RbfCon: Construct Radial Basis Function Neural Networks with Grammatical Evolution
Abstract
1. Introduction
- The vector represents the input pattern with dimension d.
- The parameter k stands for the number of weights of the model. These weights are represented by vector .
- The vectors represent the so-called centers of the network.
- The final output of the model for the input pattern is denoted by .
- The function in most cases is represented by the Gaussian function defined asThis function is selected as the output function since the output value uses only the distance of vectors x and c.
- The method can efficiently construct the structure of RBF networks and achieve optimal adjustment of their parameters.
- The resulting networks do not have the numerical problems caused by the traditional training technique of RBF networks.
- The created software provides an easy user interface for performing experiments and can be installed on almost any operating system.
2. Materials and Methods
2.1. The Original Training Procedure of RBF Neural Networks
- Set the matrix of k weights, , and , where are the expected values for input patterns .
- SolveThe matrix denotes the the pseudo-inverse of , with
| Algorithm 1 The used K-means Algorithm |
|
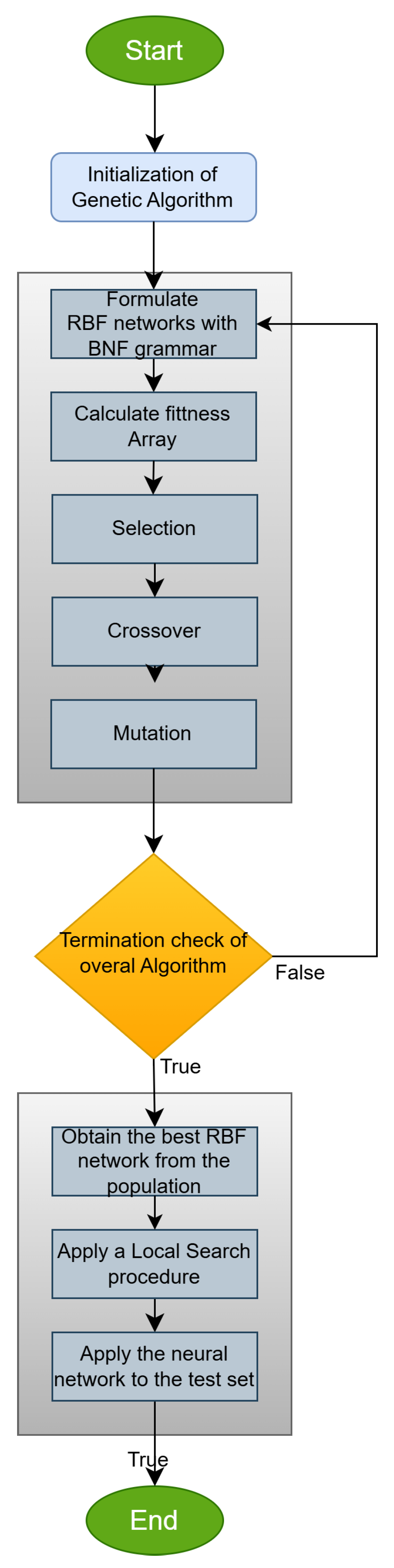
2.2. The Used Construction Procedure
- The symbol N stands for the set of non-terminal symbols;
- The symbol T represents the set of terminal symbols;
- S is a non-terminal symbol, used as the start symbol of the grammar;
- P is the set of production rules that are used to produce terminal symbols from non-terminal symbols.
- Obtain the next element V from the under-processing chromosome.
- Select the next production rule according to the equation, Rule = V mod , where the quantity stands for the total number of production rules for the non-terminal symbol that is under processing.
- Initialization Step.
- (a)
- Set the number of chromosomes and the number of allowed generations .
- (b)
- Set the selection rate of the genetic algorithm, denoted as where .
- (c)
- Set the mutation rate of the genetic algorithm, denoted as , with 1.
- (d)
- Initialize the chromosomes as sets of positive random integers.
- (e)
- Set k = 0, the generation number.
- Main loop step.
- (a)
- Evaluate fitness.
- For every chromosome create the corresponding neural network using the grammar of Figure 1. Denote this network as .
- Compute the fitness for the train set asThe set represents the train dataset of the objective problem.
- (b)
- Apply the selection operator. A sorting of the chromosomes is performed according to their fitness values. The best of them are copied without changes to the next generation. The rest of the chromosomes will be replaced by new chromosomes produced during the crossover procedure.
- (c)
- Apply the crossover operator. During this procedure, a series of offsprings will be produced. For each pair of offsprings denoted as and , two chromosomes should be selected from the original population using tournament selection. The production of the new chromosomes is conducted using the one-point crossover procedure. An example of this process is graphically outlined in Figure 2.
- (d)
- Perform mutation. For each element of every chromosome, randomly draw a number . If , then the corresponding element is replaced by a new randomly produced element.
- (e)
- Set k = k + 1.
- Check for the termination step. If , then go to step 2.
