] [CrossRef]
- Olerinyova, A.; Sonn-Segev, A.; Gault, J.; Eichmann, C.; Schimpf, J.; Kopf, A.H.; Rudden, L.S.P.; Ashkinadze, D.; Bomba, R.; Frey, L.; et al. Mass Photometry of Membrane Proteins. Chem 2021, 7, 224–236. [Google Scholar] [CrossRef]
- Schlick, K.H.; Lange, C.K.; Gillispie, G.D.; Cloninger, M.J. Characterization of protein aggregation via intrinsic fluorescence lifetime. J. Am. Chem. Soc. 2009, 131, 16608–16609. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Zhang, M.; Bertheleme, N.; Kara, E.; Strange, P.G.; Byrne, B. Radioligand binding analysis as a tool for quality control of GPCR production for structural characterization: Adenosine A2aR as a template for study. Curr. Protoc. Protein Sci. 2012, 29, 29.3. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Ghosh, S.; Jana, S.; Robertson, N.; Tate, C.G.; Vaidehi, N. How Do Branched Detergents Stabilize GPCRs in Micelles? Biochemistry 2020, 59, 2125–2134. [Google Scholar] [CrossRef]
- Holdgate, G.; Embrey, K.; Milbradt, A.; Davies, G. Biophysical methods in early drug discovery. ADMET DMPK 2019, 7, 222–241. [Google Scholar] [CrossRef] [PubMed]
- van der Velden, W.J.C.; Heitman, L.H.; Rosenkilde, M.M. Perspective: Implications of Ligand-Receptor Binding Kinetics for Therapeutic Targeting of G Protein-Coupled Receptors. ACS Pharmacol. Transl. Sci. 2020, 3, 179–189. [Google Scholar] [CrossRef]
- Copeland, R.A. The drug-target residence time model: A 10-year retrospective. Nat. Rev. Drug Discov. 2016, 15, 87–95. [Google Scholar] [CrossRef]
- Guo, D.; Heitman, L.H.; IJzerman, A.P. Kinetic Aspects of the Interaction between Ligand and G Protein-Coupled Receptor: The Case of the Adenosine Receptors. Chem. Rev. 2017, 117, 38–66. [Google Scholar] [CrossRef] [PubMed]
- IJzerman, A.P.; Guo, D. Drug-Target Association Kinetics in Drug Discovery. Trends Biochem. Sci. 2019, 44, 861–871. [Google Scholar] [CrossRef]
- Rich, R.L.; Myszka, D.G. Advances in surface plasmon resonance biosensor analysis. Curr. Opin. Biotechnol. 2000, 11, 54–61. [Google Scholar] [CrossRef]
- Locatelli-Hoops, S.; Yeliseev, A.A.; Gawrisch, K.; Gorshkova, I. Surface plasmon resonance applied to G protein-coupled receptors. Biomed. Spectrosc. Imaging 2013, 2, 155–181. [Google Scholar] [CrossRef] [PubMed]
- Patching, S.G. Surface plasmon resonance spectroscopy for characterisation of membrane protein-ligand interactions and its potential for drug discovery. Biochim. Biophys. Acta 2014, 1838, 43–55. [Google Scholar] [CrossRef]
- Aristotelous, T.; Hopkins, A.L.; Navratilova, I. Surface plasmon resonance analysis of seven-transmembrane receptors. Methods Enzymol. 2015, 556, 499–525. [Google Scholar] [CrossRef]
- Olaru, A.; Bala, C.; Jaffrezic-Renault, N.; Aboul-Enein, H.Y. Surface plasmon resonance (SPR) biosensors in pharmaceutical analysis. Crit. Rev. Anal. Chem. 2015, 45, 97–105. [Google Scholar] [CrossRef]
- Giannetti, A.M. From experimental design to validated hits a comprehensive walk-through of fragment lead identification using surface plasmon resonance. Methods Enzymol. 2011, 493, 169–218. [Google Scholar] [CrossRef] [PubMed]
- Rich, R.L.; Errey, J.; Marshall, F.; Myszka, D.G. Biacore analysis with stabilized G-protein-coupled receptors. Anal. Biochem. 2011, 409, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, C.A.; Hopkins, A.L.; Navratilova, I. Fragment screening by SPR and advanced application to GPCRs. Prog. Biophys. Mol. Biol. 2014, 116, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Robertson, N.; Jazayeri, A.; Errey, J.; Baig, A.; Hurrell, E.; Zhukov, A.; Langmead, C.J.; Weir, M.; Marshall, F.H. The properties of thermostabilised G protein-coupled receptors (StaRs) and their use in drug discovery. Neuropharmacology 2011, 60, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Congreve, M.; Rich, R.L.; Myszka, D.G.; Figaroa, F.; Siegal, G.; Marshall, F.H. Fragment screening of stabilized G-protein-coupled receptors using biophysical methods. Methods Enzymol. 2011, 493, 115–136. [Google Scholar] [CrossRef]
- Congreve, M.; Andrews, S.P.; Doré, A.S.; Hollenstein, K.; Hurrell, E.; Langmead, C.J.; Mason, J.S.; Ng, I.W.; Tehan, B.; Zhukov, A.; et al. Discovery of 1,2,4-triazine derivatives as adenosine A2A antagonists using structure based drug design. J. Med. Chem. 2012, 55, 1898–1903. [Google Scholar] [CrossRef]
- Zhukov, A.; Andrews, S.P.; Errey, J.C.; Robertson, N.; Tehan, B.; Mason, J.S.; Marshall, F.H.; Weir, M.; Congreve, M. Biophysical mapping of the adenosine A2A receptor. J. Med. Chem. 2011, 54, 4312–4323. [Google Scholar] [CrossRef]
- Roos, H.; Karlsson, R.; Nilshans, H.; Persson, A. Thermodynamic analysis of protein interactions with biosensor technology. J. Mol. Recognit. 1998, 11, 204–210. [Google Scholar] [CrossRef]
- Babazada, H.; Alekberli, T.; Hajieva, P.; Farajov, E. Biosensor-based kinetic and thermodynamic characterization of opioids interaction with human μ-opioid receptor. Eur. J. Pharm. Sci. 2019, 138, 105017. [Google Scholar] [CrossRef]
- Deganutti, G.; Zhukov, A.; Deflorian, F.; Federico, S.; Spalluto, G.; Cooke, R.M.; Moro, S.; Mason, J.S.; Bortolato, A. Impact of protein-ligand solvation and desolvation on transition state thermodynamic properties of adenosine A2A ligand binding kinetics. Silico Pharmacol. 2017, 5, 16. [Google Scholar] [CrossRef]
- Coyle, J.; Walser, R. Applied Biophysical Methods in Fragment-Based Drug Discovery. SLAS Discov. 2020, 25, 471–490. [Google Scholar] [CrossRef]
- Lu, Y.; Qin, S.; Zhang, B.; Dai, A.; Cai, X.; Ma, M.; Gao, Z.-G.; Yang, D.; Stevens, R.C.; Jacobson, K.A.; et al. Accelerating the Throughput of Affinity Mass Spectrometry-Based Ligand Screening toward a G Protein-Coupled Receptor. Anal. Chem. 2019, 91, 8162–8169. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, C.; Robinson, S.; Berizzi, A.; Thompson, L.E.J.; Bird, L.; Culurgioni, S.; Varzandeh, S.; Rawlins, P.B.; Olsen, R.H.J.; Navratilova, I.H. Surface Plasmon Resonance Screening to Identify Active and Selective Adenosine Receptor Binding Fragments. ACS Med. Chem. Lett. 2022, 13, 1172–1181. [Google Scholar] [CrossRef]
- Koretz, K.S.; McGraw, C.E.; Stradley, S.; Elbaradei, A.; Malmstadt, N.; Robinson, A.S. Characterization of binding kinetics of A2AR to Gαs protein by surface plasmon resonance. Biophys. J. 2021, 120, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Damian, L. Isothermal titration calorimetry for studying protein-ligand interactions. Methods Mol. Biol. 2013, 1008, 103–118. [Google Scholar] [CrossRef]
