1. Introduction
Cardiovascular disease (CVD) contributes significantly to mortality worldwide, accounting for 31% of deaths in 2019 [
1]. The presence of CVD is indicated by heart sound abnormality. In order to manage CVD, prompt and accurate diagnosis must be performed. Despite this, traditional heart auscultation approaches have low accuracy rates [
2,
3,
4]. Cardiac auscultation is a cost-effective pre-screening method that involves a physician’s interpretation of heart sounds. However, the frequencies of heart sounds are close to the boundaries of human hearing, inhibiting their accurate interpretation, thereby risking misdiagnosis [
5].
Computer-aided auscultation screening presents an opportunity to detect CVD earlier than traditional approaches. Although there are many approaches to detect abnormal heart sounds, most perform poorly due to the noisy and limited datasets available. Noisy data provides challenges as classifiers rely on extracting key information to distinguish classes. Noise within the data can then hide or distort this information. Classifiers trained on limited datasets are prone to overfitting, causing poor performance on real-world data.
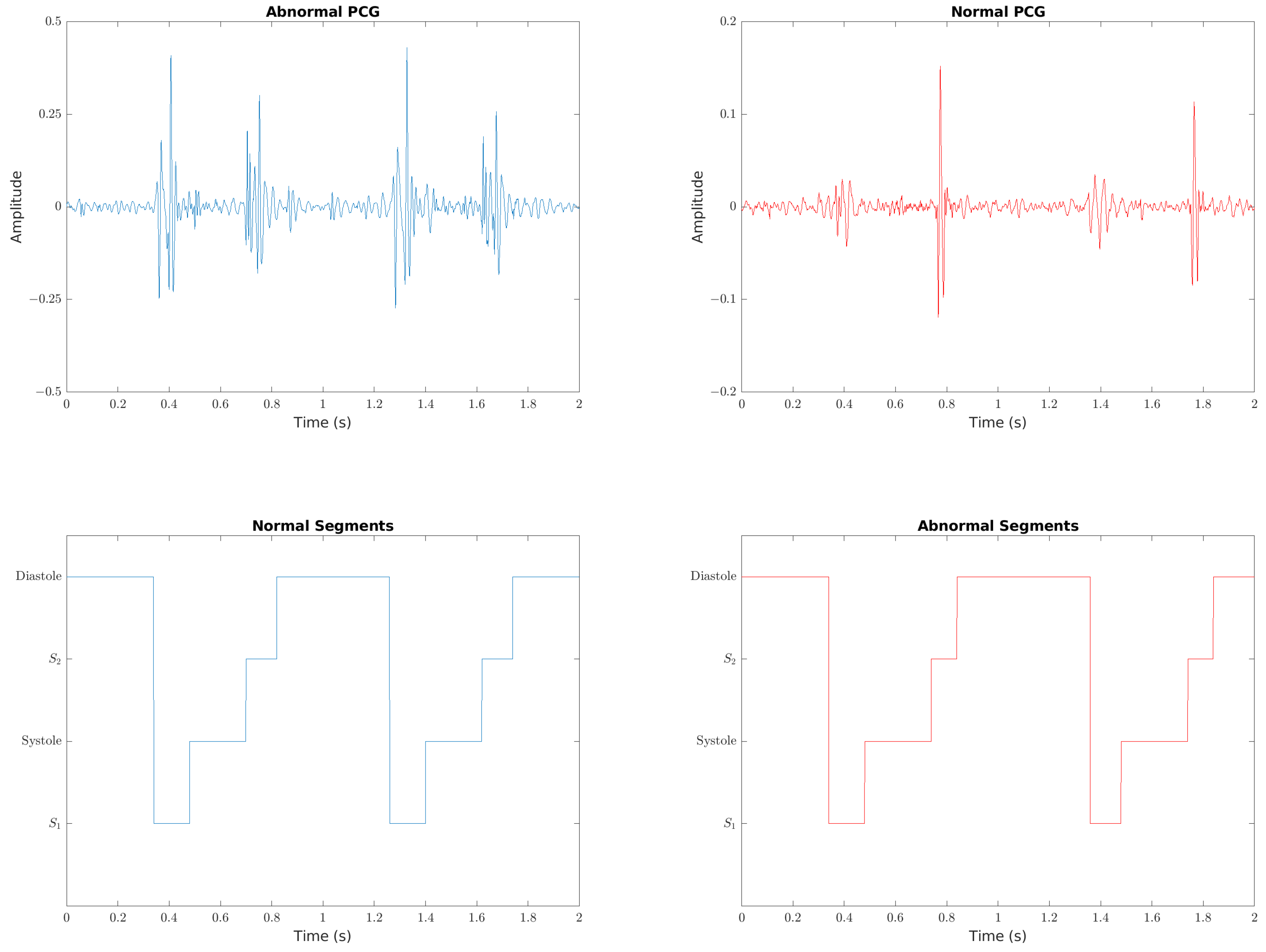
The complex and non-stationary nature of heart sounds makes analysis difficult. Pre-processing heart sounds can provide a cleaner input signal to the classifier. Cleaner signals lead to more accurate results. Segmenting the signal into heart sound cycles assists in detecting abnormalities such as murmurs, which are highly periodic. Segmentation is particularly important for use in clinical environments, where low signal-to-noise ratios pose a significant problem for detection. As noise from the friction of handheld stethoscopes and background sources is difficult to avoid, pre-processing methods may be used to overcome challenges to abnormal heart sound detection.
Existing state-of-the-art methods are not accurate enough for use in real-world scenarios. Recently, a start-up company, Ticking Heart, and researchers at Curtin University [
6,
7] worked together to create a wearable device for use in the pre-screening of CVD. The device combines up to seven phonocardiogram (PCG) signals, and one lead-I electrocardiogram (ECG) signal to provide more robust and accurate classification when compared to a single signal.
In this study, we integrate established pre-processing and segmentation techniques with transfer learning using pre-trained Convolutional Neural Network (CNN) models to enhance the performance of abnormal heart sound classification. Moreover, we delve into the interpretation of the features learned by the model. The paper is structured as follows: We begin with the ’Background’, providing context and introducing existing methodologies. Following this is the ’Materials and Methodology’ section, detailing our adopted approach. In the ’Results’ section, we present the performance of our classifier, complemented by interpretability images. Subsequent discussions about these findings are covered in the ’Discussion’ section. The paper concludes with the ’Conclusion’, summarising our key takeaways and ’Further Work’ to guide future research.
3. Materials and Methods
The models are trained using an EVGA Nvidia RTX 3090 sourced from Perth, Australia.
Figure 3 shows the flow of data through the system. Our methodology consisted of the following steps:
Figure 3.
Classification process.
Figure 3.
Classification process.
3.1. Dataset
A subset of the CinC 2016 dataset was used for training and testing. The CinC 2016 dataset contains data from clinical and isolated environments from multiple research groups worldwide. The databases collectively offered 2435 recordings from 1297 distinct patients [
8]. After breaking down longer recordings into shorter snippets and leaving out the maternal-fetal database, the competition utilised 4430 samples gathered from 1072 individuals. These samples correspond to 233,512 individual heart sounds, 116,865 cardiac cycles, and almost 30 h of PCG data.
Our work examines the classification of abnormal heart sounds by combining PCG and ECG data. As such, only the training-a database, which provides combined PCG and ECG data, is used. The training database contains 409 recordings, 4 of which do not include ECG signals, so they have been excluded. Hence, 405 different PCG recordings with an associated ECG recording are used. The dataset was split 60-20-20 train-validation-test.