- Local Optimization step. Obtain the chromosome and produce the corresponding RBF neural network . The parameters of this neural network are obtained by minimizing with some local search procedure the training error defined as
- Application in test set. Apply the final model to the test dataset and report the test error.
2.3. Installation Procedure
- unzip RbfCon-master.zip;
- cd RbfCon-master;
- qmake (or qmake-qt5 in some Linux installations);
- make.
2.4. The Main Executable RbfCon
- trainfile=<filename> The string parameter filename determines the name of the file containing the training dataset for the software. The user is required to provide this parameter to start the process. The format for this file is shown in Figure 4. The first number denoted as D in this figure determines the dimension of the input dataset, and the second number denoted as M represents the number of input patterns. Every line in the file contains a pattern and the required desired output.
- testfile=<filename> The string parameter filename stands for the name of the test dataset used by the software. The user should provide at least the parameters trainfile and testfile in order to initiate the method. The format of the test dataset is the same as the training dataset.
- chromosome_count=<count> This integer parameter denotes the number of chromosomes in the genetic algorithm (parameter of the algorithm). The default value for this parameter is 500.
- chromosome_size=<size> The integer parameter size determines the size of each chromosome for the Grammatical Evolution process. The default value for this parameter is 100.
- selection_rate=<rate> The double precision parameter rate stands for the selection rate of the Grammatical Evolution process. The default value for this parameter is 0.10 (10%).
- mutation_rate=<rate> The double precision parameter rate represents the mutation rate for the Grammatical Evolution process. The default value for this parameter is 0.05 (5%).
- generations=<gens> The integer parameter gens stands for the maximum number of allowed generations for the Grammatical Evolution procedure (parameter of the current algorithm). The original value is 500.
- local_method=<method> The used local optimization method will be applied to the parameters of the RBF model when the Grammatical Evolution procedure finished. The available local optimization methods are as follows:
- (a)
- lbfgs. The method L-BFGS can be considered a variation in the Broyden–Fletcher–Goldfarb–Shanno (BFGS) optimization technique [53], which utilizes a minimum amount of memory. This optimization method was used in many cases, such as image reconstruction [54], inverse eigenvalue problems [55], seismic problems [56], training of deep neural networks [57], etc. Also, the series of modifications that take advantage of modern parallel computing systems were proposed [58,59,60].
- (b)
- bfgs. The BFGS variant of Powell was used here [61], when this option is enabled.
- (c)
- adam. This option denotes the application of the Adam optimizer [62] as the local optimization algorithm.
- (d)
- gradient. This option represents the usage of the Gradient Descent method [63] as the local optimization algorithm.
- (e)
- none. With this option, no local optimization method will be used after the Genetic Algorithm is terminated.
- iterations=<iters> This integer parameter determines the number of consecutive applications of the proposed technique to the original dataset. The default value is 30.
2.5. Example of Execution
./RbfCon --trainfile=EXAMPLES/wdbc.train --testfile=EXAMPLES/wdbc.test --iterations=2
| Algorithm 2 The output of the example run |
| Iteration: 1 TRAIN ERROR: 20.40039584 Iteration: 1 TEST ERROR: 0.07733519754 Iteration: 1 CLASS ERROR: 9.12% Iteration: 1 SOLUTION: ((6.711)*(exp((−((pow(x13 − (−97.73),2))+ ((pow(x23 − (43.7),2))+((pow(x23 − (84.7),2))+ (pow(x23 − (91.3),2))))))/(2*pow((60.1),2))))+ (−0.0099969))+((−0.39)*(exp((−((pow(x25 − (9.336),2))+ ((pow(x18 − (6.8),2))+(pow(x4 − (6.52),2)))))/(2*pow((711.5),2))))+ (0.00185452))+((3.7)*(exp((−(pow(x3 − (−6.8),2)))/(2*pow((2.3),2))))+ (−0.001))+((−5.8)*(exp((−(pow(x2 − (−99969.10),2)))/ (2*pow((6342.3747),2))))+(−0.002)) Iteration: 2 TRAIN ERROR: 16.06705417 Iteration: 2 TEST ERROR: 0.05654528154 Iteration: 2 CLASS ERROR: 4.56% Iteration: 2 SOLUTION: ((4.4)*(exp((−((pow(x27 − (3.6),2))+ (pow(x23 − (8.9893),2))))/(2*pow((49.9),2))))+(−0.004))+ ((−2.09)*(exp((−((pow(x23 − (−9.1),2))+ ((pow(x30 − (−40.4),2))+((pow(x4 − (98.5),2))+ (pow(x22 − (−4.4),2))))))/(2*pow((189.9),2))))+(0.007))+ ((−9.8)*(exp((−((pow(x2 − (24.4),2))+((pow(x28 − (2.04),2))+ (pow(x27 − (9.11),2)))))/(2*pow((3.1),2))))+(0.009))+ ((−67.49)*(exp((−(pow(x10 − (−2629940.4),2)))/(2*pow((098.5),2))))+(0.0094)) Average Train Error: 18.23372501 Average Test Error: 0.06694023954 Average Class Error: 6.84% |
3. Results
- The UCI repository, https://archive.ics.uci.edu/ (accessed on 14 November 2024) [65];
- The Keel repository, https://sci2s.ugr.es/keel/datasets.php (accessed on 14 November 2024) [66];
- The Statlib URL ftp://lib.stat.cmu.edu/datasets/index.html (accessed on 14 November 2024).