- Pierce, M.M.; Raman, C.S.; Nall, B.T. Isothermal titration calorimetry of protein-protein interactions. Methods 1999, 19, 213–221. [Google Scholar] [CrossRef]
- Draczkowski, P.; Matosiuk, D.; Jozwiak, K. Isothermal titration calorimetry in membrane protein research. J. Pharm. Biomed. Anal. 2014, 87, 313–325. [Google Scholar] [CrossRef]
- Rajarathnam, K.; Rösgen, J. Isothermal titration calorimetry of membrane proteins-progress and challenges. Biochim. Biophys. Acta 2014, 1838, 69–77. [Google Scholar] [CrossRef]
- Magalhaes, A.C.; Dunn, H.; Ferguson, S.S. Regulation of GPCR activity, trafficking and localization by GPCR-interacting proteins. Br. J. Pharmacol. 2012, 165, 1717–1736. [Google Scholar] [CrossRef] [PubMed]
- Klaasse, E.C.; IJzerman, A.P.; de Grip, W.J.; Beukers, M.W. Internalization and desensitization of adenosine receptors. Purinergic Signal. 2008, 4, 21–37. [Google Scholar] [CrossRef]
- Burgueño, J.; Blake, D.J.; Benson, M.A.; Tinsley, C.L.; Esapa, C.T.; Canela, E.I.; Penela, P.; Mallol, J.; Mayor, F.; Lluis, C.; et al. The adenosine A2A receptor interacts with the actin-binding protein alpha-actinin. J. Biol. Chem. 2003, 278, 37545–37552. [Google Scholar] [CrossRef]
- Woods, A.S.; Marcellino, D.; Jackson, S.N.; Franco, R.; Ferré, S.; Agnati, L.F.; Fuxe, K. How calmodulin interacts with the adenosine A2A and the dopamine D2 receptors. J. Proteome Res. 2008, 7, 3428–3434. [Google Scholar] [CrossRef]
- Milojevic, T.; Reiterer, V.; Stefan, E.; Korkhov, V.M.; Dorostkar, M.M.; Ducza, E.; Ogris, E.; Boehm, S.; Freissmuth, M.; Nanoff, C. The ubiquitin-specific protease Usp4 regulates the cell surface level of the A2A receptor. Mol. Pharmacol. 2006, 69, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
- Canela, L.; Luján, R.; Lluís, C.; Burgueño, J.; Mallol, J.; Canela, E.I.; Franco, R.; Ciruela, F. The neuronal Ca2+-binding protein 2 (NECAB2) interacts with the adenosine A2A receptor and modulates the cell surface expression and function of the receptor. Mol. Cell. Neurosci. 2007, 36, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.-N.; Cheng, H.-C.; Chou, J.-L.; Lee, S.-Y.; Lin, Y.-W.; Lai, H.-L.; Chen, H.-M.; Chern, Y. Rescue of p53 blockage by the A2A adenosine receptor via a novel interacting protein, translin-associated protein X. Mol. Pharmacol. 2006, 70, 454–466. [Google Scholar] [CrossRef]
- Gsandtner, I.; Charalambous, C.; Stefan, E.; Ogris, E.; Freissmuth, M.; Zezula, J. Heterotrimeric G protein-independent signaling of a G protein-coupled receptor. Direct binding of ARNO/cytohesin-2 to the carboxyl terminus of the A2A adenosine receptor is necessary for sustained activation of the ERK/MAP kinase pathway. J. Biol. Chem. 2005, 280, 31898–31905. [Google Scholar] [CrossRef]
- Navarro, G.; Hradsky, J.; Lluís, C.; Casadó, V.; McCormick, P.J.; Kreutz, M.R.; Mikhaylova, M. NCS-1 associates with adenosine A2A receptors and modulates receptor function. Front. Mol. Neurosci. 2012, 5, 53. [Google Scholar] [CrossRef] [PubMed]
- Piirainen, H.; Hellman, M.; Tossavainen, H.; Permi, P.; Kursula, P.; Jaakola, V.-P. Human adenosine A2A receptor binds calmodulin with high affinity in a calcium-dependent manner. Biophys. J. 2015, 108, 903–917. [Google Scholar] [CrossRef] [PubMed]
- Piirainen, H.; Taura, J.; Kursula, P.; Ciruela, F.; Jaakola, V.-P. Calcium modulates calmodulin/α-actinin 1 interaction with and agonist-dependent internalization of the adenosine A2A receptor. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 674–686. [Google Scholar] [CrossRef]
- Quast, R.B.; Margeat, E. Studying GPCR conformational dynamics by single molecule fluorescence. Mol. Cell. Endocrinol. 2019, 493, 110469. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Fürstenberg, A.; Huber, T. Labeling and Single-Molecule Methods To Monitor G Protein-Coupled Receptor Dynamics. Chem. Rev. 2017, 117, 186–245. [Google Scholar] [CrossRef]
- Hoffmann, C.; Gaietta, G.; Bünemann, M.; Adams, S.R.; Oberdorff-Maass, S.; Behr, B.; Vilardaga, J.-P.; Tsien, R.Y.; Ellisman, M.H.; Lohse, M.J. A FlAsH-based FRET approach to determine G protein-coupled receptor activation in living cells. Nat. Methods 2005, 2, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Stoddart, L.A.; Kindon, N.D.; Otun, O.; Harwood, C.R.; Patera, F.; Veprintsev, D.B.; Woolard, J.; Briddon, S.J.; Franks, H.A.; Hill, S.J.; et al. Ligand-directed covalent labelling of a GPCR with a fluorescent tag in live cells. Commun. Biol. 2020, 3, 722. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Thakur, N.; Ray, A.P.; Jin, B.; Obeng, S.; McCurdy, C.R.; McMahon, L.R.; Gutiérrez-de-Terán, H.; Eddy, M.T.; Lamichhane, R. Slow conformational dynamics of the human A2A adenosine receptor are temporally ordered. Structure 2022, 30, 329–337.e5. [Google Scholar] [CrossRef]
- Fernandes, D.D.; Neale, C.; Gomes, G.-N.W.; Li, Y.; Malik, A.; Pandey, A.; Orazietti, A.P.; Wang, X.; Ye, L.; Scott Prosser, R.; et al. Ligand modulation of the conformational dynamics of the A2A adenosine receptor revealed by single-molecule fluorescence. Sci. Rep. 2021, 11, 5910. [Google Scholar] [CrossRef] [PubMed]
- Zemella, A.; Richter, T.; Thoring, L.; Kubick, S. A Combined Cell-Free Protein Synthesis and Fluorescence-Based Approach to Investigate GPCR Binding Properties. Methods Mol. Biol. 2019, 1947, 57–77. [Google Scholar] [CrossRef]
- Raingeval, C.; Krimm, I. NMR investigation of protein-ligand interactions for G-protein coupled receptors. Future Med. Chem. 2019, 11, 1811–1825. [Google Scholar] [CrossRef]
- Stauch, B.; Orts, J.; Carlomagno, T. The description of protein internal motions aids selection of ligand binding poses by the INPHARMA method. J. Biomol. NMR 2012, 54, 245–256. [Google Scholar] [CrossRef]
- Yen, H.-Y.; Hoi, K.K.; Liko, I.; Hedger, G.; Horrell, M.R.; Song, W.; Wu, D.; Heine, P.; Warne, T.; Lee, Y.; et al. PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature 2018, 559, 423–427. [Google Scholar] [CrossRef]
- Agasid, M.T.; Sørensen, L.; Urner, L.H.; Yan, J.; Robinson, C.V. The Effects of Sodium Ions on Ligand Binding and Conformational States of G Protein-Coupled Receptors-Insights from Mass Spectrometry. J. Am. Chem. Soc. 2021, 143, 4085–4089. [Google Scholar] [CrossRef] [PubMed]
