Identification of the Recently Described Avian Hepatitis E Genotype 7 in an Outbreak of Hepatitis-Splenomegaly Syndrome (HSS) with High Mortality and Severe Drop in Egg Production in a Parent Stock Flock in Bangladesh
Abstract
1. Introduction
2. Materials and Methods
2.1. Description of Field Case and Clinical Investigations
2.2. Histopathology and Immunohistochemistry
2.3. RNA Extraction and RT-PCR Investigations
2.4. Next-Generation Sequencing
2.5. Phylogenetic Analysis
3. Results
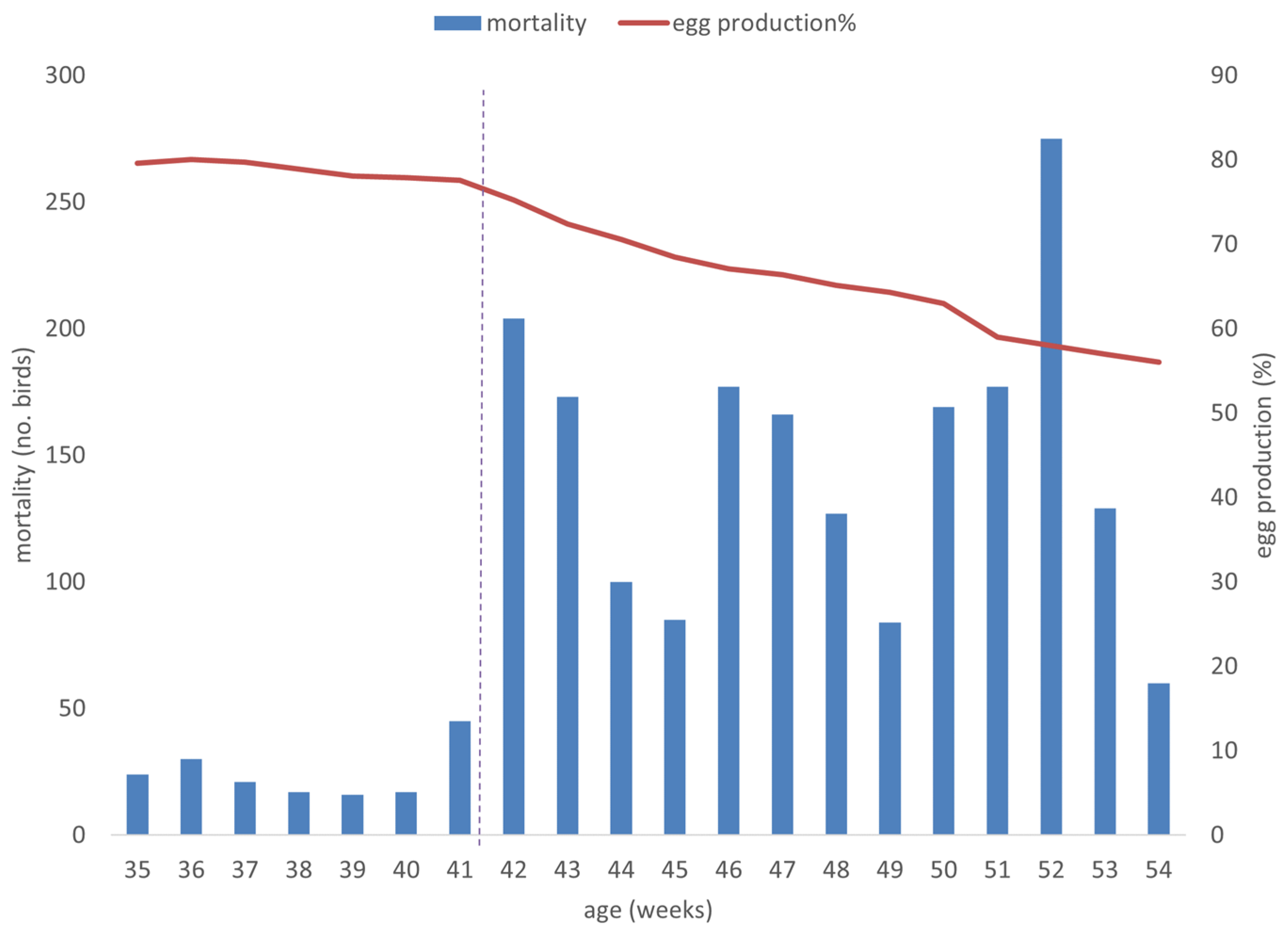
3.1. Clinical Features of the HSS Outbreak
3.2. Histological Assessment
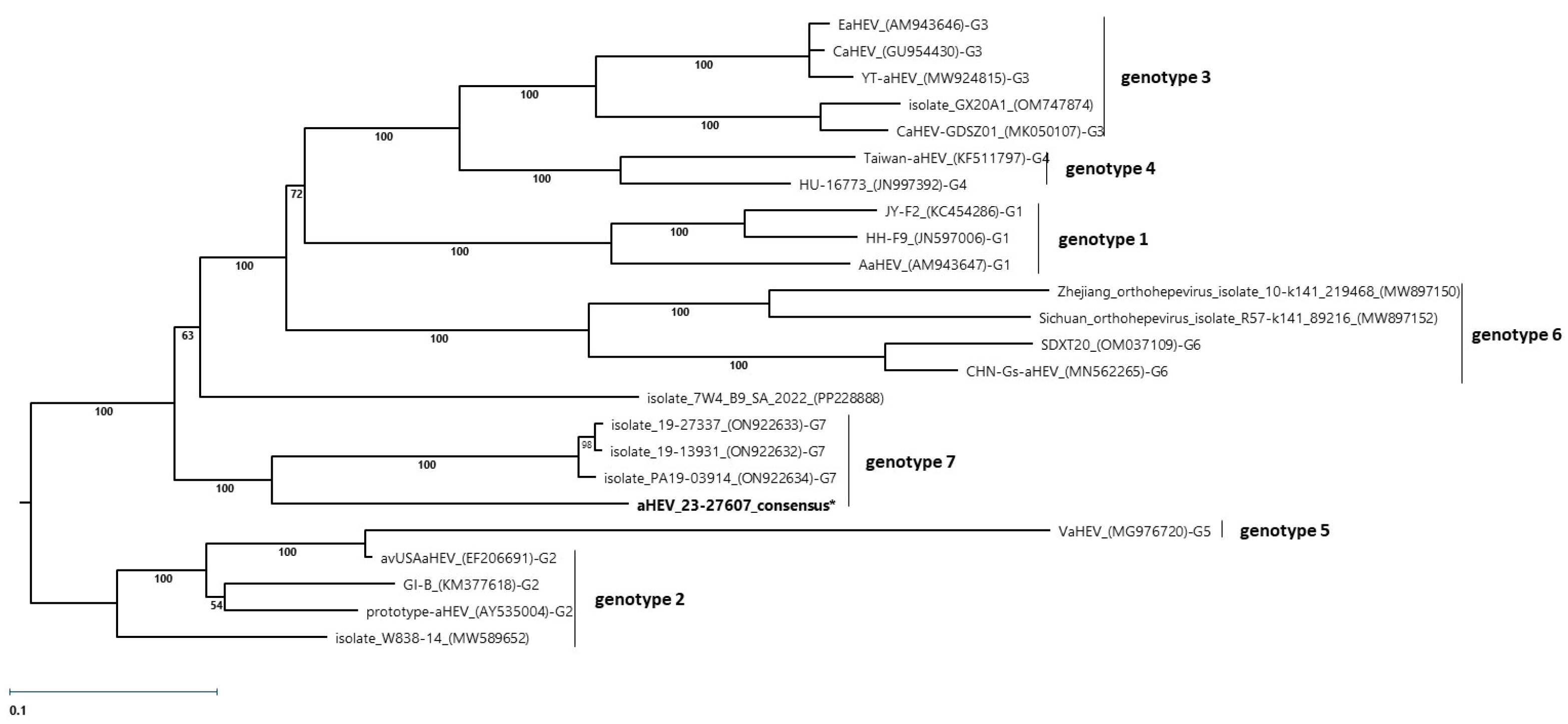
3.3. RT-PCR Investigations, Sequencing, and Phylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meng, X.J.; Shivaprasad, H.L. Avian Hepatitis E Virus Infections. In Diseases of Poultry; Swayne, D.E., Boulianne, M., Logue, C.M., McDougald, L.R., Nair, V., Suarez, D.L., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2020; pp. 528–534. [Google Scholar]
- Handlinger, J.H.; Williams, W. An Egg Drop Associated with Splenomegaly in Broiler Breeders. Avian Dis. 1988, 32, 773–778. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Lin, S.; He, S.; Zhou, E.; Zhao, Q. Avian Hepatitis E Virus: With the Trend of Genotypes and Host Expansion. Front. Microbiol. 2019, 10, 1696. [Google Scholar] [CrossRef]
- Matos, M.; Bilic, I.; Tvarogová, J.; Palmieri, N.; Furmanek, D.; Gotowiecka, M.; Liebhart, D.; Hess, M. A Novel Genotype of Avian Hepatitis E Virus Identified in Chickens and Common Pheasants (Phasianus colchicus), Extending Its Host Range. Sci. Rep. 2022, 12, 21743. [Google Scholar] [CrossRef]
- Siedlecka, M.; Kublicka, A.; Wieliczko, A.; Matczuk, A.K. Molecular Detection of Avian Hepatitis E Virus (Orthohepevirus B) in Chickens, Ducks, Geese, and Western Capercaillies in Poland. PLoS ONE 2022, 17, e0269854. [Google Scholar] [CrossRef]
- Zhang, X.; Bilic, I.; Troxler, S.; Hess, M. Evidence of Genotypes 1 and 3 of Avian Hepatitis E Virus in Wild Birds. Virus Res. 2017, 228, 75–78. [Google Scholar] [CrossRef]