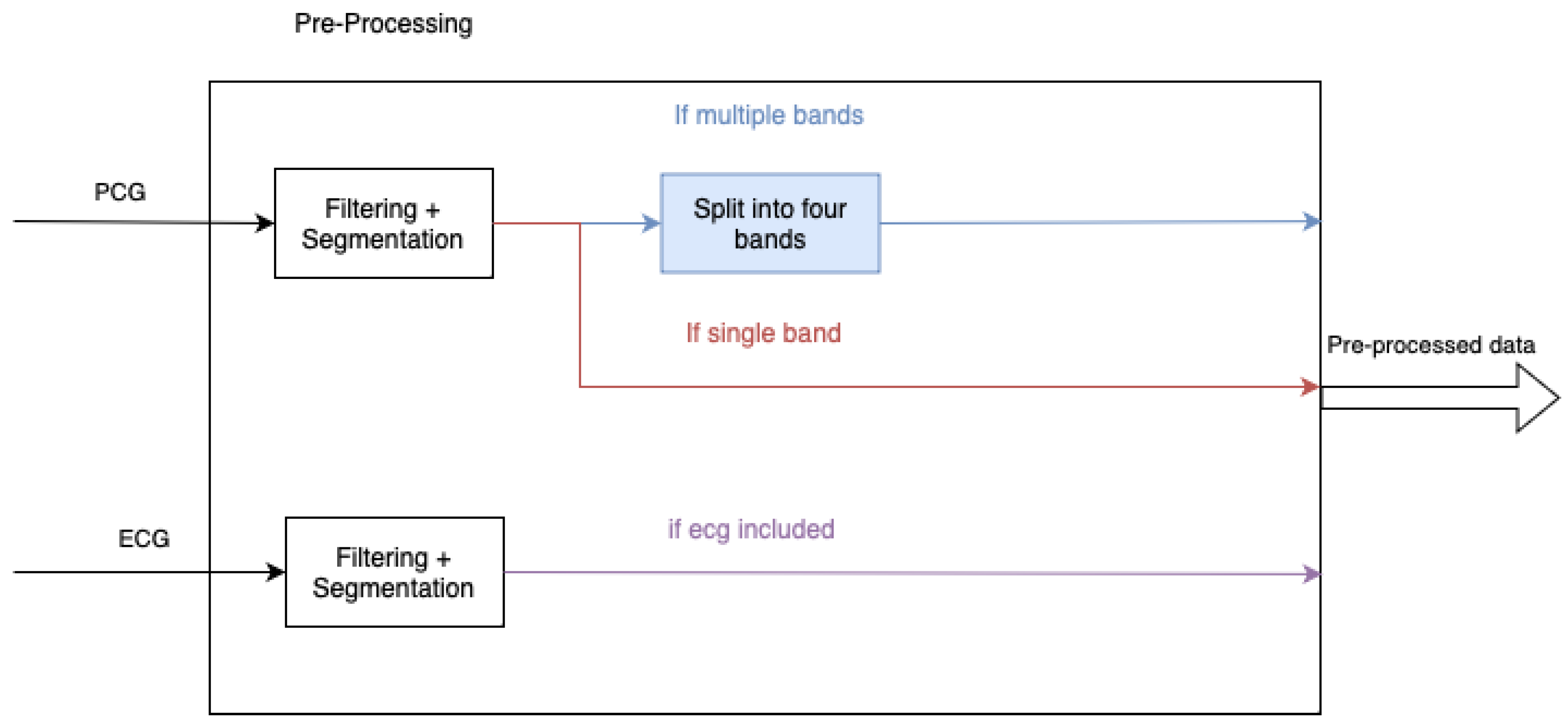
3.2. Pre-Processing
Pre-processing of the data, summarised in
Figure 4, is used to reduce noise and segment heart cycles. The pre-processing involved is the same as the work of Rong et al., [
6] which is derived from Potes et al. [
16] The PCG signal is first resampled down to 1k Hz before being bandpass filtered between 25 Hz and 400 Hz. Spikes are removed using the approach from Schmidt et al. [
27]. If used, the ECG signal is resampled to 1k Hz and bandpass filtered between 2 Hz and 60 Hz. The HSMM-based approach by Springer et al. is used to segment the heart sounds, which are then used to split the data into heart cycles. The ECG data is split at the same samples as the PCG. The first 10 heart cycles of each patient are then extracted, with the 10 fragments used as separate inputs to train the model. For the approaches where the PCG data is to be split into multiple bands, the bands used are 25 Hz–45 Hz, 45 Hz–80 Hz, 80 Hz–200 Hz and 200 Hz–400 Hz. The PCG and ECG are then zero-padded to a size of 2500 samples, and the result used to create the corresponding image representation of a spectrogram or scalogram.
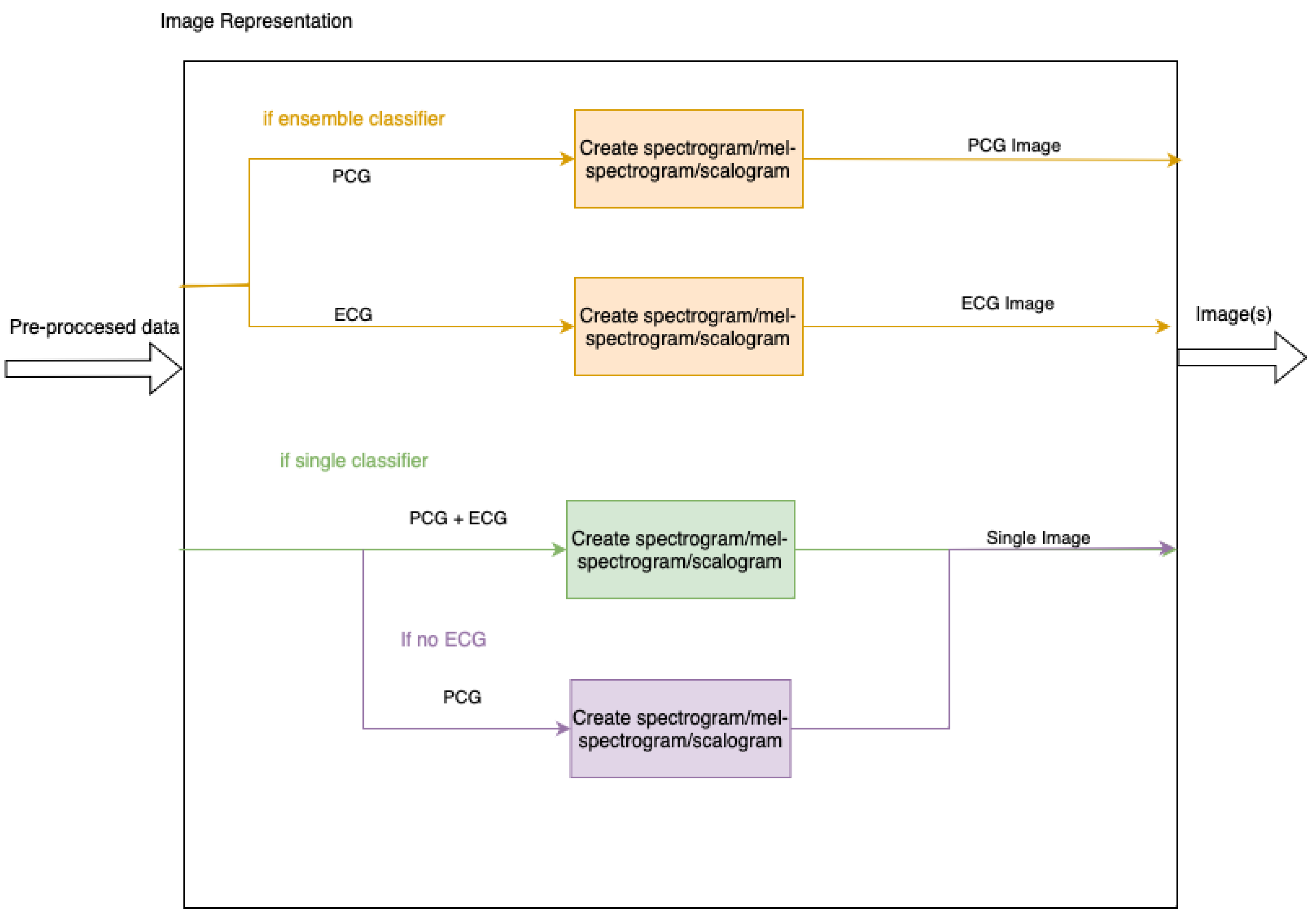
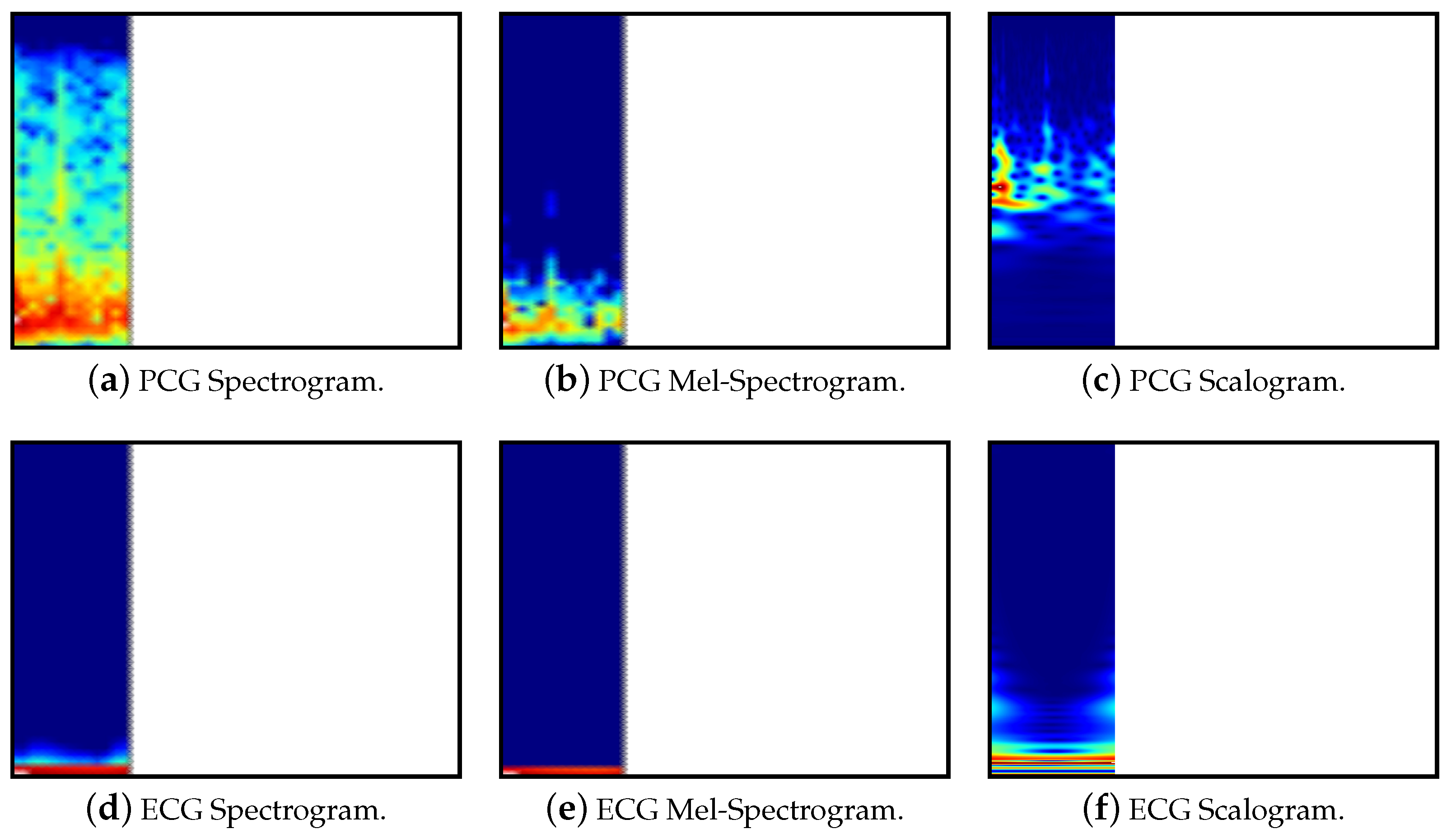
3.3. Image Representation
As each of the models uses an existing pre-trained model which takes images as input, the pre-processed data must be transformed into this format. Input images may include both PCG and ECG signals or have them as separate inputs to an ensemble of two models. The ensemble will be achieved by utilising two models, one for each image, and then adding a linear layer to combine the two. In addition to this, the PCG signal may be split into separate bands, as specified in
Section 3.2. To show a clear separation between multiple bands, the images display zero-padding as white space.