3.1. The Used Datasets
- Alcohol, a dataset related to alcohol consumption [69]. This dataset contains four distinct classes.
- Australian, an economic dataset [70]. The number of classes for this dataset is two.
- Bands, related to problems that occur in printing [71]. It has two classes.
- Dermatology, which is also a medical dataset originated in [74]. It has six classes.
- Ecoli, that is related to problems regarding proteins [75]. It has eight classes.
- Fert, used in demographics. It has two distinct classes.
- Haberman, a medical dataset related to the detection of breast cancer. This dataset has two classes.
- Hayes-roth dataset [76]. This dataset has three classes.
- Heart, a medical dataset about heart diseases [77]. This dataset has two classes.
- HeartAttack, which is a medical dataset related to heart diseases. It has two distinct classes.
- Hepatitis, a dataset used for the detection of hepatitis.
- Housevotes, used for the Congressional voting in USA [78].
- Lymography [83], which has four classes.
- Magic, this dataset contains generated data to simulate registration of high energy gamma particles [84]. It has two classes.
- Mammographic, a medical dataset related to the presence of breast cancer [85].
- Pima, a medical dataset that was useful for the detection of diabetes [88].
- Popfailures, a dataset contains climate measurements [89]. It has two classes.
- Regions2, a medical dataset that contains measurements from liver biopsy images [90]. It has five classes.
- Ring, which is a problem with 20 dimensions and two classes, related to a series of multivariate normal distributions.
- Saheart, a medical dataset related to heart diseases with two classes [91].
- Statheart, which is a also a medical dataset related to heart diseases.
- Spambase, a dataset used to detect spam emails from a large database. The dataset has two distinct classes.
- Spiral, an artificial dataset with two classes.
- Student, a dataset contains measurements from various experiments in schools [92].
- Tae, this dataset consist of evaluations of teaching performance. It has three classes.
- Transfusion, which is a medical dataset [93].
- Zoo, which used to detect the class of some animals [100]. It contains seven classes.
- Abalone, a dataset that used to predict the age of abalones with 8 features.
- Airfoil, a dataset provided by NASA [101] with 5 features.
- Auto, a dataset used to predict the fuel consumption with 7 features.
- BK, that contains measurements from a series of basketball games with 4 features.
- BL, a dataset used to record electricity experiments with 7 features.
- Baseball, a dataset with 16 features used to estimate the income of baseball players.
- Concrete, a dataset with 8 features used in civil engineering [102].
- DEE, a dataset with 6 featured that was to predict the electricity cost.
- FA, that contains measurements about the body fat with 18 features.
- HO, a dataset originated in the STATLIB repository with 13 features.
- Housing, used to predict the price of houses [103] with 13 features.
- Laser, which is a dataset with 4 features. It has been used in various laser experiments.
- LW, a dataset with 9 features used to record the weight of babies.
- MB, a dataset provide by from Smoothing Methods in Statistics [104] with 2 features.
- Mortgage, an economic dataset from USA with 15 features.
- NT, a dataset with 2 features used to record body temperatures [105].
- Plastic, a dataset with 2 features used to detect the pressure on plastics.
- PL, a dataset with 2 features provided by the STATLIB repository.
- Quake, a dataset used to measure the strength of earthquakes with 3 features.
- SN, a dataset that provides experimental measurements related to trellising and pruning. This dataset has 11 features.
- Stock, a dataset with 9 features used to approximate the prices of various stocks.
- Treasury, an economic dataset from USA that contains 15 features.
- TZ, which is a dataset originated in the STATLIB repository. It has 60 features.