- Olsen, R.H.J.; English, J.G. Advancements in G protein-coupled receptor biosensors to study GPCR-G protein coupling. Br. J. Pharmacol. 2022. [Google Scholar] [CrossRef]
- Zhou, Y.; Meng, J.; Xu, C.; Liu, J. Multiple GPCR Functional Assays Based on Resonance Energy Transfer Sensors. Front. Cell Dev. Biol. 2021, 9, 611443. [Google Scholar] [CrossRef] [PubMed]
- Milligan, G. Principles: Extending the utility of [35S]GTPγS binding assays. Trends Pharmacol. Sci. 2003, 24, 87–90. [Google Scholar] [CrossRef]
- McEwen, D.P.; Gee, K.R.; Kang, H.C.; Neubig, R.R. Fluorescent BODIPY-GTP analogs: Real-time measurement of nucleotide binding to G proteins. Anal. Biochem. 2001, 291, 109–117. [Google Scholar] [CrossRef]
- Ravyn, V.; Bostwick, J.R. Functional coupling of the Galpha(olf) variant XLGalpha(olf) with the human adenosine A2A receptor. J. Recept. Signal Transduct. Res. 2006, 26, 241–258. [Google Scholar] [CrossRef] [PubMed]
- Beckner, R.L.; Zoubak, L.; Hines, K.G.; Gawrisch, K.; Yeliseev, A.A. Probing thermostability of detergent-solubilized CB2 receptor by parallel G protein-activation and ligand-binding assays. J. Biol. Chem. 2020, 295, 181–190. [Google Scholar] [CrossRef]
- Arcemisbéhère, L.; Sen, T.; Boudier, L.; Balestre, M.-N.; Gaibelet, G.; Detouillon, E.; Orcel, H.; Mendre, C.; Rahmeh, R.; Granier, S.; et al. Leukotriene BLT2 receptor monomers activate the G(i2) GTP-binding protein more efficiently than dimers. J. Biol. Chem. 2010, 285, 6337–6347. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, S.; Ghirlando, R.; White, J.F.; Gvozdenovic-Jeremic, J.; Northup, J.K.; Grisshammer, R. Modulation of the interaction between neurotensin receptor NTS1 and Gq protein by lipid. J. Mol. Biol. 2012, 417, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Damian, M.; Marie, J.; Leyris, J.-P.; Fehrentz, J.-A.; Verdié, P.; Martinez, J.; Banères, J.-L.; Mary, S. High constitutive activity is an intrinsic feature of ghrelin receptor protein: A study with a functional monomeric GHS-R1a receptor reconstituted in lipid discs. J. Biol. Chem. 2012, 287, 3630–3641. [Google Scholar] [CrossRef]
- Gregorio, G.G.; Masureel, M.; Hilger, D.; Terry, D.S.; Juette, M.; Zhao, H.; Zhou, Z.; Perez-Aguilar, J.M.; Hauge, M.; Mathiasen, S.; et al. Single-molecule analysis of ligand efficacy in β2AR-G-protein activation. Nature 2017, 547, 68–73. [Google Scholar] [CrossRef]
- Damian, M.; Louet, M.; Gomes, A.A.S.; M’Kadmi, C.; Denoyelle, S.; Cantel, S.; Mary, S.; Bisch, P.M.; Fehrentz, J.-A.; Catoire, L.J.; et al. Allosteric modulation of ghrelin receptor signaling by lipids. Nat. Commun. 2021, 12, 3938. [Google Scholar] [CrossRef] [PubMed]
- Bayburt, T.H.; Vishnivetskiy, S.A.; McLean, M.A.; Morizumi, T.; Huang, C.-C.; Tesmer, J.J.G.; Ernst, O.P.; Sligar, S.G.; Gurevich, V.V. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J. Biol. Chem. 2011, 286, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Baidya, M.; Dwivedi-Agnihotri, H.; Shukla, A.K. Site-directed labeling of β-arrestin with monobromobimane for measuring their interaction with G protein-coupled receptors. Methods Enzymol. 2020, 633, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Sommer, M.E.; Farrens, D.L. Arrestin can act as a regulator of rhodopsin photochemistry. Vis. Res. 2006, 46, 4532–4546. [Google Scholar] [CrossRef] [PubMed]
- Rahmeh, R.; Damian, M.; Cottet, M.; Orcel, H.; Mendre, C.; Durroux, T.; Sharma, K.S.; Durand, G.; Pucci, B.; Trinquet, E.; et al. Structural insights into biased G protein-coupled receptor signaling revealed by fluorescence spectroscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 6733–6738. [Google Scholar] [CrossRef]
- Kumari, P.; Srivastava, A.; Banerjee, R.; Ghosh, E.; Gupta, P.; Ranjan, R.; Chen, X.; Gupta, B.; Gupta, C.; Jaiman, D.; et al. Functional competence of a partially engaged GPCR-β-arrestin complex. Nat. Commun. 2016, 7, 13416. [Google Scholar] [CrossRef]
- Staus, D.P.; Wingler, L.M.; Choi, M.; Pani, B.; Manglik, A.; Kruse, A.C.; Lefkowitz, R.J. Sortase ligation enables homogeneous GPCR phosphorylation to reveal diversity in β-arrestin coupling. Proc. Natl. Acad. Sci. USA 2018, 115, 3834–3839. [Google Scholar] [CrossRef]
- Prosser, R.S.; Ye, L.; Pandey, A.; Orazietti, A. Activation processes in ligand-activated G protein-coupled receptors: A case study of the adenosine A2A receptor. Bioessays 2017, 39, 1700072. [Google Scholar] [CrossRef] [PubMed]
- Welihinda, A.A.; Kaur, M.; Greene, K.; Zhai, Y.; Amento, E.P. The adenosine metabolite inosine is a functional agonist of the adenosine A2A receptor with a unique signaling bias. Cell. Signal. 2016, 28, 552–560. [Google Scholar] [CrossRef]
- Kolb, P.; Kenakin, T.; Alexander, S.P.H.; Bermudez, M.; Bohn, L.M.; Breinholt, C.S.; Bouvier, M.; Hill, S.J.; Kostenis, E.; Martemyanov, K.A.; et al. Community guidelines for GPCR ligand bias: IUPHAR review 32. Br. J. Pharmacol. 2022, 179, 3651–3674. [Google Scholar] [CrossRef]
- Benleulmi-Chaachoua, A.; Chen, L.; Sokolina, K.; Wong, V.; Jurisica, I.; Emerit, M.B.; Darmon, M.; Espin, A.; Stagljar, I.; Tafelmeyer, P.; et al. Protein interactome mining defines melatonin MT1 receptors as integral component of presynaptic protein complexes of neurons. J. Pineal Res. 2016, 60, 95–108. [Google Scholar] [CrossRef]
- Overton, M.C.; Blumer, K.J. The extracellular N-terminal domain and transmembrane domains 1 and 2 mediate oligomerization of a yeast G protein-coupled receptor. J. Biol. Chem. 2002, 277, 41463–41472. [Google Scholar] [CrossRef] [PubMed]
- Mahesh, G.; Jaiswal, P.; Dey, S.; Sengupta, J.; Mukherjee, S. Cloning, Expression, Purification and Characterization of Oligomeric States of the Native 5HT2A G-Protein-Coupled Receptor. Protein Pept. Lett. 2018, 25, 390–397. [Google Scholar] [CrossRef]
- Fernández-Dueñas, V.; Bonaventura, J.; Aso, E.; Luján, R.; Ferré, S.; Ciruela, F. Overcoming the Challenges of Detecting GPCR Oligomerization in the Brain. Curr. Neuropharmacol. 2022, 20, 1035–1045. [Google Scholar] [CrossRef]