Figure 5 contains a high level summary of the image creation. The representations are shown below in
Table 1, along with
Table 2.
Spectrograms are created using the Short Time Fourier Transform (STFT). The STFT can be seen as taking Fourier transforms over potentially overlapping segments of the signal. This results in a two-dimensional representation of the signal showing the spectral components over time [
28]. Our approach uses a window length of 100 and an overlap of 50 between each segment.
Mel-spectrograms differ from linear spectrograms in that they use the mel-scale for frequency. This more accurately models humans hearing perception. This transformation can be performed by combining the STFT with a mel filter bank, applying the transformation through the use of multiple non-uniform triangular filters [
29]. Our approach uses 64 mel bins, a window length of 100 and an overlap of 50.
Scalograms show how the continuous wavelet transform (CWT) changes over time. One axis of the scalogram represents time and the other represents the reciprocal of the frequency, known as the scale [
28]. By using scales instead of a fixed window length, the CWT avoids the trade-off between time resolution and frequency resolution. Although the CWT results in better time and frequency resolution than the STFT, it is more computationally complex and often results in blurry images. This can be overcome using a method called synchrosqueezing [
30]. Our approach uses the synchrosqueeze CWT algorithm with a Morlet mother wavelet.
3.4. Model Training
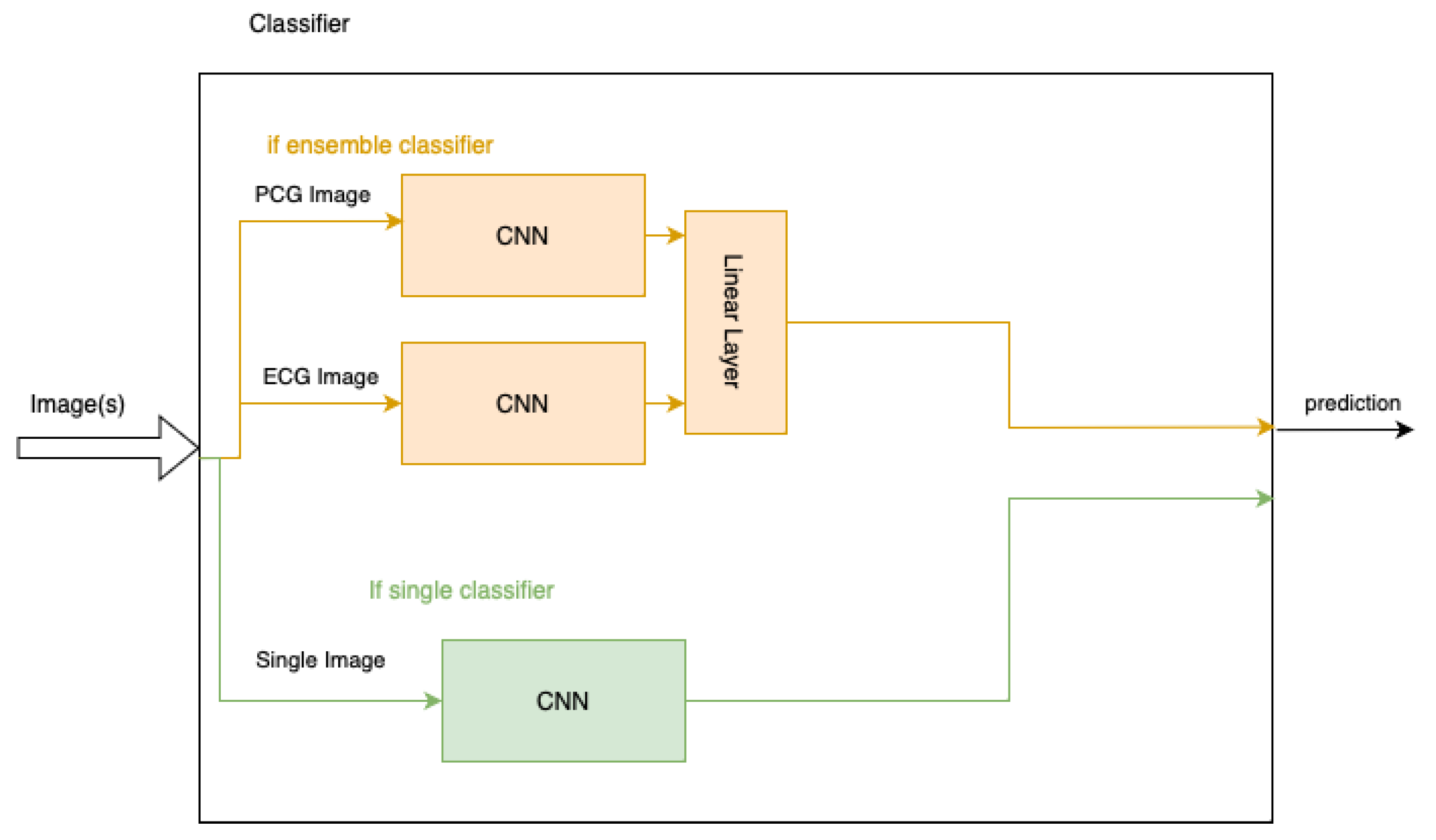
Various pre-trained models are used as the basis of the classifier, altered to take the necessary input images and output a binary classification. This is summarised in
Table 3. In addition to this, the image formats used are summarised in
Figure 6.
The particular variations used for each model include ResNet34, VGG19 and inceptionv3. All of these models correspond to PyTorch models trained on the ImageNet dataset. Each model has also been modified by changing the final layer to a size of two, with one neuron representing the abnormal case and the other the normal.
Where two images are required, the CNN is duplicated to have one model per image. One CNN takes the PCG image as input and the other the ECG image. A fully-connected layer is added to combine the outputs into one classification.
Figure 6 illustrates the layout of these ensemble models in orange.
All models have been trained with a stochastic gradient descent optimiser, using a learning rate of 0.001 and a momentum of 0.9. We also use cross entropy loss for each model. A learning rate scheduler with a step size of 7 and multiplicative learning rate decay factor of 0.1 is used. The ResNet models use a batch size of 64 and 4 workers. Both the VGG and inceptionv3 models use a batch size of 32 and 2 workers. The ensemble models use the same batch size as the models that comprise them.
As discussed earlier, the first 10 recording fragments for each patient are used for training. For determining a patient’s case, a decision rule is applied to all 10 fragments. As a result, there is a fragment and patient accuracy associated with each model. The fragment accuracy will be optimised during training as a higher fragment accuracy generally results in a higher patient accuracy. In addition to this, measures derived from the confusion matrix for each patient are used, with the most important being accuracy, sensitivity, and specificity [
8].