- Yang, C.; Wang, L.; Shen, H.; Zheng, Y.; Gauger, P.C.; Chen, Q.; Zhang, J.; Yoon, K.J.; Harmon, K.M.; Main, R.G.; et al. Detection and Genomic Characterization of New Avian-like Hepatitis E Virus in a Sparrow in the United States. Arch. Virol. 2018, 163, 2861–2864. [Google Scholar] [CrossRef] [PubMed]
- Purdy, M.A.; Drexler, J.F.; Meng, X.J.; Norder, H.; Okamoto, H.; Van der Poel, W.H.M.; Reuter, G.; de Souza, W.M.; Ulrich, R.G.; Smith, D.B. ICTV Virus Taxonomy Profile: Hepeviridae 2022. J. Gen. Virol. 2022, 103, 001778. [Google Scholar] [CrossRef]
- Liu, B.; Chen, Y.; Zhao, L.; Zhang, M.; Ren, X.; Zhang, Y.; Zhang, B.; Fan, M.; Zhao, Q.; Zhou, E.M. Identification and Pathogenicity of a Novel Genotype Avian Hepatitis E Virus from Silkie Fowl (Gallus Gallus). Vet. Microbiol. 2020, 245, 108688. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Zhang, Z.; Zhang, Y.; Cui, Z.; Chang, S.; Zhao, P. Complete Genome Analysis of Avian Hepatitis E Virus from Chicken with Hepatic Rupture Hemorrhage Syndrome. Vet. Microbiol. 2020, 242, 108577. [Google Scholar] [CrossRef]
- Bilic, I.; Jaskulska, B.; Basic, A.; Morrow, C.J.; Hess, M. Sequence Analysis and Comparison of Avian Hepatitis E Viruses from Australia and Europe Indicate the Existence of Different Genotypes. J. Gen. Virol. 2009, 90, 863–873. [Google Scholar] [CrossRef]
- Martin, M. Marcel Martin Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Haqshenas, G.; Shivaprasad, H.L.; Woolcock, P.R.; Read, D.H.; Meng, X.J. Genetic Identification and Characterization of a Novel Virus Related to Human Hepatitis E Virus from Chickens with Hepatitis-Splenomegaly Syndrome in the United States. J. Gen. Virol. 2001, 82, 2449–2462. [Google Scholar] [CrossRef] [PubMed]
- Payne, C.J.; Ellisa, T.M.; Planta, S.L.; Gregorya, A.R.; Wilcoxb, G.E. Sequence Data Suggests Big Liver and Spleen Disease Virus (BLSV) Is Genetically Related to Hepatitis E Virus. Vet. Microbiol. 1999, 68, 119–125. [Google Scholar] [CrossRef]