- Mansoor, S.; Kayık, G.; Durdagi, S.; Sensoy, O. Mechanistic insight into the impact of a bivalent ligand on the structure and dynamics of a GPCR oligomer. Comput. Struct. Biotechnol. J. 2022, 20, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Canals, M.; Burgueño, J.; Marcellino, D.; Cabello, N.; Canela, E.I.; Mallol, J.; Agnati, L.; Ferré, S.; Bouvier, M.; Fuxe, K.; et al. Homodimerization of adenosine A2A receptors: Qualitative and quantitative assessment by fluorescence and bioluminescence energy transfer. J. Neurochem. 2004, 88, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Ciruela, F.; Casadó, V.; Rodrigues, R.J.; Luján, R.; Burgueño, J.; Canals, M.; Borycz, J.; Rebola, N.; Goldberg, S.R.; Mallol, J.; et al. Presynaptic control of striatal glutamatergic neurotransmission by adenosine A1-A2A receptor heteromers. J. Neurosci. 2006, 26, 2080–2087. [Google Scholar] [CrossRef]
- Franco, R.; Cordomí, A.; Del Llinas Torrent, C.; Lillo, A.; Serrano-Marín, J.; Navarro, G.; Pardo, L. Structure and function of adenosine receptor heteromers. Cell. Mol. Life Sci. 2021, 78, 3957–3968. [Google Scholar] [CrossRef] [PubMed]
- Carriba, P.; Ortiz, O.; Patkar, K.; Justinova, Z.; Stroik, J.; Themann, A.; Müller, C.; Woods, A.S.; Hope, B.T.; Ciruela, F.; et al. Striatal adenosine A2A and cannabinoid CB1 receptors form functional heteromeric complexes that mediate the motor effects of cannabinoids. Neuropsychopharmacology 2007, 32, 2249–2259. [Google Scholar] [CrossRef] [PubMed]
- Hillion, J.; Canals, M.; Torvinen, M.; Casado, V.; Scott, R.; Terasmaa, A.; Hansson, A.; Watson, S.; Olah, M.E.; Mallol, J.; et al. Coaggregation, cointernalization, and codesensitization of adenosine A2A receptors and dopamine D2 receptors. J. Biol. Chem. 2002, 277, 18091–18097. [Google Scholar] [CrossRef] [PubMed]
- Canals, M.; Marcellino, D.; Fanelli, F.; Ciruela, F.; de Benedetti, P.; Goldberg, S.R.; Neve, K.; Fuxe, K.; Agnati, L.F.; Woods, A.S.; et al. Adenosine A2A-dopamine D2 receptor-receptor heteromerization: Qualitative and quantitative assessment by fluorescence and bioluminescence energy transfer. J. Biol. Chem. 2003, 278, 46741–46749. [Google Scholar] [CrossRef]
- Kamiya, T.; Saitoh, O.; Yoshioka, K.; Nakata, H. Oligomerization of adenosine A2A and dopamine D2 receptors in living cells. Biochem. Biophys. Res. Commun. 2003, 306, 544–549. [Google Scholar] [CrossRef]
- Torvinen, M.; Marcellino, D.; Canals, M.; Agnati, L.F.; Lluis, C.; Franco, R.; Fuxe, K. Adenosine A2A receptor and dopamine D3 receptor interactions: Evidence of functional A2A/D3 heteromeric complexes. Mol. Pharmacol. 2005, 67, 400–407. [Google Scholar] [CrossRef]
- Cabello, N.; Gandía, J.; Bertarelli, D.C.G.; Watanabe, M.; Lluís, C.; Franco, R.; Ferré, S.; Luján, R.; Ciruela, F. Metabotropic glutamate type 5, dopamine D2 and adenosine A2a receptors form higher-order oligomers in living cells. J. Neurochem. 2009, 109, 1497–1507. [Google Scholar] [CrossRef] [PubMed]
- Trifilieff, P.; Rives, M.-L.; Urizar, E.; Piskorowski, R.A.; Vishwasrao, H.D.; Castrillon, J.; Schmauss, C.; Slättman, M.; Gullberg, M.; Javitch, J.A. Detection of antigen interactions ex vivo by proximity ligation assay: Endogenous dopamine D2-adenosine A2A receptor complexes in the striatum. Biotechniques 2011, 51, 111–118. [Google Scholar] [CrossRef]
- Fernández-Dueñas, V.; Gómez-Soler, M.; Valle-León, M.; Watanabe, M.; Ferrer, I.; Ciruela, F. Revealing Adenosine A2A-Dopamine D2 Receptor Heteromers in Parkinson’s Disease Post-Mortem Brain through a New AlphaScreen-Based Assay. Int. J. Mol. Sci. 2019, 20, 3600. [Google Scholar] [CrossRef] [PubMed]
- Beggiato, S.; Antonelli, T.; Tomasini, M.C.; Borelli, A.C.; Agnati, L.F.; Tanganelli, S.; Fuxe, K.; Ferraro, L. Adenosine A2A-D2 receptor-receptor interactions in putative heteromers in the regulation of the striato-pallidal gaba pathway: Possible relevance for parkinson’s disease and its treatment. Curr. Protein Pept. Sci. 2014, 15, 673–680. [Google Scholar] [CrossRef]
- Ferré, S.; Bonaventura, J.; Tomasi, D.; Navarro, G.; Moreno, E.; Cortés, A.; Lluís, C.; Casadó, V.; Volkow, N.D. Allosteric mechanisms within the adenosine A2A-dopamine D2 receptor heterotetramer. Neuropharmacology 2016, 104, 154–160. [Google Scholar] [CrossRef]
- Romero-Fernandez, W.; Taura, J.J.; Crans, R.A.J.; Lopez-Cano, M.; Fores-Pons, R.; Narváez, M.; Carlsson, J.; Ciruela, F.; Fuxe, K.; Borroto-Escuela, D.O. The mGlu5 Receptor Protomer-Mediated Dopamine D2 Receptor Trans-Inhibition Is Dependent on the Adenosine A2A Receptor Protomer: Implications for Parkinson’s Disease. Mol. Neurobiol. 2022, 59, 5955–5969. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.A.; Denisov, I.G.; Sligar, S.G. Nanodiscs as a new tool to examine lipid-protein interactions. Methods Mol. Biol. 2013, 974, 415–433. [Google Scholar] [CrossRef] [PubMed]
- White, J.F.; Grodnitzky, J.; Louis, J.M.; Trinh, L.B.; Shiloach, J.; Gutierrez, J.; Northup, J.K.; Grisshammer, R. Dimerization of the class A G protein-coupled neurotensin receptor NTS1 alters G protein interaction. Proc. Natl. Acad. Sci. USA 2007, 104, 12199–12204. [Google Scholar] [CrossRef] [PubMed]
- Bayburt, T.H.; Leitz, A.J.; Xie, G.; Oprian, D.D.; Sligar, S.G. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J. Biol. Chem. 2007, 282, 14875–14881. [Google Scholar] [CrossRef]
- El Moustaine, D.; Granier, S.; Doumazane, E.; Scholler, P.; Rahmeh, R.; Bron, P.; Mouillac, B.; Banères, J.-L.; Rondard, P.; Pin, J.-P. Distinct roles of metabotropic glutamate receptor dimerization in agonist activation and G-protein coupling. Proc. Natl. Acad. Sci. USA 2012, 109, 16342–16347. [Google Scholar] [CrossRef]
- Mary, S.; Fehrentz, J.-A.; Damian, M.; Gaibelet, G.; Orcel, H.; Verdié, P.; Mouillac, B.; Martinez, J.; Marie, J.; Banères, J.-L. Heterodimerization with Its splice variant blocks the ghrelin receptor 1a in a non-signaling conformation: A study with a purified heterodimer assembled into lipid discs. J. Biol. Chem. 2013, 288, 24656–24665. [Google Scholar] [CrossRef]
- Mesnier, D.; Banères, J.-L. Cooperative conformational changes in a G-protein-coupled receptor dimer, the leukotriene B(4) receptor BLT1. J. Biol. Chem. 2004, 279, 49664–49670. [Google Scholar] [CrossRef]
- Damian, M.; Pons, V.; Renault, P.; M’Kadmi, C.; Delort, B.; Hartmann, L.; Kaya, A.I.; Louet, M.; Gagne, D.; Ben Haj Salah, K.; et al. GHSR-D2R heteromerization modulates dopamine signaling through an effect on G protein conformation. Proc. Natl. Acad. Sci. USA 2018, 115, 4501–4506. [Google Scholar] [CrossRef]