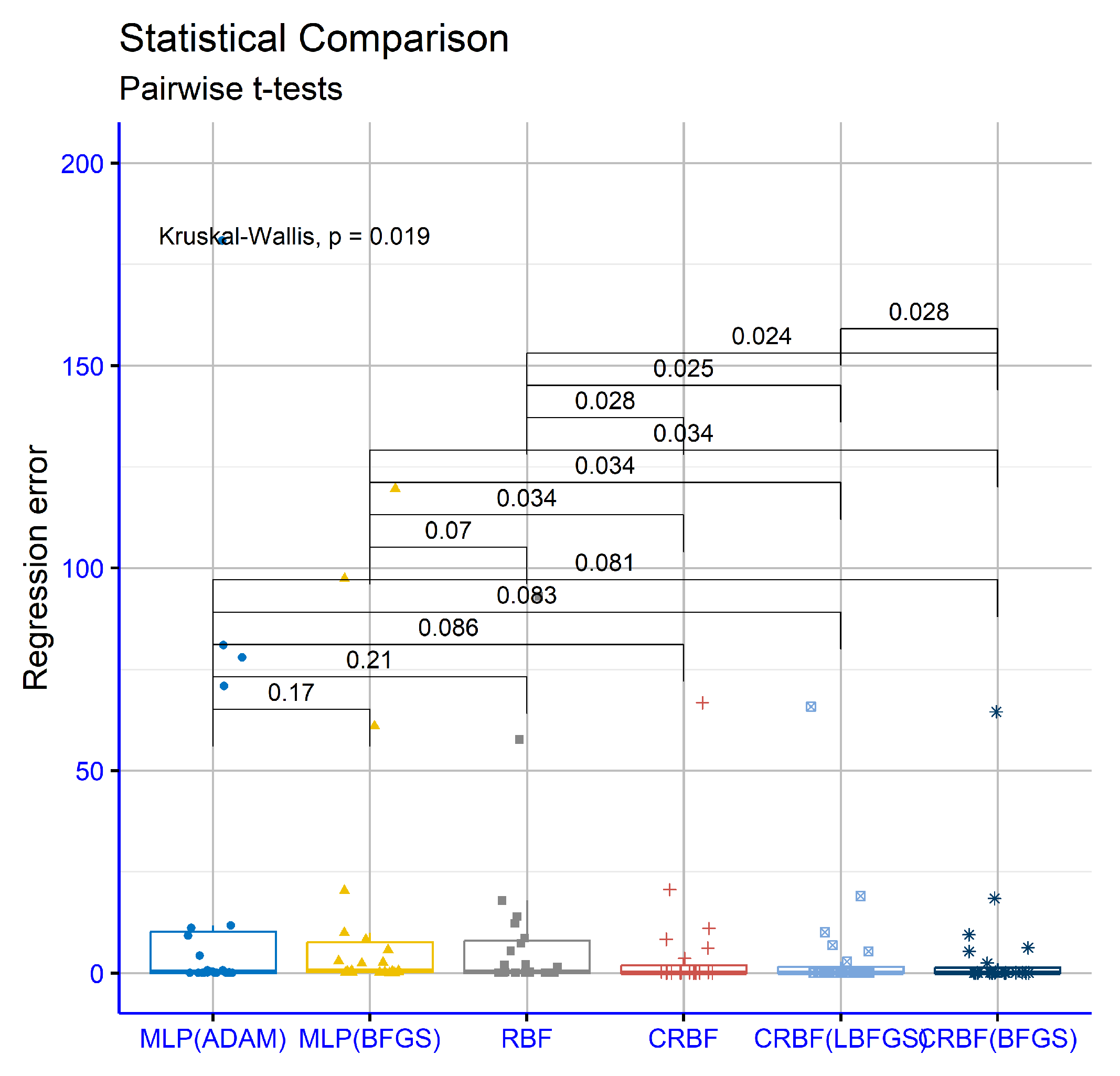
3.2. Experimental Results
- The column MLP(ADAM) represents the usage of the ADAM optimization method [62] to train an artificial neural network with processing nodes.
- The column MLP(BFGS) stands for the usage of the BFGS optimization method [61] in the training process of an artificial neural network with processing nodes.
- The column RBF denotes the usage of the original two-phase method for the training of an RBF neural network with weights. This method was described previously in Section 2.1.
- The column CRBF stands for the utilization of the current software using the parameters of Table 1, and as local search procedure, the none option was selected.
- The column CRBF(LBFGS) denotes the application of the current method using as local search procedure the L-Bfgs method.
- The column CRBF(BFGS) stands for the application of the current work using as local search procedure the Bfgs method.
- The row denoted as AVERAGE outlines the average classification or regression error for all datasets in the corresponding table.
- The row denoted as STDEV depicts the standard deviation for all datasets in the corresponding table.