- Agunos, A.C.; Yoo, D.; Youssef, S.A.; Ran, D.; Binnington, B.; Hunter, D.B. Avian Hepatitis E Virus in an Outbreak of Hepatitis-Splenomegaly Syndrome and Fatty Liver Haemorrhage Syndrome in Two Flaxseed-Fed Layer Flocks in Ontario. Avian Pathol. 2006, 35, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.F.; Haqshenas, G.; Shivaprasad, H.L.; Guenette, D.K.; Woolcock, P.R.; Larsen, C.T.; Pierson, F.W.; Elvinger, F.; Toth, T.E.; Meng, X.J. Heterogeneity and Seroprevalence of a Newly Identified Avian Hepatitis E Virus from Chickens in the United States. J. Clin. Microbiol. 2002, 40, 4197–4202. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.F.; Larsen, C.T.; Dunlop, A.; Huang, F.F.; Pierson, F.W.; Toth, T.E.; Meng, X.J. Genetic Identification of Avian Hepatitis E Virus (HEV) from Healthy Chicken Flocks and Characterization of the Capsid Gene of 14 Avian HEV Isolates from Chickens with Hepatitis-Splenomegaly Syndrome in Different Geographical Regions of the United States. J. Gen. Virol. 2004, 85, 693–700. [Google Scholar] [CrossRef]
- Johne, R.; Dremsek, P.; Reetz, J.; Heckel, G.; Hess, M.; Ulrich, R.G. Hepeviridae: An Expanding Family of Vertebrate Viruses. Infect. Genet. Evol. 2014, 27, 212–229. [Google Scholar] [CrossRef]
- Iqbal, T.; Rashid, U.; Idrees, M.; Afroz, A.; Kamili, S.; Purdy, M.A. A Novel Avian Isolate of Hepatitis E Virus from Pakistan. Virol. J. 2019, 16, 142. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Li, Y.; Meng, F.; Cui, Z.; Chang, S.; Zhao, P. Hepatic Rupture Hemorrhage Syndrome in Chickens Caused by a Novel Genotype Avian Hepatitis E Virus. Vet. Microbiol. 2018, 222, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Chi, Z.; Zhang, Y.; Xue, Q.; Zhai, T.; Chang, S.; Wang, J.; Zhao, P. Natural Co-Infection of Fowl Adenovirus Type E-8b and Avian Hepatitis E Virus in Parental Layer Breeders in Hebei, China. Virol. Sin. 2023, 38, 317–320. [Google Scholar] [CrossRef]