- Tsukamoto, H.; Sinha, A.; DeWitt, M.; Farrens, D.L. Monomeric rhodopsin is the minimal functional unit required for arrestin binding. J. Mol. Biol. 2010, 399, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Piirainen, H.; Ashok, Y.; Nanekar, R.T.; Jaakola, V.-P. Structural features of adenosine receptors: From crystal to function. Biochim. Biophys. Acta 2011, 1808, 1233–1244. [Google Scholar] [CrossRef]
- Langelaan, D.N.; Ngweniform, P.; Rainey, J.K. Biophysical characterization of G-protein coupled receptor-peptide ligand binding. Biochem. Cell Biol. 2011, 89, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Marcoux, J.; Cianférani, S. Towards integrative structural mass spectrometry: Benefits from hybrid approaches. Methods 2015, 89, 4–12. [Google Scholar] [CrossRef]
- Dafun, A.S.; Marcoux, J. Structural mass spectrometry of membrane proteins. Biochim. Biophys. Acta Proteins Proteom. 2022, 1870, 140813. [Google Scholar] [CrossRef] [PubMed]
- Khanal, A.; Pan, Y.; Brown, L.S.; Konermann, L. Pulsed hydrogen/deuterium exchange mass spectrometry for time-resolved membrane protein folding studies. J. Mass Spectrom. 2012, 47, 1620–1626. [Google Scholar] [CrossRef]
- Masson, G.R.; Burke, J.E.; Ahn, N.G.; Anand, G.S.; Borchers, C.; Brier, S.; Bou-Assaf, G.M.; Engen, J.R.; Englander, S.W.; Faber, J.; et al. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods 2019, 16, 595–602. [Google Scholar] [CrossRef]
- Möller, I.R.; Slivacka, M.; Hausner, J.; Nielsen, A.K.; Pospíšilová, E.; Merkle, P.S.; Lišková, R.; Polák, M.; Loland, C.J.; Kádek, A.; et al. Improving the Sequence Coverage of Integral Membrane Proteins during Hydrogen/Deuterium Exchange Mass Spectrometry Experiments. Anal. Chem. 2019, 91, 10970–10978. [Google Scholar] [CrossRef]
- Trabjerg, E.; Nazari, Z.E.; Rand, K.D. Conformational analysis of complex protein states by hydrogen/deuterium exchange mass spectrometry (HDX-MS): Challenges and emerging solutions. TrAC Trends Anal. Chem. 2018, 106, 125–138. [Google Scholar] [CrossRef]
- Martens, C.; Politis, A. A glimpse into the molecular mechanism of integral membrane proteins through hydrogen-deuterium exchange mass spectrometry. Protein Sci. 2020, 29, 1285–1301. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rempel, D.L.; Zhang, J.; Sharma, A.K.; Mirica, L.M.; Gross, M.L. Pulsed hydrogen-deuterium exchange mass spectrometry probes conformational changes in amyloid beta (Aβ) peptide aggregation. Proc. Natl. Acad. Sci. USA 2013, 110, 14604–14609. [Google Scholar] [CrossRef]
- Du, Y.; Duc, N.M.; Rasmussen, S.G.F.; Hilger, D.; Kubiak, X.; Wang, L.; Bohon, J.; Kim, H.R.; Wegrecki, M.; Asuru, A.; et al. Assembly of a GPCR-G Protein Complex. Cell 2019, 177, 1232–1242.e11. [Google Scholar] [CrossRef]
- Konermann, L.; Tong, X.; Pan, Y. Protein structure and dynamics studied by mass spectrometry: H/D exchange, hydroxyl radical labeling, and related approaches. J. Mass Spectrom. 2008, 43, 1021–1036. [Google Scholar] [CrossRef]
- Wang, L.; Chance, M.R. Protein Footprinting Comes of Age: Mass Spectrometry for Biophysical Structure Assessment. Mol. Cell. Proteom. 2017, 16, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, C.; Piotrowski, C.; Aebersold, R.; Amaral, B.C.; Andrews, P.; Bernfur, K.; Borchers, C.; Brodie, N.I.; Bruce, J.E.; Cao, Y.; et al. First Community-Wide, Comparative Cross-Linking Mass Spectrometry Study. Anal. Chem. 2019, 91, 6953–6961. [Google Scholar] [CrossRef]
- Steigenberger, B.; Albanese, P.; Heck, A.J.R.; Scheltema, R.A. To Cleave or Not To Cleave in XL-MS? J. Am. Soc. Mass Spectrom. 2020, 31, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Ma, Z.; Tong, J.; Tang, Y.; Li, S.; Qin, S.; Lou, R.; Zhao, S.; Lei, X.; Shui, W. Evaluation of chemical cross-linkers for in-depth structural analysis of G protein-coupled receptors through cross-linking mass spectrometry. Anal. Chim. Acta 2020, 1102, 53–62. [Google Scholar] [CrossRef]
- Jones, A.X.; Cao, Y.; Tang, Y.-L.; Wang, J.-H.; Ding, Y.-H.; Tan, H.; Chen, Z.-L.; Fang, R.-Q.; Yin, J.; Chen, R.-C.; et al. Improving mass spectrometry analysis of protein structures with arginine-selective chemical cross-linkers. Nat. Commun. 2019, 10, 3911. [Google Scholar] [CrossRef]
- Rosenbaum, D.M.; Cherezov, V.; Hanson, M.A.; Rasmussen, S.G.F.; Thian, F.S.; Kobilka, T.S.; Choi, H.-J.; Yao, X.-J.; Weis, W.I.; Stevens, R.C.; et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science 2007, 318, 1266–1273. [Google Scholar] [CrossRef]
- Magnani, F.; Serrano-Vega, M.J.; Shibata, Y.; Abdul-Hussein, S.; Lebon, G.; Miller-Gallacher, J.; Singhal, A.; Strege, A.; Thomas, J.A.; Tate, C.G. A mutagenesis and screening strategy to generate optimally thermostabilized membrane proteins for structural studies. Nat. Protoc. 2016, 11, 1554–1571. [Google Scholar] [CrossRef] [PubMed]
- Ciancetta, A.; Rubio, P.; Lieberman, D.I.; Jacobson, K.A. A3 adenosine receptor activation mechanisms: Molecular dynamics analysis of inactive, active, and fully active states. J. Comput. Aided Mol. Des. 2019, 33, 983–996. [Google Scholar] [CrossRef] [PubMed]
- Venkatakrishnan, A.J.; Deupi, X.; Lebon, G.; Tate, C.G.; Schertler, G.F.; Babu, M.M. Molecular signatures of G-protein-coupled receptors. Nature 2013, 494, 185–194. [Google Scholar] [CrossRef]
- Rucktooa, P.; Cheng, R.K.Y.; Segala, E.; Geng, T.; Errey, J.C.; Brown, G.A.; Cooke, R.M.; Marshall, F.H.; Doré, A.S. Towards high throughput GPCR crystallography: In Meso soaking of Adenosine A2A Receptor crystals. Sci. Rep. 2018, 8, 41. [Google Scholar] [CrossRef]
- García-Nafría, J.; Lee, Y.; Bai, X.; Carpenter, B.; Tate, C.G. Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. eLife 2018, 7, e35946. [Google Scholar] [CrossRef]
- Amelia, T.; van Veldhoven, J.P.D.; Falsini, M.; Liu, R.; Heitman, L.H.; van Westen, G.J.P.; Segala, E.; Verdon, G.; Cheng, R.K.Y.; Cooke, R.M.; et al. Crystal Structure and Subsequent Ligand Design of a Nonriboside Partial Agonist Bound to the Adenosine A2A Receptor. J. Med. Chem. 2021, 64, 3827–3842. [Google Scholar] [CrossRef] [PubMed]
- Venkatakrishnan, A.J.; Deupi, X.; Lebon, G.; Heydenreich, F.M.; Flock, T.; Miljus, T.; Balaji, S.; Bouvier, M.; Veprintsev, D.B.; Tate, C.G.; et al. Diverse activation pathways in class A GPCRs converge near the G-protein-coupling region. Nature 2016, 536, 484–487. [Google Scholar] [CrossRef]