3.5. Model Evaluation
The final classification result depends on the output of the 10 fragments for each patient. For each fragment, the model outputs two values that represent the expectation of belonging to the abnormal and normal classes. These values are then averaged separately across the abnormal and normal cases before the softmax function is applied. The resulting values, between 0 and 1, represent the expected probability that the patient belongs to either class. The patient is classified as an abnormal case if the probability is greater than a threshold value of 0.4. This value was found through experimentation on the validation set, achieving a balance between accuracy, sensitivity, and specificity. Our primary evaluation is accuracy, followed by specificity. Specificity was chosen over sensitivity due to the data imbalance against normal cases.
3.6. Model Interpretation
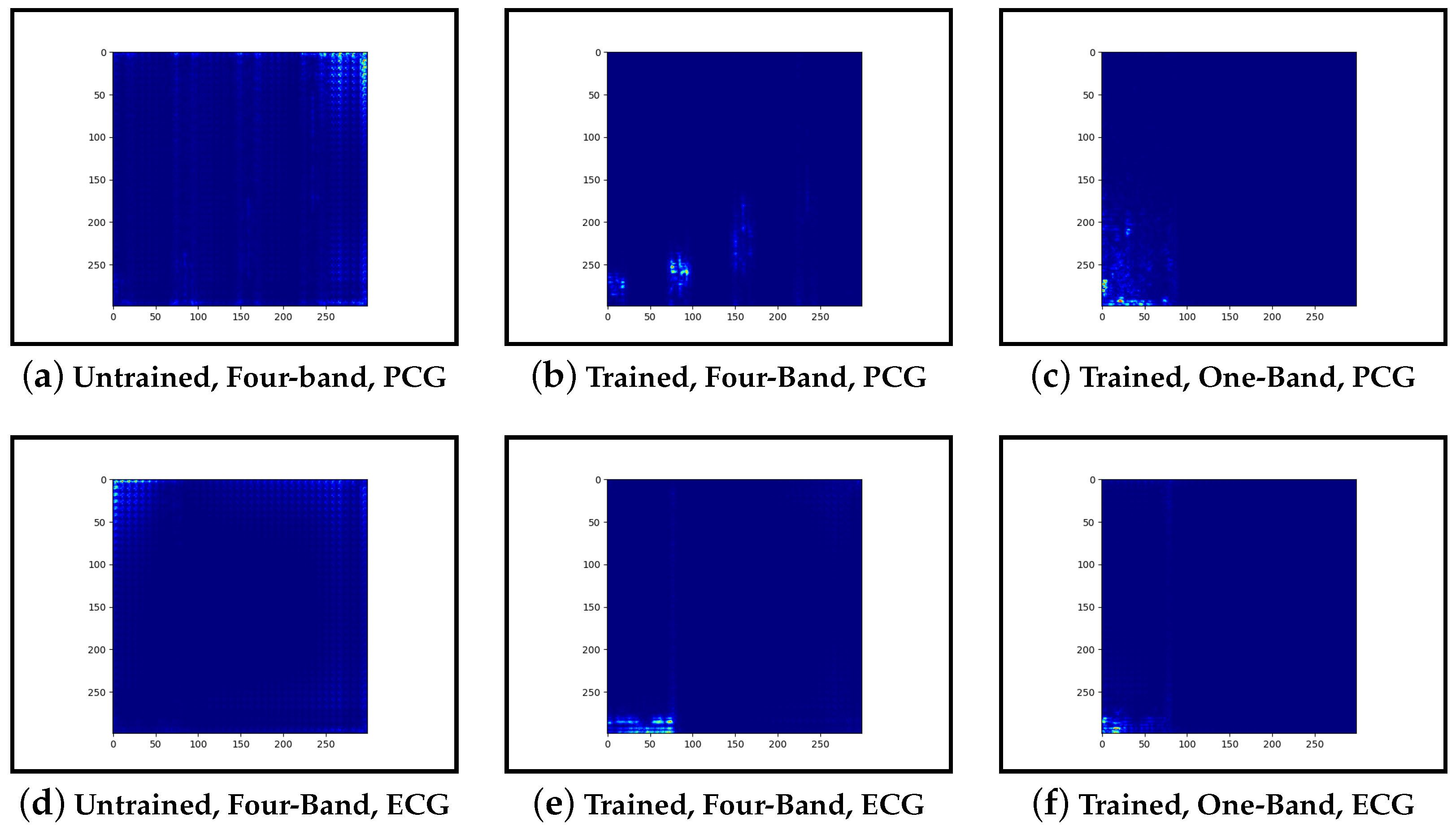
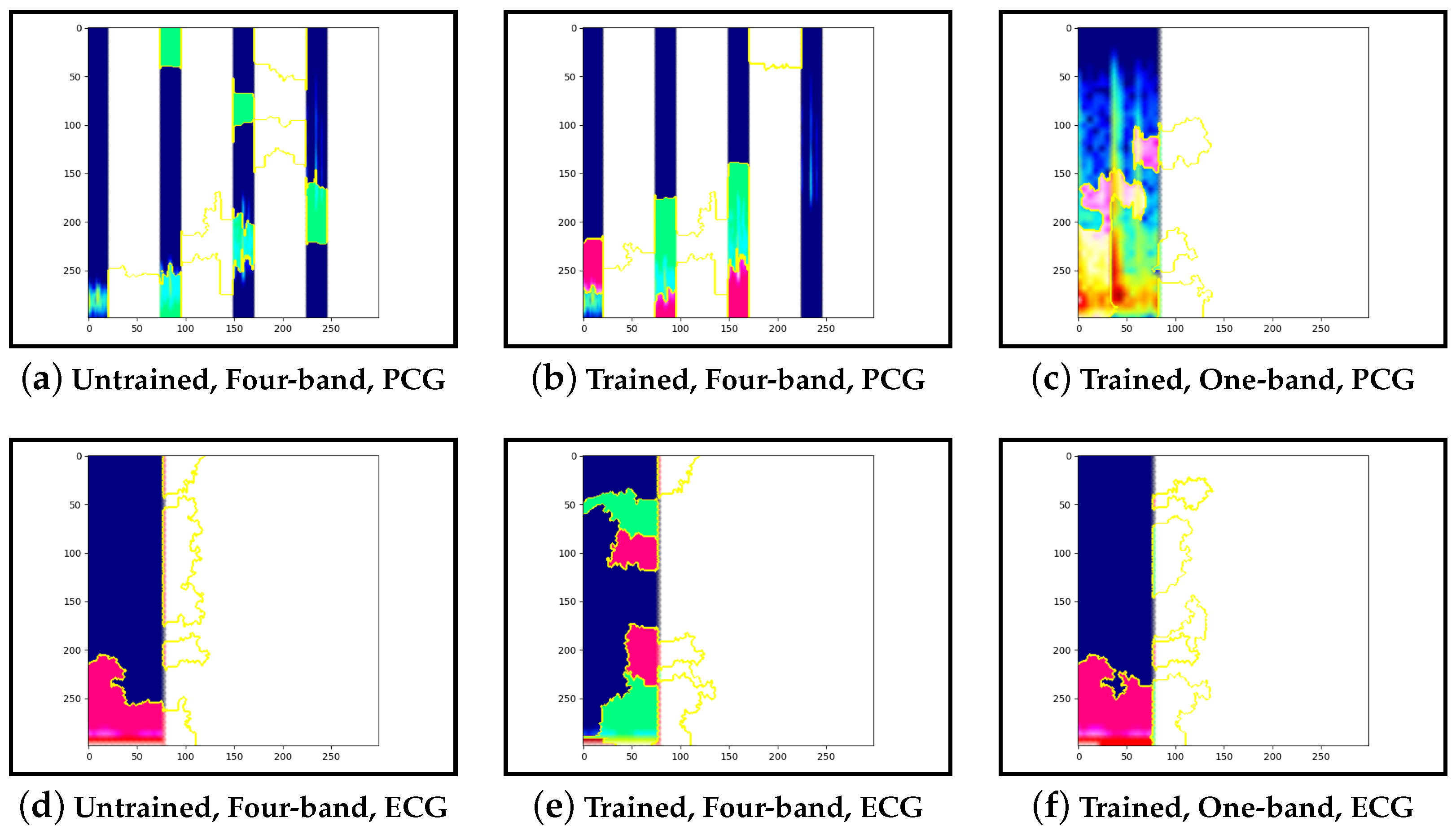
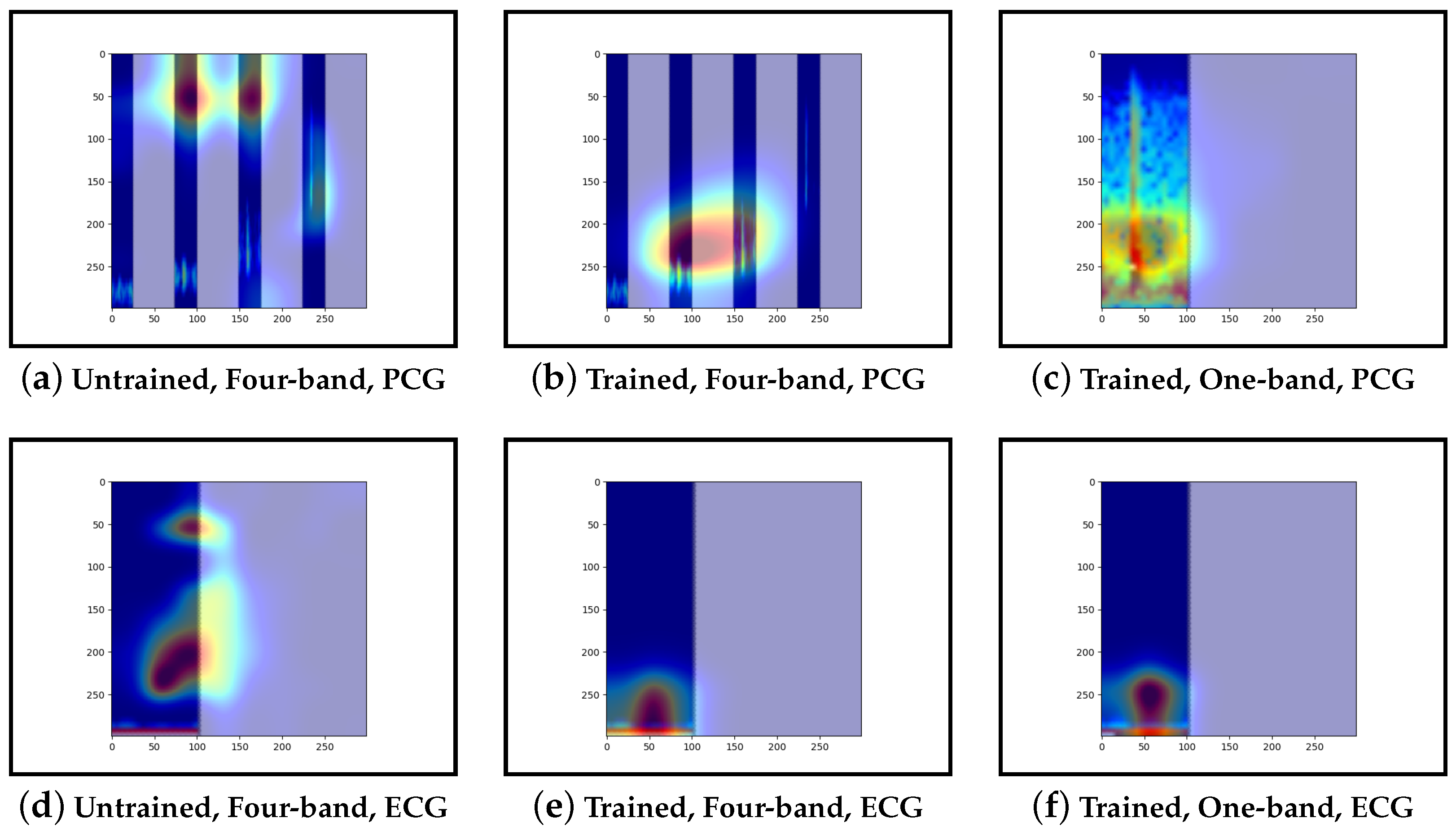
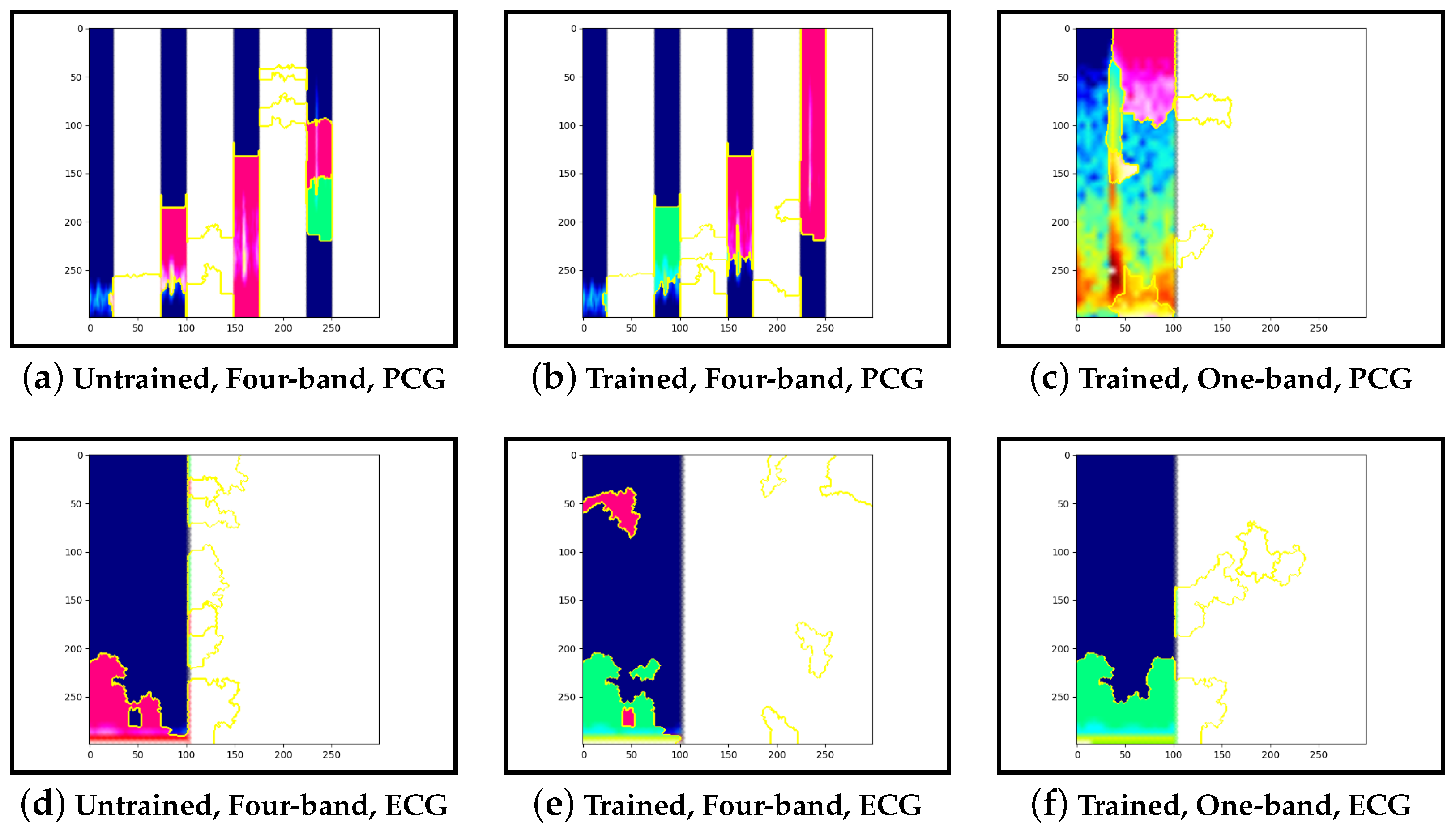
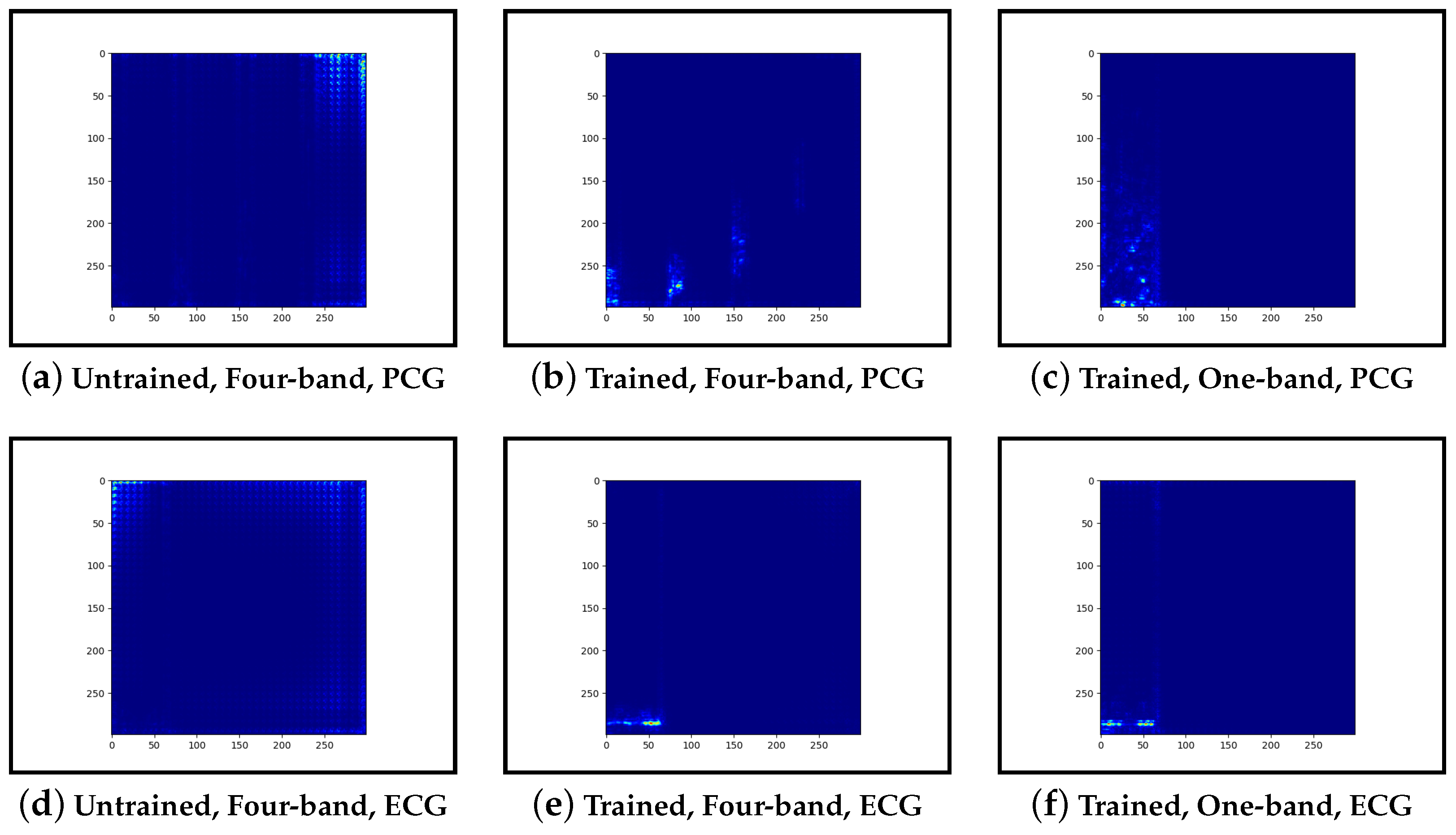
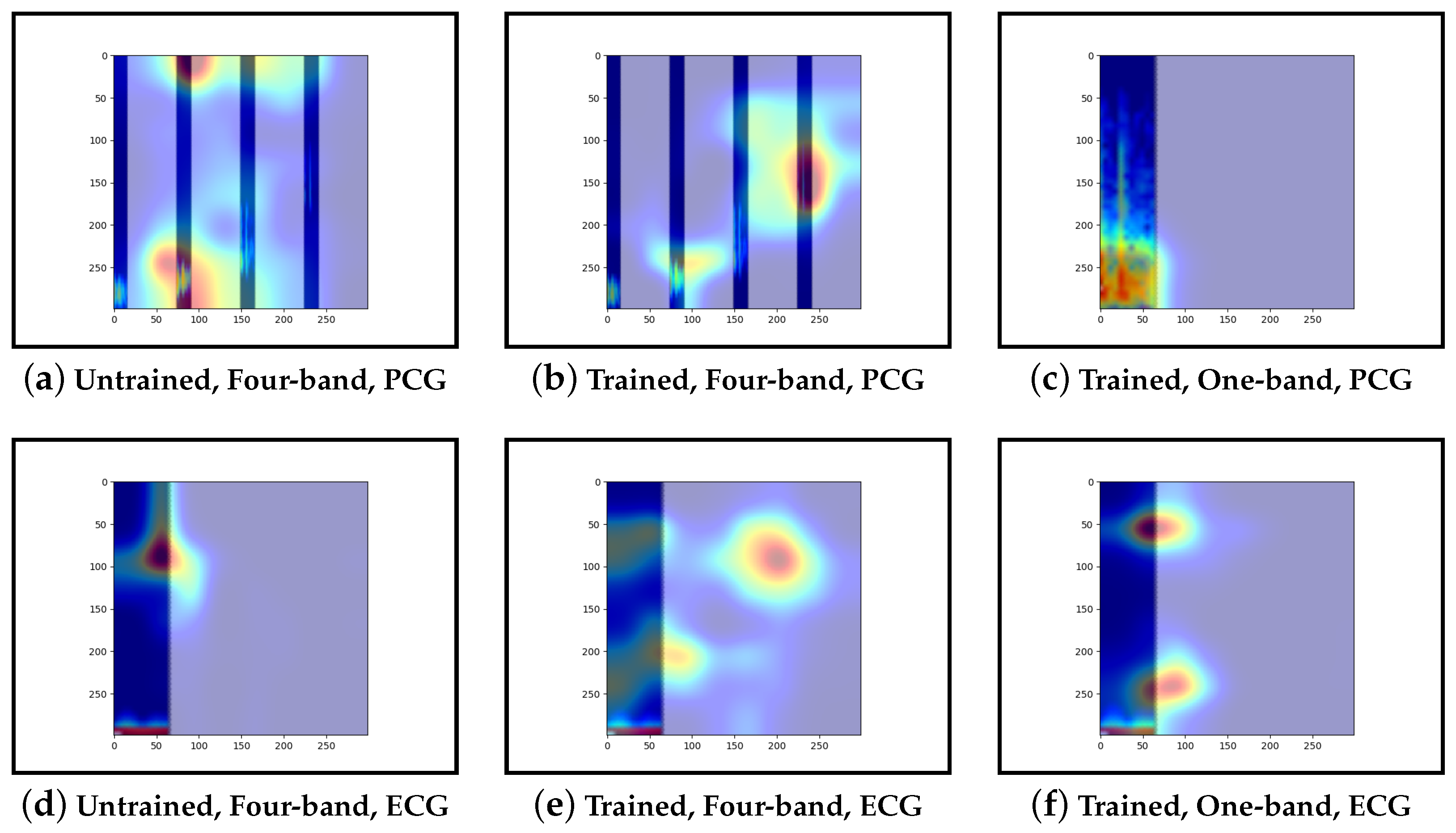
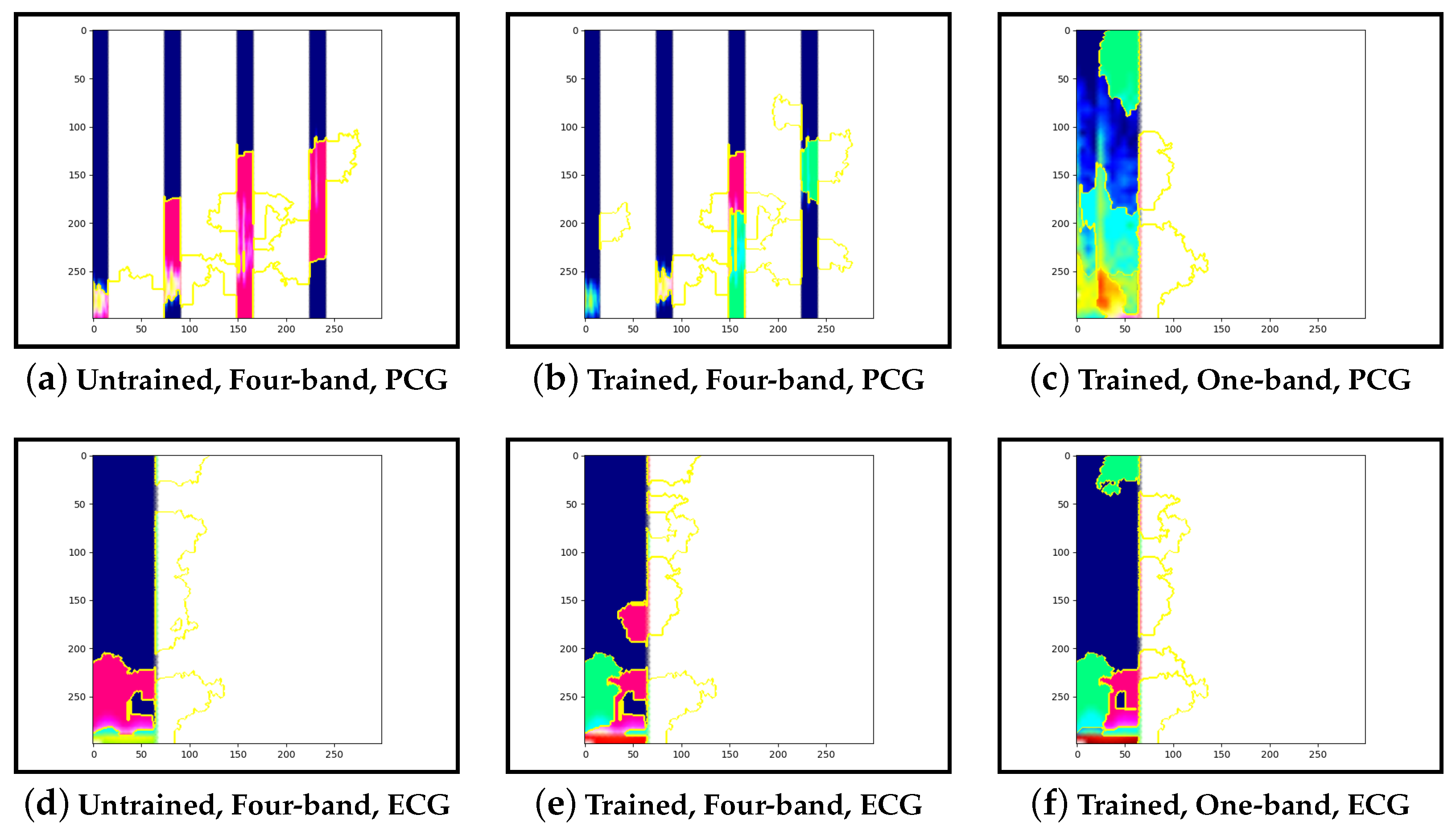
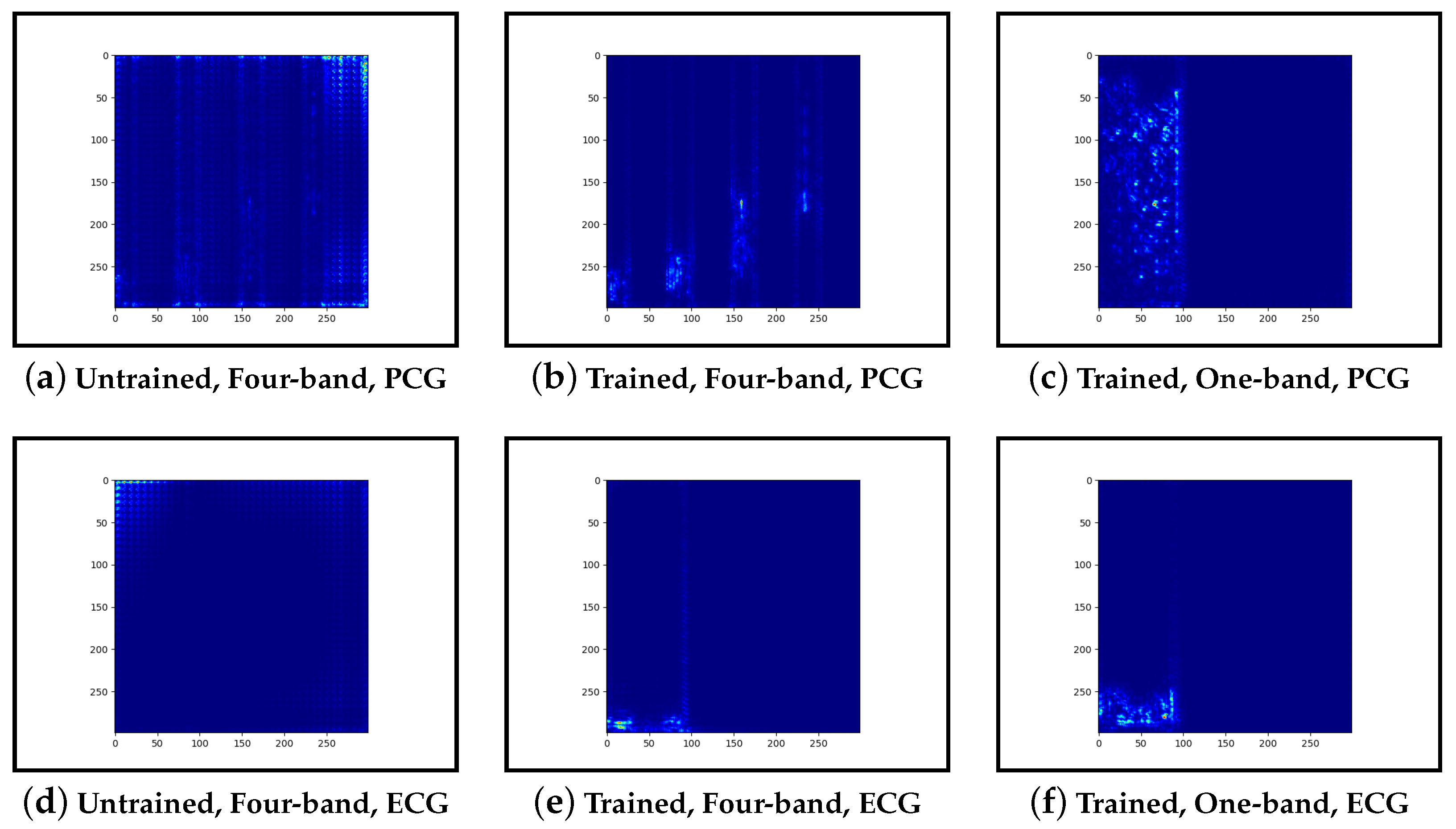
Guided-backpropagation, Grad-CAM, and LIME are used to provide interpretation of our results. Each method uses a different approach to provide a unique local interpretation. We use each method on two abnormal cases and two normal cases. These cases are taken from the test set of the model that achieved the best performance. This is then repeated for the same type of model but instead with a single band of PCG in the input image. Lastly, we compare the results against pre-trained models that are not fine-tuned. From this, we expect to see a progression from the features learnt from the ImageNet dataset to the features learnt from our training set.