| Dataset | MLP (ADAM) | MLP (BFGS) | RBF | CRBF | CRBF (LBFGS) | CRBF(BFGS) |
|---|---|---|---|---|---|---|
| APPENDICITIS | 16.50% | 18.00% | 12.23% | 13.60% | 14.10% | 13.60% |
| ALCOHOL | 57.78% | 41.50% | 49.38% | 51.24% | 49.32% | 45.11% |
| AUSTRALIAN | 35.65% | 38.13% | 34.89% | 14.14% | 14.26% | 14.23% |
| BANDS | 36.92% | 36.67% | 37.17% | 35.75% | 36.58% | 36.03% |
| CLEVELAND | 67.55% | 77.55% | 67.10% | 49.14% | 49.52% | 50.28% |
| DERMATOLOGY | 26.14% | 52.92% | 62.34% | 45.20% | 38.43% | 36.66% |
| ECOLI | 64.43% | 69.52% | 59.48% | 54.18% | 54.39% | 53.03% |
| FERT | 23.98% | 23.20% | 15.00% | 15.20% | 15.50% | 15.90% |
| HABERMAN | 29.00% | 29.34% | 25.10% | 26.27% | 27.07% | 26.40% |
| HAYES-ROTH | 59.70% | 37.33% | 64.36% | 34.54% | 39.00% | 36.76% |
| HEART | 38.53% | 39.44% | 31.20% | 17.22% | 17.44% | 16.96% |
| HEARTATTACK | 45.55% | 46.67% | 29.00% | 22.60% | 21.83% | 21.53% |
| HEPATITIS | 68.13% | 72.47% | 64.63% | 54.25% | 47.50% | 48.75% |
| HOUSEVOTES | 7.48% | 7.13% | 6.13% | 3.05% | 3.65% | 3.05% |
| IONOSPHERE | 16.64% | 15.29% | 16.22% | 14.32% | 13.40% | 12.37% |
| LIVERDISORDER | 41.53% | 42.59% | 30.84% | 32.21% | 31.71% | 31.35% |
| LYMOGRAPHY | 39.79% | 35.43% | 25.50% | 26.36% | 25.71% | 21.49% |
| MAGIC | 40.55% | 17.30% | 21.28% | 22.18% | 19.62% | 20.35% |
| MAMMOGRAPHIC | 46.25% | 17.24% | 21.38% | 17.67% | 19.02% | 17.30% |
| PARKINSONS | 24.06% | 27.58% | 17.41% | 12.79% | 13.37% | 12.47% |
| PIMA | 34.85% | 35.59% | 25.78% | 24.13% | 24.90% | 24.15% |
| POPFAILURES | 5.18% | 5.24% | 7.04% | 6.98% | 6.94% | 6.80% |
| REGIONS2 | 29.85% | 36.28% | 38.29% | 26.34% | 26.81% | 26.55% |
| RING | 28.80% | 29.24% | 21.67% | 11.08% | 10.13% | 10.33% |
| SAHEART | 34.04% | 37.48% | 32.19% | 29.52% | 29.28% | 29.19% |
| SPAMBASE | 48.05% | 18.16% | 29.35% | 16.95% | 15.53% | 15.60% |
| SPIRAL | 47.67% | 47.99% | 44.87% | 42.14% | 43.88% | 43.05% |
| STATHEART | 44.04% | 39.65% | 31.36% | 19.22% | 19.15% | 18.19% |
| STUDENT | 5.13% | 7.14% | 5.49% | 7.63% | 6.33% | 4.32% |
| TAE | 60.20% | 51.58% | 60.02% | 56.73% | 56.33% | 56.40% |
| TRANSFUSION | 25.68% | 25.84% | 26.41% | 25.15% | 24.70% | 24.36% |
| WDBC | 35.35% | 29.91% | 7.27% | 6.77% | 6.52% | 6.36% |
| WINE | 29.40% | 59.71% | 31.41% | 11.00% | 11.65% | 10.71% |
| Z_F_S | 47.81% | 39.37% | 13.16% | 11.13% | 11.47% | 10.57% |
| Z_O_N_F_S | 78.79% | 65.67% | 48.70% | 52.34% | 49.32% | 46.22% |
| ZO_NF_S | 47.43% | 43.04% | 9.02% | 11.08% | 11.18% | 8.90% |
| ZONF_S | 11.99% | 15.62% | 4.03% | 4.14% | 3.94% | 3.58% |
| ZOO | 14.13% | 10.70% | 21.93% | 11.60% | 9.00% | 10.90% |
| AVERAGE | 37.23% | 35.36% | 30.23% | 24.63% | 24.17% | 23.42% |
| STDEV | 18.25% | 18.36% | 18.41% | 15.92% | 15.46% | 15.25% |
| Dataset | MLP (ADAM) | MLP (BFGS) | RBF | CRBF | CRBF (LBFGS) | CRBF (BFGS) |
|---|---|---|---|---|---|---|
| ABALONE | 4.30 | 5.69 | 7.37 | 6.14 | 5.35 | 5.32 |
| AIRFOIL | 0.005 | 0.003 | 0.27 | 0.004 | 0.004 | 0.002 |
| AUTO | 70.84 | 60.97 | 17.87 | 11.03 | 10.03 | 9.45 |
| BK | 0.025 | 0.28 | 0.02 | 0.02 | 0.02 | 0.03 |
| BL | 0.62 | 2.55 | 0.013 | 0.04 | 0.024 | 0.01 |
| BASEBALL | 77.90 | 119.63 | 93.02 | 66.74 | 65.80 | 64.52 |
| CONCRETE | 0.078 | 0.066 | 0.011 | 0.012 | 0.010 | 0.009 |
| DEE | 0.63 | 2.36 | 0.17 | 0.23 | 0.22 | 0.20 |
| FA | 0.048 | 0.43 | 0.015 | 0.013 | 0.014 | 0.011 |
| HO | 0.035 | 0.62 | 0.03 | 0.013 | 0.015 | 0.012 |
| HOUSING | 81.00 | 97.38 | 57.68 | 20.60 | 19.02 | 18.40 |
| LASER | 0.03 | 0.015 | 0.03 | 0.06 | 0.05 | 0.03 |
| LW | 0.028 | 2.98 | 0.03 | 0.011 | 0.011 | 0.011 |
| MB | 0.06 | 0.129 | 5.43 | 0.055 | 0.06 | 0.12 |
| MORTGAGE | 9.24 | 8.23 | 1.45 | 0.165 | 0.18 | 0.074 |
| NT | 0.006 | 0.129 | 13.97 | 0.006 | 0.006 | 0.006 |
| PLASTIC | 11.71 | 20.32 | 8.62 | 3.58 | 2.86 | 2.43 |
| PL | 0.32 | 0.58 | 2.118 | 0.064 | 0.026 | 0.024 |
| QUAKE | 0.117 | 0.29 | 0.07 | 0.036 | 0.036 | 0.036 |