- Meng, K.; Liu, M.; Zhang, Y.; Yuan, X.; Xu, H. A Genetically Novel Avian Hepatitis E Virus in China. Virus Genes. 2022, 58, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Peralta, B.; Biarnés, M.; Ordóñez, G.; Porta, R.; Martín, M.; Mateu, E.; Pina, S.; Meng, X.J. Evidence of Widespread Infection of Avian Hepatitis E Virus (Avian HEV) in Chickens from Spain. Vet. Microbiol. 2009, 137, 31–36. [Google Scholar] [CrossRef]
- Sprygin, A.V.; Nikonova, Z.B.; Zinyakov, N.G. Avian Hepatitis E Virus Identified in Russian Chicken Flocks Exhibits High Genetic Divergence Based on the ORF2 Capsid Gene. Avian Pathol. 2012, 41, 459–463. [Google Scholar] [CrossRef]
- Troxler, S.; Pać, K.; Prokofieva, I.; Liebhart, D.; Chodakowska, B.; Furmanek, D.; Hess, M. Subclinical Circulation of Avian Hepatitis E Virus within a Multiple-Age Rearing and Broiler Breeder Farm Indicates Persistence and Vertical Transmission of the Virus. Avian Pathol. 2014, 43, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Nwokorogu, V.C.; Pillai, S.; San, J.E.; Pillay, C.; Nyaga, M.M.; Sabiu, S. A Metagenomic Investigation of the Faecal RNA Virome Structure of Asymptomatic Chickens Obtained from a Commercial Farm in Durban, KwaZulu-Natal Province, South Africa. BMC Genom. 2024, 25, 629. [Google Scholar] [CrossRef]
- Chen, Y.-M.; Sadiq, S.; Tian, J.-H.; Chen, X.; Lin, X.-D.; Shen, J.-J.; Chen, H.; Hao, Z.-Y.; Wille, M.; Zhou, Z.-C.; et al. RNA Viromes from Terrestrial Sites across China Expand Environmental Viral Diversity. Nat. Microbiol. 2022, 7, 1312–1323. [Google Scholar] [CrossRef]
- Clarke, J.K.; Allan, G.M.; Bryson, D.G.; Todd, D.; MacKie, D.P.; McFerran, J.B.; Williams, W. Big Liver and Spleen Disease of Broiler Breeders. Avian Pathol. 1990, 19, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Morrow, C.J.; Samu, G.; Mátrai, E.; Klausz, Á.; Wood, A.M.; Richter, S.; Jaskulska, B.; Hess, M. Avian Hepatitis E Virus Infection and Possible Associated Clinical Disease in Broiler Breeder Flocks in Hungary. Avian Pathol. 2008, 37, 527–535. [Google Scholar] [CrossRef] [PubMed]