5. Discussion
Our results show that the models that include both PCG and ECG data provided the most accurate results. The accuracy was further improved when the PCG was split into four bands. Furthermore, as shown in
Table 4, with the spectrogram input data, ensemble approaches performed better than those combining the PCG and ECG data into one image. From
Table A1,
Table A2 and
Table A3, we can see that across each configuration, providing only the PCG data led to lower accuracy than PCG and ECG, with a difference of up to 13%. In addition to this, the spectrogram representation led to greater accuracy, followed by the mel-spectrogram. This is a desirable outcome as the spectrogram is less computationally expensive than the other representations.
Despite being less computationally demanding, the ResNet and inceptionv3-based models performed better than the VGG architecture. The inceptionv3-based ensemble model had the best overall performance, achieving an accuracy of 91.25% and specificity of 70%. This was 10% greater accuracy and almost 25% greater specificity than the CNN from Rong et al. [
6], shown in
Table 5. This significant difference shows improvements from the utilisation of pre-trained models.
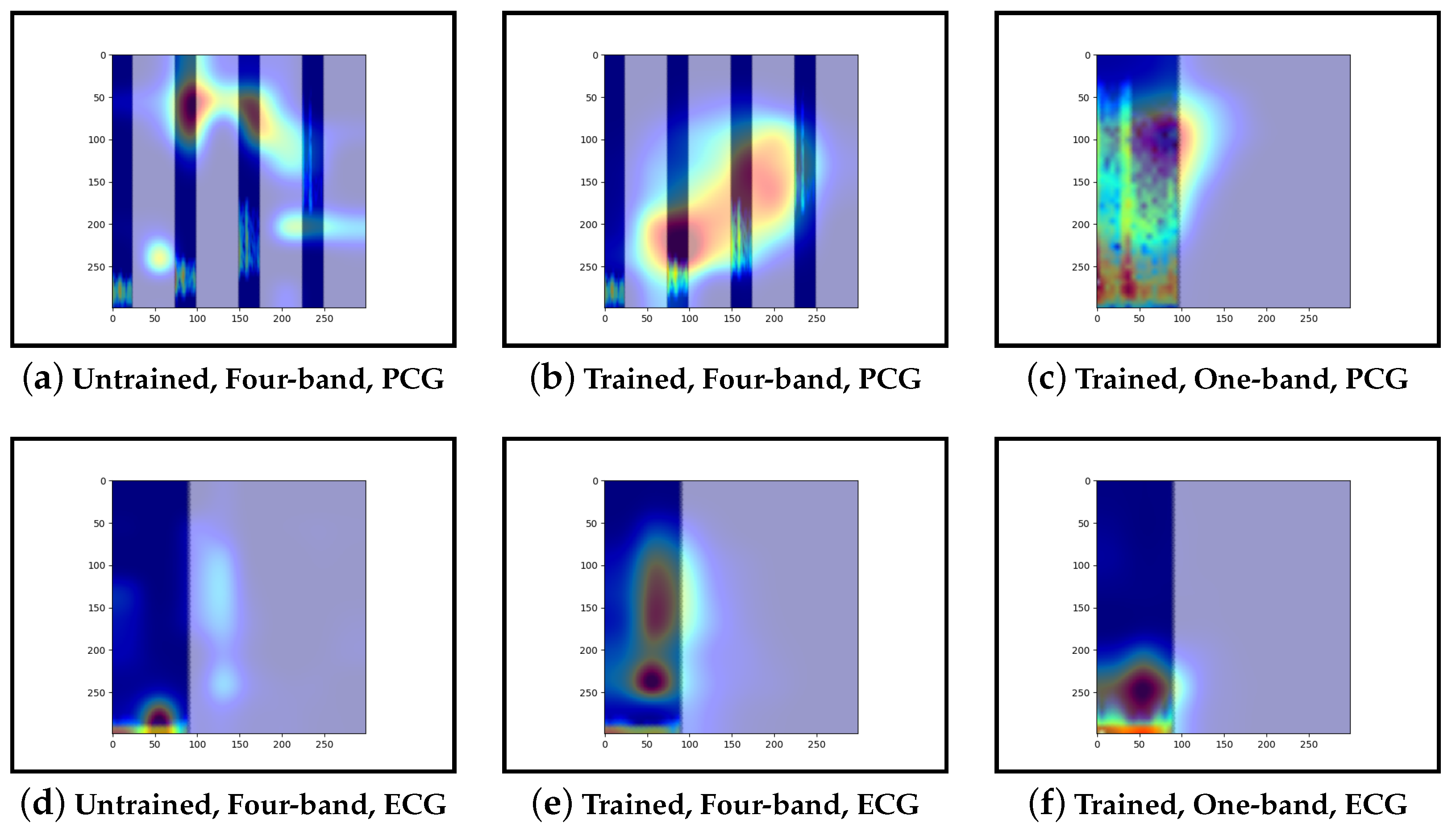
Our interpretability results suggest that an abnormal prediction favours more energy in higher frequency PCG bands. These results are expected given that this is where murmur sounds are commonly found. This can be seen through Grad-CAM, in
Figure 16b and