| SN | 0.026 | 0.4 | 0.027 | 0.025 | 0.026 | 0.025 |
| STOCK | 180.89 | 302.43 | 12.23 | 8.31 | 6.90 | 6.25 |
| TREASURY | 11.16 | 9.91 | 2.02 | 0.0027 | 0.12 | 0.10 |
| TZ | 0.43 | 0.22 | 0.036 | 0.036 | 0.036 | 0.035 |
| AVERAGE | 19.54 | 27.64 | 9.67 | 5.10 | 4.82 | 4.66 |
| STDEV | 43.67 | 68.11 | 22.01 | 14.34 | 14.05 | 13.76 |
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mjahed, M. The use of clustering techniques for the classification of high energy physics data, Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers. Detect. Assoc. Equip. 2006, 559, 199–202. [Google Scholar] [CrossRef]
- Andrews, M.; Paulini, M.; Gleyzer, S.; Poczos, B. End-to-End Event Classification of High-Energy Physics Data. J. Physics Conf. Ser. 2018, 1085, 042022. [Google Scholar] [CrossRef]
- He, P.; Xu, C.J.; Liang, Y.Z.; Fang, K.T. Improving the classification accuracy in chemistry via boosting technique. Chemom. Intell. Lab. Syst. 2004, 70, 39–46. [Google Scholar] [CrossRef]
- Aguiar, J.A.; Gong, M.L.; Tasdizen, T. Crystallographic prediction from diffraction and chemistry data for higher throughput classification using machine learning. Comput. Mater. Sci. 2020, 173, 109409. [Google Scholar] [CrossRef]
- Kaastra, I.; Boyd, M. Designing a neural network for forecasting financial and economic time series. Neurocomputing 1996, 10, 215–236. [Google Scholar] [CrossRef]
- Hafezi, R.; Shahrabi, J.; Hadavandi, E. A bat-neural network multi-agent system (BNNMAS) for stock price prediction: Case study of DAX stock price. Appl. Soft Comput. 2015, 29, 196–210. [Google Scholar] [CrossRef]
- Yadav, S.S.; Jadhav, S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J. Big Data 2019, 6, 113. [Google Scholar] [CrossRef]
- Qing, L.; Linhong, W.; Xuehai, D. A Novel Neural Network-Based Method for Medical Text Classification. Future Internet 2019, 11, 255. [Google Scholar] [CrossRef]
- Park, J.; Sandberg, I.W. Universal Approximation Using Radial-Basis-Function Networks. Neural Comput. 1991, 3, 246–257. [Google Scholar] [CrossRef]
- Montazer, G.A.; Giveki, D.; Karami, M.; Rastegar, H. Radial basis function neural networks: A review. Comput. Rev. J. 2018, 1, 52–74. [Google Scholar]
- Gorbachenko, V.I.; Zhukov, M.V. Solving boundary value problems of mathematical physics using radial basis function networks. Comput. Math. Math. Phys. 2017, 57, 145–155. [Google Scholar] [CrossRef]
- Määttä, J.; Bazaliy, V.; Kimari, J.; Djurabekova, F.; Nordlund, K.; Roos, T. Gradient-based training and pruning of radial basis function networks with an application in materials physics. Neural Netw. 2021, 133, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Lian, R.-J. Adaptive Self-Organizing Fuzzy Sliding-Mode Radial Basis-Function Neural-Network Controller for Robotic Systems. IEEE Trans. Ind. Electron. 2014, 61, 1493–1503. [Google Scholar] [CrossRef]
- Vijay, M.; Jena, D. Backstepping terminal sliding mode control of robot manipulator using radial basis functional neural networks. Comput. Electr. Eng. 2018, 67, 690–707. [Google Scholar] [CrossRef]
- Ravale, U.; Marathe, N.; Padiya, P. Feature Selection Based Hybrid Anomaly Intrusion Detection System Using K Means and RBF Kernel Function. Procedia Comput. Sci. 2015, 45, 428–435. [Google Scholar] [CrossRef]
- Lopez-Martin, M.; Sanchez-Esguevillas, A.; Arribas, J.I.; Carro, B. Network Intrusion Detection Based on Extended RBF Neural Network With Offline Reinforcement Learning. IEEE Access 2021, 9, 153153–153170. [Google Scholar] [CrossRef]
- Foody, G.M. Supervised image classification by MLP and RBF neural networks with and without an exhaustively defined set of classes. Int. J. Remote Sens. 2004, 25, 3091–3104. [Google Scholar] [CrossRef]
- Er, M.J.; Wu, S.; Lu, J.; Toh, H.L. Face recognition with radial basis function (RBF) neural networks. IEEE Trans. Neural Netw. 2002, 13, 697–710. [Google Scholar]