| Primer | Sequence (5′-3′) | Annealing Temperature (°C) | Product Size (bp) |
|---|---|---|---|
| N_term_aHEV-F1 | CCTACCTTACCCAACAGCAG | 49 | 1076 |
| N_term_aHEV-R1 | CCTCACACCACGAGCTATAC | ||
| N_term_aHEV-F2 | GGTACTCAGCCGGATTCTATC | 53 | 1051 |
| N_term_aHEV-R2 | CGGCCTTCGTTTAGTACAAAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matos, M.; Bilic, I.; Kőrösi, L.; Hasan, R.; Liebhart, D.; Palmieri, N.; Hess, M. Identification of the Recently Described Avian Hepatitis E Genotype 7 in an Outbreak of Hepatitis-Splenomegaly Syndrome (HSS) with High Mortality and Severe Drop in Egg Production in a Parent Stock Flock in Bangladesh. Poultry 2025, 4, 16. https://doi.org/10.3390/poultry4020016
Matos M, Bilic I, Kőrösi L, Hasan R, Liebhart D, Palmieri N, Hess M. Identification of the Recently Described Avian Hepatitis E Genotype 7 in an Outbreak of Hepatitis-Splenomegaly Syndrome (HSS) with High Mortality and Severe Drop in Egg Production in a Parent Stock Flock in Bangladesh. Poultry. 2025; 4(2):16. https://doi.org/10.3390/poultry4020016
Chicago/Turabian StyleMatos, Miguel, Ivana Bilic, László Kőrösi, Rakibul Hasan, Dieter Liebhart, Nicola Palmieri, and Michael Hess. 2025. "Identification of the Recently Described Avian Hepatitis E Genotype 7 in an Outbreak of Hepatitis-Splenomegaly Syndrome (HSS) with High Mortality and Severe Drop in Egg Production in a Parent Stock Flock in Bangladesh" Poultry 4, no. 2: 16. https://doi.org/10.3390/poultry4020016
APA StyleMatos, M., Bilic, I., Kőrösi, L., Hasan, R., Liebhart, D., Palmieri, N., & Hess, M. (2025). Identification of the Recently Described Avian Hepatitis E Genotype 7 in an Outbreak of Hepatitis-Splenomegaly Syndrome (HSS) with High Mortality and Severe Drop in Egg Production in a Parent Stock Flock in Bangladesh. Poultry, 4(2), 16. https://doi.org/10.3390/poultry4020016