Figure 19b, where we see very high activation for an abnormal case. Through Grad-CAM in a normal case, it is shown that there is fewer high-frequency activations and more low-frequency activations, as shown in
Figure 10b and
Figure 13b. Our results with LIME also convey these findings, with abnormal cases having more features within the high-frequency bands, as in
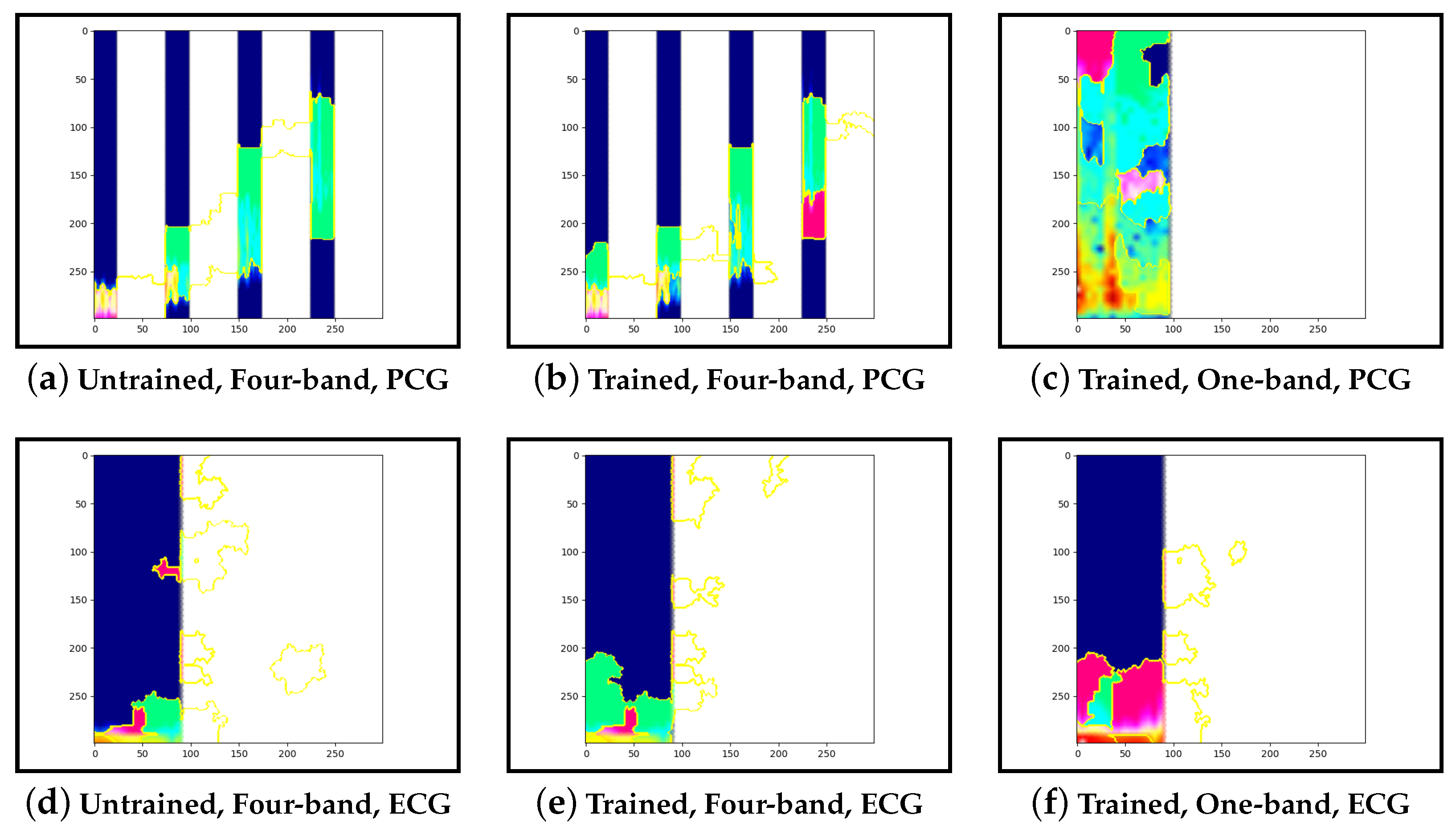
Figure 17b and
Figure 20b.
Figure 11b and
Figure 14b show normal cases with more features within low-frequency bands. Lastly, guided-backpropagation generally agrees with the previous results, as shown with the abnormal case in
Figure 18b and the normal cases in
Figure 9b and
Figure 12b. However,
Figure 15b shows lower activations in the high-frequency bands than in the other abnormal case. The results generally indicate that energy in higher frequency PCG bands is associated with abnormal features, which aligns with the literature.
The ECG band was a prominent feature shown by the high activations across all interpretability methods. As mentioned, models that included ECG as input performed better than those with only PCG. These results were expected as more information was provided to the model. The particular features within the ECG band were difficult to discern and, as such, were not explored in this work.
Examining the model that contains a single PCG band and ECG within its image, abnormal cases were associated with greater high-frequency PCG activation for both LIME and Grad-CAM. Guided-backpropagation, however, shows that one of the normal predictions is mostly based on high-frequency PCG. There was a difference in performance between the single-band PCG and four-band PCG. This suggests that splitting the PCG into multiple bands may help the model learn to extract features associated with abnormal cases, such as murmurs.