- Kuncheva, L.I. Initializing of an RBF network by a genetic algorithm. Neurocomputing 1997, 14, 273–288. [Google Scholar] [CrossRef]
- Ros, F.; Pintore, M.; Deman, A.; Chrétien, J.R. Automatical initialization of RBF neural networks. Chemom. Intell. Lab. Syst. 2007, 87, 26–32. [Google Scholar] [CrossRef]
- Wang, D.; Zeng, X.J.; Keane, J.A. A clustering algorithm for radial basis function neural network initialization. Neurocomputing 2012, 77, 144–155. [Google Scholar] [CrossRef]
- Ricci, E.; Perfetti, R. Improved pruning strategy for radial basis function networks with dynamic decay adjustment. Neurocomputing 2006, 69, 1728–1732. [Google Scholar] [CrossRef]
- Huang, G.; Saratchandran, P.; Sundararajan, N. A generalized growing and pruning RBF (GGAP-RBF) neural network for function approximation. IEEE Trans. Neural Netw. 2005, 16, 57–67. [Google Scholar] [CrossRef]
- Bortman, M.; Aladjem, M. A Growing and Pruning Method for Radial Basis Function Networks. IEEE Trans. Neural Netw. 2009, 20, 1039–1045. [Google Scholar] [CrossRef]
- Chen, J.Y.; Qin, Z.; Jia, J. A PSO-Based Subtractive Clustering Technique for Designing RBF Neural Networks. In Proceedings of the 2008 IEEE Congress on Evolutionary Computation (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–6 June 2008; pp. 2047–2052. [Google Scholar]
- Esmaeili, A.; Mozayani, N. Adjusting the parameters of radial basis function networks using Particle Swarm Optimization. In Proceedings of the 2009 IEEE International Conference on Computational Intelligence for Measurement Systems and Applications, Hong Kong, China, 11–13 May 2009; pp. 179–181. [Google Scholar]
- O’Hora, B.; Perera, J.; Brabazon, A. Designing Radial Basis Function Networks for Classification Using Differential Evolution. In Proceedings of the 2006 IEEE International Joint Conference on Neural Network Proceedings, Vancouver, BC, USA, 16–21 July 2006; pp. 2932–2937. [Google Scholar]
- Benoudjit, N.; Verleysen, M. On the Kernel Widths in Radial-Basis Function Networks. Neural Process. Lett. 2003, 18, 139–154. [Google Scholar] [CrossRef]
- Paetz, J. Reducing the number of neurons in radial basis function networks with dynamic decay adjustment. Neurocomputing 2004, 62, 79–91. [Google Scholar] [CrossRef]
- Yu, H.; Reiner, P.D.; Xie, T.; Bartczak, T.; Wilamowski, B.M. An Incremental Design of Radial Basis Function Networks. IEEE Trans. Neural Netw. Learn. Syst. 2014, 25, 1793–1803. [Google Scholar] [CrossRef]
- Alexandridis, A.; Chondrodima, E.; Sarimveis, H. Cooperative learning for radial basis function networks using particle swarm optimization. Appl. Soft Comput. 2016, 49, 485–497. [Google Scholar] [CrossRef]
- Neruda, R.; Kudova, P. Learning methods for radial basis function networks. Future Gener. Comput. Syst. 2005, 21, 1131–1142. [Google Scholar] [CrossRef]
- Yokota, R.; Barba, L.A.; Knepley, M.G. PetRBF—A parallel O(N) algorithm for radial basis function interpolation with Gaussians. Comput. Methods Appl. Mech. Eng. 2010, 199, 1793–1804. [Google Scholar] [CrossRef]
- Lu, C.; Ma, N.; Wang, Z. Fault detection for hydraulic pump based on chaotic parallel RBF network. EURASIP J. Adv. Signal Process. 2011, 2011, 49. [Google Scholar] [CrossRef]
- MacQueen, J. Some methods for classification and analysis of multivariate observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Berkeley, CA, USA, 27 December 1965–7 January 1966; Volume 1, pp. 281–297. [Google Scholar]
- O’Neill, M.; Ryan, C. Grammatical evolution. IEEE Trans. Evol. Comput. 2001, 5, 349–358. [Google Scholar] [CrossRef]
- Backus, J.W. The Syntax and Semantics of the Proposed International Algebraic Language of the Zurich ACM-GAMM Conference. In Proceedings of the International Conference on Information Processing, Pris, France, 15–20 June 1959; pp. 125–132. [Google Scholar]