- Shiriaeva, A.; Park, D.; Kim, G.; Lee, Y.; Hou, X.; Jarhad, D.B.; Kim, G.; Yu, J.; Hyun, Y.E.; Kim, W.; et al. GPCR Agonist-to-Antagonist Conversion: Enabling the Design of Nucleoside Functional Switches for the A2A Adenosine Receptor. J. Med. Chem. 2022, 65, 11648–11657. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Bachhawat, P.; Chu, M.L.-H.; Wood, M.; Ceska, T.; Sands, Z.A.; Mercier, J.; Lebon, F.; Kobilka, T.S.; Kobilka, B.K. Crystal structure of the adenosine A2A receptor bound to an antagonist reveals a potential allosteric pocket. Proc. Natl. Acad. Sci. USA 2017, 114, 2066–2071. [Google Scholar] [CrossRef]
- Martin-Garcia, J.M.; Conrad, C.E.; Nelson, G.; Stander, N.; Zatsepin, N.A.; Zook, J.; Zhu, L.; Geiger, J.; Chun, E.; Kissick, D.; et al. Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation. IUCrJ 2017, 4, 439–454. [Google Scholar] [CrossRef] [PubMed]
- Weinert, T.; Olieric, N.; Cheng, R.; Brünle, S.; James, D.; Ozerov, D.; Gashi, D.; Vera, L.; Marsh, M.; Jaeger, K.; et al. Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons. Nat. Commun. 2017, 8, 542. [Google Scholar] [CrossRef]
- Martin-Garcia, J.M.; Zhu, L.; Mendez, D.; Lee, M.-Y.; Chun, E.; Li, C.; Hu, H.; Subramanian, G.; Kissick, D.; Ogata, C.; et al. High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source. IUCrJ 2019, 6, 412–425. [Google Scholar] [CrossRef]
- Ishchenko, A.; Stauch, B.; Han, G.W.; Batyuk, A.; Shiriaeva, A.; Li, C.; Zatsepin, N.; Weierstall, U.; Liu, W.; Nango, E.; et al. Toward G protein-coupled receptor structure-based drug design using X-ray lasers. IUCrJ 2019, 6, 1106–1119. [Google Scholar] [CrossRef] [PubMed]
- Shimazu, Y.; Tono, K.; Tanaka, T.; Yamanaka, Y.; Nakane, T.; Mori, C.; Terakado Kimura, K.; Fujiwara, T.; Sugahara, M.; Tanaka, R.; et al. High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure. J. Appl. Crystallogr. 2019, 52, 1280–1288. [Google Scholar] [CrossRef]
- Ihara, K.; Hato, M.; Nakane, T.; Yamashita, K.; Kimura-Someya, T.; Hosaka, T.; Ishizuka-Katsura, Y.; Tanaka, R.; Tanaka, T.; Sugahara, M.; et al. Isoprenoid-chained lipid EROCOC17+4: A new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography. Sci. Rep. 2020, 10, 19305. [Google Scholar] [CrossRef]
- García-Nafría, J.; Tate, C.G. Structure determination of GPCRs: Cryo-EM compared with X-ray crystallography. Biochem. Soc. Trans. 2021, 49, 2345–2355. [Google Scholar] [CrossRef] [PubMed]
- Vénien-Bryan, C.; Li, Z.; Vuillard, L.; Boutin, J.A. Cryo-electron microscopy and X-ray crystallography: Complementary approaches to structural biology and drug discovery. Acta Crystallogr. F Struct. Biol. Commun. 2017, 73, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Boutin, J.A.; Li, Z.; Vuillard, L.; Vénien-Bryan, C. La cryo-microscopie, une alternative à la cristallographie aux rayons X? Med. Sci. 2016, 32, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, M.G.; Battisti, A.J.; Plevka, P. Future prospects. Adv. Protein Chem. Struct. Biol. 2011, 82, 101–121. [Google Scholar] [CrossRef]
- Blundell, T.L.; Chaplin, A.K. The resolution revolution in X-ray diffraction, Cryo-EM and other Technologies. Prog. Biophys. Mol. Biol. 2021, 160, 2–4. [Google Scholar] [CrossRef]
- Glukhova, A.; Draper-Joyce, C.J.; Sunahara, R.K.; Christopoulos, A.; Wootten, D.; Sexton, P.M. Rules of Engagement: GPCRs and G Proteins. ACS Pharmacol. Transl. Sci. 2018, 1, 73–83. [Google Scholar] [CrossRef]
- Liang, Y.-L.; Khoshouei, M.; Radjainia, M.; Zhang, Y.; Glukhova, A.; Tarrasch, J.; Thal, D.M.; Furness, S.G.B.; Christopoulos, G.; Coudrat, T.; et al. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature 2017, 546, 118–123. [Google Scholar] [CrossRef]
- Martynowycz, M.W.; Shiriaeva, A.; Ge, X.; Hattne, J.; Nannenga, B.L.; Cherezov, V.; Gonen, T. MicroED structure of the human adenosine receptor determined from a single nanocrystal in LCP. Proc. Natl. Acad. Sci. USA 2021, 118, e2106041118. [Google Scholar] [CrossRef]
- Frauenfelder, H.; Sligar, S.G.; Wolynes, P.G. The energy landscapes and motions of proteins. Science 1991, 254, 1598–1603. [Google Scholar] [CrossRef]
- Kitevski-LeBlanc, J.L.; Prosser, R.S. Current applications of 19F NMR to studies of protein structure and dynamics. Prog. Nucl. Magn. Reson. Spectrosc. 2012, 62, 1–33. [Google Scholar] [CrossRef]
- Picard, L.-P.; Prosser, R.S. Advances in the study of GPCRs by 19F NMR. Curr. Opin. Struct. Biol. 2021, 69, 169–176. [Google Scholar] [CrossRef]
- Liu, D.; Wüthrich, K. Ring current shifts in (19)F-NMR of membrane proteins. J. Biomol. NMR 2016, 65, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Hagn, F.; Etzkorn, M.; Raschle, T.; Wagner, G. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J. Am. Chem. Soc. 2013, 135, 1919–1925. [Google Scholar] [CrossRef]
- Pozza, A.; Giraud, F.; Cece, Q.; Casiraghi, M.; Point, E.; Damian, M.; Le Bon, C.; Moncoq, K.; Banères, J.-L.; Lescop, E.; et al. Exploration of the dynamic interplay between lipids and membrane proteins by hydrostatic pressure. Nat. Commun. 2022, 13, 1780. [Google Scholar] [CrossRef] [PubMed]
- Casiraghi, M.; Damian, M.; Lescop, E.; Point, E.; Moncoq, K.; Morellet, N.; Levy, D.; Marie, J.; Guittet, E.; Banères, J.-L.; et al. Functional Modulation of a G Protein-Coupled Receptor Conformational Landscape in a Lipid Bilayer. J. Am. Chem. Soc. 2016, 138, 11170–11175. [Google Scholar] [CrossRef]
- Nygaard, R.; Frimurer, T.M.; Holst, B.; Rosenkilde, M.M.; Schwartz, T.W. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 2009, 30, 249–259. [Google Scholar] [CrossRef]
- Eddy, M.T.; Gao, Z.-G.; Mannes, P.; Patel, N.; Jacobson, K.A.; Katritch, V.; Stevens, R.C.; Wüthrich, K. Extrinsic Tryptophans as NMR Probes of Allosteric Coupling in Membrane Proteins: Application to the A2A Adenosine Receptor. J. Am. Chem. Soc. 2018, 140, 8228–8235. [Google Scholar] [CrossRef]
- Eddy, M.T.; Martin, B.T.; Wüthrich, K. A2A Adenosine Receptor Partial Agonism Related to Structural Rearrangements in an Activation Microswitch. Structure 2021, 29, 170–176.e3. [Google Scholar] [CrossRef] [PubMed]
- Casiraghi, M.; Damian, M.; Lescop, E.; Banères, J.-L.; Catoire, L.J. Illuminating the Energy Landscape of GPCRs: The Key Contribution of Solution-State NMR Associated with Escherichia coli as an Expression Host. Biochemistry 2018, 57, 2297–2307. [Google Scholar] [CrossRef]
- Sekhar, A.; Kay, L.E. NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc. Natl. Acad. Sci. USA 2013, 110, 12867–12874. [Google Scholar] [CrossRef]