Examining the inceptionv3 model with pre-trained weights, it was observed that the model does not appear to examine the relevant clinical features. With guided-backpropagation, the model behaves as an edge detector in some cases whilst not discerning any features in others. For Grad-CAM, the model is activated across the entire image instead of within the frequency bands. The LIME images do not appear to look at specific features, instead examining broad regions of the image. Comparing these results to the trained models, they have learnt clinically-significant features. Further, it suggests that these features may be responsible for achieving high accuracy.
Our work improves on Rong et al. [
6], as shown by an increase in accuracy of 10%. In addition to this, we have provided local interpretation of select samples, which indicated that the learned features were clinically-significant. However, the use of deep pre-trained models introduces additional complexity as images need to be created before classification can occur. The introduction of ECG signals reduces the amount of data available compared to the CinC 2016 challenge models. By using less data for training, our model is more likely to overfit. In addition to this, using less data for testing, it is more difficult to identify overfitting. This may lead to a less robust model as compared to the works of Potes et al. [
16].
Deep learning offers potential benefits for cardiology, as shown in our work. The use of deep learning, however, raises substantial ethical and operational concerns. Ensuring the diversity of training data is paramount to prevent biases, as witnessed in other deep learning applications [
31]. Overfitting remains a technical concern, where algorithms might be overly tailored to specific datasets, compromising their broader applicability. Additionally, societal unease about deep-learning-driven decisions in healthcare emphasises the need for human oversight and transparent accountability.
Furthermore, ethical concerns related to patient privacy and confidentiality present a challenge for the collection of data. As a result, only limited medical datasets are available. Extensive processes are required to anonymise medical data and make it available for use in deep learning models. Despite these challenges, AI’s supportive role, aiming to enhance, not replace, human expertise, presents promising advancements in medical practice.
6. Conclusions
Our work has extended the performance of machine learning models in classifying abnormal heart sounds using PCG and ECG signals from the CinC 2016 training-a dataset. The audio is pre-processed to remove spikes and segment the data into heart cycles. After training the model on image transformations of these fragments, the results from 10 fragments are combined to predict the patient’s case.
We achieved 91.25% accuracy by fine-tuning deep image-based CNN models with spectrograms. In addition to this, it was found that using feature engineering to split the PCG into four bands and combining this with the ECG achieved the highest accuracy.
Interpretability results indicated that our models learnt clinically-significant features. These features included murmurs in high-frequency PCG bands. Moreover, splitting the data into these bands appears to significantly improve each model’s ability to learn these features.