- Gunkel, M.; Schöneberg, J.; Alkhaldi, W.; Irsen, S.; Noé, F.; Kaupp, U.B.; Al-Amoudi, A. Higher-order architecture of rhodopsin in intact photoreceptors and its implication for phototransduction kinetics. Structure 2015, 23, 628–638. [Google Scholar] [CrossRef]
- Fotiadis, D.; Liang, Y.; Filipek, S.; Saperstein, D.A.; Engel, A.; Palczewski, K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature 2003, 421, 127–128. [Google Scholar] [CrossRef]
- Sapra, K.T.; Spoerri, P.M.; Engel, A.; Alsteens, D.; Müller, D.J. Seeing and sensing single G protein-coupled receptors by atomic force microscopy. Curr. Opin. Cell Biol. 2019, 57, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, K.A.; Tosh, D.K.; Jain, S.; Gao, Z.-G. Historical and Current Adenosine Receptor Agonists in Preclinical and Clinical Development. Front. Cell. Neurosci. 2019, 13, 124. [Google Scholar] [CrossRef]
- Melman, A.; Gao, Z.-G.; Kumar, D.; Wan, T.C.; Gizewski, E.; Auchampach, J.A.; Jacobson, K.A. Design of (N)-methanocarba adenosine 5′-uronamides as species-independent A3 receptor-selective agonists. Bioorg. Med. Chem. Lett. 2008, 18, 2813–2819. [Google Scholar] [CrossRef]
- Ivanov, A.A.; Jacobson, K.A. Molecular modeling of a PAMAM-CGS21680 dendrimer bound to an A2A adenosine receptor homodimer. Bioorg. Med. Chem. Lett. 2008, 18, 4312–4315. [Google Scholar] [CrossRef] [PubMed]
- Dal Ben, D.; Lambertucci, C.; Buccioni, M.; Martí Navia, A.; Marucci, G.; Spinaci, A.; Volpini, R. Non-Nucleoside Agonists of the Adenosine Receptors: An Overview. Pharmaceuticals 2019, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.E.; Jacobson, K.A. Xanthines as adenosine receptor antagonists. Handb. Exp. Pharmacol. 2011, 200, 151–199. [Google Scholar] [CrossRef]
- Ukena, D.; Padgett, W.L.; Hong, O.; Daly, J.W.; Daly, D.T.; Olsson, R.A. N6-substituted 9-methyladenines: A new class of adenosine receptor antagonists. FEBS Lett. 1987, 215, 203–208. [Google Scholar] [CrossRef]
- Zhou, X.; Khanapur, S.; Huizing, A.P.; Zijlma, R.; Schepers, M.; Dierckx, R.A.J.O.; van Waarde, A.; de Vries, E.F.J.; Elsinga, P.H. Synthesis and preclinical evaluation of 2-(2-furanyl)-7-2-4-4-(2-11Cmethoxyethoxy)phenyl-1-piperazinylethyl7H-pyrazolo4,3-e1,2,4triazolo1,5-cpyrimidine-5-amine (11CPreladenant) as a PET tracer for the imaging of cerebral adenosine A2A receptors. J. Med. Chem. 2014, 57, 9204–9210. [Google Scholar] [CrossRef]
- Shinkre, B.A.; Kumar, T.S.; Gao, Z.-G.; Deflorian, F.; Jacobson, K.A.; Trenkle, W.C. Synthesis and evaluation of 1,2,4-triazolo1,5-cpyrimidine derivatives as A2A receptor-selective antagonists. Bioorg. Med. Chem. Lett. 2010, 20, 5690–5694. [Google Scholar] [CrossRef] [PubMed]
- Todde, S.; Moresco, R.M.; Simonelli, P.; Baraldi, P.G.; Cacciari, B.; Spalluto, G.; Varani, K.; Monopoli, A.; Matarrese, M.; Carpinelli, A.; et al. Design, radiosynthesis, and biodistribution of a new potent and selective ligand for in vivo imaging of the adenosine A2A receptor system using positron emission tomography. J. Med. Chem. 2000, 43, 4359–4362. [Google Scholar] [CrossRef] [PubMed]
- Zocchi, C.; Ongini, E.; Conti, A.; Monopoli, A.; Negretti, A.; Baraldi, P.G.; Dionisotti, S. The non-xanthine heterocyclic compound SCH 58261 is a new potent and selective A2a adenosine receptor antagonist. J. Pharmacol. Exp. Ther. 1996, 276, 398–404. [Google Scholar] [PubMed]
- Marques, T.R.; Natesan, S.; Rabiner, E.A.; Searle, G.E.; Gunn, R.; Howes, O.D.; Kapur, S. Adenosine A2A receptor in schizophrenia: An in vivo brain PET imaging study. Psychopharmacology 2021, 239, 3439–3445. [Google Scholar] [CrossRef]
- Boutin, J.A.; Witt-Enderby, P.A.; Sotriffer, C.; Zlotos, D.P. Melatonin receptor ligands: A pharmaco-chemical perspective. J. Pineal Res. 2020, 69, e12672. [Google Scholar] [CrossRef]
- Bojarski, A.J. Pharmacophore models for metabotropic 5-HT receptor ligands. Curr. Top. Med. Chem. 2006, 6, 2005–2026. [Google Scholar] [CrossRef]
- Gao, Z.-G.; Jacobson, K.A. Allosteric modulation and functional selectivity of G protein-coupled receptors. Drug Discov. Today Technol. 2013, 10, e237–e243. [Google Scholar] [CrossRef]
- Lütjens, R.; Rocher, J.-P. Recent advances in drug discovery of GPCR allosteric modulators for neurodegenerative disorders. Curr. Opin. Pharmacol. 2017, 32, 91–95. [Google Scholar] [CrossRef]
- van der Westhuizen, E.T.; Valant, C.; Sexton, P.M.; Christopoulos, A. Endogenous allosteric modulators of G protein-coupled receptors. J. Pharmacol. Exp. Ther. 2015, 353, 246–260. [Google Scholar] [CrossRef]
- Madsen, J.J.; Ye, L.; Frimurer, T.M.; Olsen, O.H. Mechanistic basis of GPCR activation explored by ensemble refinement of crystallographic structures. Protein Sci. 2022, 31, e4456. [Google Scholar] [CrossRef]
- Swaminath, G.; Lee, T.W.; Kobilka, B. Identification of an allosteric binding site for Zn2+ on the beta2 adrenergic receptor. J. Biol. Chem. 2003, 278, 352–356. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, H.; Yang, D.; Zhong, L.; Xin, Y.; Zhao, S.; Wang, M.-W.; Zhou, Q.; Shui, W. Affinity Mass Spectrometry-Based Fragment Screening Identified a New Negative Allosteric Modulator of the Adenosine A2A Receptor Targeting the Sodium Ion Pocket. ACS Chem. Biol. 2021, 16, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Korkutata, M.; Saitoh, T.; Cherasse, Y.; Ioka, S.; Duo, F.; Qin, R.; Murakoshi, N.; Fujii, S.; Zhou, X.; Sugiyama, F.; et al. Enhancing endogenous adenosine A2A receptor signaling induces slow-wave sleep without affecting body temperature and cardiovascular function. Neuropharmacology 2019, 144, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Welihinda, A.A.; Amento, E.P. Positive allosteric modulation of the adenosine A2a receptor attenuates inflammation. J. Inflamm. 2014, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Welihinda, A.A.; Kaur, M.; Raveendran, K.S.; Amento, E.P. Enhancement of inosine-mediated A2AR signaling through positive allosteric modulation. Cell. Signal. 2018, 42, 227–235. [Google Scholar] [CrossRef]
- Salovich, J.M.; Sheffler, D.J.; Vinson, P.N.; Lamsal, A.; Utley, T.J.; Blobaum, A.L.; Bridges, T.M.; Le, U.; Jones, C.K.; Wood, M.R.; et al. Probe Reports from the NIH Molecular Libraries Program: Discovery of a Novel Structural Class of M4 Positive Allosteric Modulators: Characterization of ML293, N-(4-Methoxy-7-Methylbenzodthiazol-2-yl)isonicotinamide, with CNS Exposure in Rats; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2010.
- Jones, C.K.; Bubser, M.; Thompson, A.D.; Dickerson, J.W.; Turle-Lorenzo, N.; Amalric, M.; Blobaum, A.L.; Bridges, T.M.; Morrison, R.D.; Jadhav, S.; et al. The metabotropic glutamate receptor 4-positive allosteric modulator VU0364770 produces efficacy alone and in combination with L-DOPA or an adenosine 2A antagonist in preclinical rodent models of Parkinson’s disease. J. Pharmacol. Exp. Ther. 2012, 340, 404–421. [Google Scholar] [CrossRef] [PubMed]
- Pinhal-Enfield, G.; Ramanathan, M.; Hasko, G.; Vogel, S.N.; Salzman, A.L.; Boons, G.-J.; Leibovich, S.J. An Angiogenic Switch in Macrophages Involving Synergy between Toll-Like Receptors 2, 4, 7, and 9 and Adenosine A2A Receptors. Am. J. Pathol. 2003, 163, 711–721. [Google Scholar] [CrossRef]
- Jung, S.-M.; Peyton, L.; Essa, H.; Choi, D.-S. Adenosine receptors: Emerging non-opioids targets for pain medications. Neurobiol. Pain 2022, 11, 100087. [Google Scholar] [CrossRef]
- Federico, S.; Lassiani, L.; Spalluto, G. Chemical Probes for the Adenosine Receptors. Pharmaceuticals 2019, 12, 168. [Google Scholar] [CrossRef]
- Soave, M.; Briddon, S.J.; Hill, S.J.; Stoddart, L.A. Fluorescent ligands: Bringing light to emerging GPCR paradigms. Br. J. Pharmacol. 2020, 177, 978–991. [Google Scholar] [CrossRef]
- Kozma, E.; Jayasekara, P.S.; Squarcialupi, L.; Paoletta, S.; Moro, S.; Federico, S.; Spalluto, G.; Jacobson, K.A. Fluorescent ligands for adenosine receptors. Bioorg. Med. Chem. Lett. 2013, 23, 26–36. [Google Scholar] [CrossRef]
- McCabe, R.T.; Skolnick, P.; Jacobson, K.A. 2-2-4-2-2-1,3-Dihydro- 1,1-bis (4-hydroxyphenyl)-3-oxo-5-isobenzofuranthioureidylethylaminocarbonylethylphenyl ethylamino-5′-N-ethylcarboxamidoadenosine (FITC-APEC): A Fluorescent Ligand for A2a-Adenosine Receptors. J. Fluoresc. 1992, 2, 217–223. [Google Scholar] [CrossRef]
- Brand, F.; Klutz, A.M.; Jacobson, K.A.; Fredholm, B.B.; Schulte, G. Adenosine A2A receptor dynamics studied with the novel fluorescent agonist Alexa488-APEC. Eur. J. Pharmacol. 2008, 590, 36–42. [Google Scholar] [CrossRef]
- Comeo, E.; Kindon, N.D.; Soave, M.; Stoddart, L.A.; Kilpatrick, L.E.; Scammells, P.J.; Hill, S.J.; Kellam, B. Subtype-Selective Fluorescent Ligands as Pharmacological Research Tools for the Human Adenosine A2A Receptor. J. Med. Chem. 2020, 63, 2656–2672. [Google Scholar] [CrossRef]
- Toti, K.S.; Campbell, R.G.; Lee, H.; Salmaso, V.; Suresh, R.R.; Gao, Z.-G.; Jacobson, K.A. Fluorescent A2A and A3 adenosine receptor antagonists as flow cytometry probes. Purinergic Signal. 2022. [Google Scholar] [CrossRef] [PubMed]
- McNeely, P.M.; Naranjo, A.N.; Forsten-Williams, K.; Robinson, A.S. A2AR Binding Kinetics in the Ligand Depletion Regime. SLAS Discov. 2017, 22, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Rieger, J.M.; Brown, M.L.; Sullivan, G.W.; Linden, J.; Macdonald, T.L. Design, synthesis, and evaluation of novel A2A adenosine receptor agonists. J. Med. Chem. 2001, 44, 531–539. [Google Scholar] [CrossRef]
- Chen, J.-B.; Liu, E.M.; Chern, T.-R.; Yang, C.-W.; Lin, C.-I.; Huang, N.-K.; Lin, Y.-L.; Chern, Y.; Lin, J.-H.; Fang, J.-M. Design and synthesis of novel dual-action compounds targeting the adenosine A2A receptor and adenosine transporter for neuroprotection. ChemMedChem 2011, 6, 1390–1400. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.; Gedeon, N.G.; Jankins, T.C.; Jones, G.B. Novel approaches for targeting the adenosine A2A receptor. Expert Opin. Drug Discov. 2015, 10, 63–80. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.; Hosila, F.J. The effect of different extraction sites upon incisor retraction. Am. J. Orthod. 1976, 69, 388–410. [Google Scholar] [CrossRef]
- Jazayeri, A.; Andrews, S.P.; Marshall, F.H. Structurally Enabled Discovery of Adenosine A2A Receptor Antagonists. Chem. Rev. 2017, 117, 21–37. [Google Scholar] [CrossRef]
- Hu, E.; Kunz, R.K.; Rumfelt, S.; Andrews, K.L.; Li, C.; Hitchcock, S.A.; Lindstrom, M.; Treanor, J. Use of structure based design to increase selectivity of pyridyl-cinnoline phosphodiesterase 10A (PDE10A) inhibitors against phosphodiesterase 3 (PDE3). Bioorg. Med. Chem. Lett. 2012, 22, 6938–6942. [Google Scholar] [CrossRef]
- Deganutti, G.; Moro, S. Supporting the Identification of Novel Fragment-Based Positive Allosteric Modulators Using a Supervised Molecular Dynamics Approach: A Retrospective Analysis Considering the Human A2A Adenosine Receptor as a Key Example. Molecules 2017, 22, 818. [Google Scholar] [CrossRef] [PubMed]
- McGraw, C.; Yang, L.; Levental, I.; Lyman, E.; Robinson, A.S. Membrane cholesterol depletion reduces downstream signaling activity of the adenosine A2A receptor. Biochim. Biophys. Acta Biomembr. 2019, 1861, 760–767. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Hu, Z.; Filipek, S.; Vogel, H. W246(6.48) opens a gate for a continuous intrinsic water pathway during activation of the adenosine A2A receptor. Angew. Chem. Int. Ed Engl. 2015, 54, 556–559. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Pan, A.C.; Dror, R.O.; Mocking, T.; Liu, R.; Heitman, L.H.; Shaw, D.E.; IJzerman, A.P. Molecular Basis of Ligand Dissociation from the Adenosine A2A Receptor. Mol. Pharmacol. 2016, 89, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Mondal, C.; Halder, A.K.; Adhikari, N.; Jha, T. Structural findings of cinnolines as anti-schizophrenic PDE10A inhibitors through comparative chemometric modeling. Mol. Divers. 2014, 18, 655–671. [Google Scholar] [CrossRef]
- Rodríguez, D.; Gao, Z.-G.; Moss, S.M.; Jacobson, K.A.; Carlsson, J. Molecular docking screening using agonist-bound GPCR structures: Probing the A2A adenosine receptor. J. Chem. Inf. Model. 2015, 55, 550–563. [Google Scholar] [CrossRef]
- Novikov, G.V.; Sivozhelezov, V.S.; Shaĭtan, K.V. Investigation of the conformational dynamics of the adenosine A2A receptor by means of molecular dynamics simulation. Biofizika 2013, 58, 618–634. [Google Scholar]
- Kalash, L.; Winfield, I.; Safitri, D.; Bermudez, M.; Carvalho, S.; Glen, R.; Ladds, G.; Bender, A. Structure-based identification of dual ligands at the A2AR and PDE10A with anti-proliferative effects in lung cancer cell-lines. J. Cheminform. 2021, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.W.; Laughton, C.A.; Doughty, S.W. Molecular dynamics simulations of the adenosine A2a receptor: Structural stability, sampling, and convergence. J. Chem. Inf. Model. 2013, 53, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Hajduk, P.J.; Greer, J. A decade of fragment-based drug design: Strategic advances and lessons learned. Nat. Rev. Drug Discov. 2007, 6, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Ranganathan, A.; IJzerman, A.P.; Siegal, G.; Carlsson, J. Complementarity between in silico and biophysical screening approaches in fragment-based lead discovery against the A2A adenosine receptor. J. Chem. Inf. Model. 2013, 53, 2701–2714. [Google Scholar] [CrossRef]
- Chen, D.; Errey, J.C.; Heitman, L.H.; Marshall, F.H.; IJzerman, A.P.; Siegal, G. Fragment screening of GPCRs using biophysical methods: Identification of ligands of the adenosine A2A receptor with novel biological activity. ACS Chem. Biol. 2012, 7, 2064–2073. [Google Scholar] [CrossRef]
- Matricon, P.; Ranganathan, A.; Warnick, E.; Gao, Z.-G.; Rudling, A.; Lambertucci, C.; Marucci, G.; Ezzati, A.; Jaiteh, M.; Dal Ben, D.; et al. Fragment optimization for GPCRs by molecular dynamics free energy calculations: Probing druggable subpockets of the A2A adenosine receptor binding site. Sci. Rep. 2017, 7, 6398. [Google Scholar] [CrossRef]
- Tosh, D.K.; Phan, K.; Gao, Z.-G.; Gakh, A.A.; Xu, F.; Deflorian, F.; Abagyan, R.; Stevens, R.C.; Jacobson, K.A.; Katritch, V. Optimization of adenosine 5′-carboxamide derivatives as adenosine receptor agonists using structure-based ligand design and fragment screening. J. Med. Chem. 2012, 55, 4297–4308. [Google Scholar] [CrossRef]
- Zhang, M.; Fan, S.; Zhou, X.; Xie, F.; Li, S.; Zhong, W. Design, synthesis and biological evaluation of 2-hydrazinyladenosine derivatives as A2A adenosine receptor ligands. Eur. J. Med. Chem. 2019, 179, 310–324. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).