- Ryan, C.; O’Neill, M.; Collins, J.J. Grammatical Evolution: Solving Trigonometric Identities; University of Limerick: Limerick, Ireland, 1998; Volume 98. [Google Scholar]
- Puente, A.O.; Alfonso, R.S.; Moreno, M.A. Automatic composition of music by means of grammatical evolution. In Proceedings of the APL ’02: Proceedings of the 2002 Conference on APL: Array Processing Languages: Lore, Problems, and Applications, Madrid, Spain, 22–25 July 2002; pp. 148–155. [Google Scholar]
- Campo, L.M.L.; Oliveira, R.C.L.; Roisenberg, M. Optimization of neural networks through grammatical evolution and a genetic algorithm. Expert Syst. Appl. 2016, 56, 368–384. [Google Scholar] [CrossRef]
- Soltanian, K.; Ebnenasir, A.; Afsharchi, M. Modular Grammatical Evolution for the Generation of Artificial Neural Networks. Evol. Comput. 2022, 30, 291–327. [Google Scholar] [CrossRef] [PubMed]
- Galván-López, E.; Swafford, J.M.; O’Neill, M.; Brabazon, A. Evolving a Ms. PacMan Controller Using Grammatical Evolution. In Applications of Evolutionary Computation; EvoApplications 2010; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2010; Volume 6024. [Google Scholar]
- Shaker, N.; Nicolau, M.; Yannakakis, G.N.; Togelius, J.; O’Neill, M. Evolving levels for Super Mario Bros using grammatical evolution. In Proceedings of the 2012 IEEE Conference on Computational Intelligence and Games (CIG), Granada, Spain, 11–14 September 2012; pp. 304–331. [Google Scholar]
- Brabazon, A.; O’Neill, M. Credit classification using grammatical evolution. Informatica 2006, 30, 325–335. [Google Scholar]
- Şen, S.; Clark, J.A. A grammatical evolution approach to intrusion detection on mobile ad hoc networks. In Proceedings of the Second ACM Conference on Wireless Network Security, Zurich, Switzerland, 16–19 March 2009. [Google Scholar]
- O’Neill, M.; Hemberg, E.; Gilligan, C.; Bartley, E.; McDermott, J.; Brabazon, A. GEVA: Grammatical evolution in Java. ACM SIGEVOlution 2008, 3, 17–22. [Google Scholar] [CrossRef]
- Noorian, F.; de Silva, A.M.; Leong, P.H.W. gramEvol: Grammatical Evolution in R. J. Stat. Softw. 2016, 71, 1–26. [Google Scholar] [CrossRef]
- Raja, M.A.; Ryan, C. GELAB–A Matlab Toolbox for Grammatical Evolution. In Intelligent Data Engineering and Automated Learning—IDEAL 2018; Yin, H., Camacho, D., Novais, P., Tallón-Ballesteros, A., Eds.; IDEAL 2018; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11315. [Google Scholar] [CrossRef]
- Anastasopoulos, N.; Tsoulos, I.G.; Tzallas, A. GenClass: A parallel tool for data classification based on Grammatical Evolution. SoftwareX 2021, 16, 100830. [Google Scholar] [CrossRef]
- Tsoulos, I.G. QFC: A Parallel Software Tool for Feature Construction, Based on Grammatical Evolution. Algorithms 2022, 15, 295. [Google Scholar] [CrossRef]
- Goldberg, D. Genetic Algorithms in Search, Optimization and Machine Learning; Addison-Wesley Publishing Company: Reading, MA, USA, 1989. [Google Scholar]
- Michaelewicz, Z. Genetic Algorithms + Data Structures = Evolution Programs; Springer: Berlin, Germany, 1996. [Google Scholar]
- Fletcher, R. A new approach to variable metric algorithms. Comput. J. 1970, 13, 317–322. [Google Scholar] [CrossRef]
- Wang, H.; Gemmeke, H.; Hopp, T.; Hesser, J. Accelerating image reconstruction in ultrasound transmission tomography using L-BFGS algorithm. In Medical Imaging 2019: Ultrasonic Imaging and Tomography; 109550B; SPIE: San Diego, CA, USA, 2019. [Google Scholar] [CrossRef]
- Dalvand, Z.; Hajarian, M. Solving generalized inverse eigenvalue problems via L-BFGS-B method. Inverse Probl. Sci. Eng. 2020, 28, 1719–1746. [Google Scholar] [CrossRef]
- Rao, Y.; Wang, Y. Seismic waveform tomography with shot-encoding using a restarted L-BFGS algorithm. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, M.; Martínez Calomardo, Á. A Stochastic Modified Limited Memory BFGS for Training Deep Neural Networks. In Intelligent Computing; Arai, K., Ed.; SAI 2022 Lecture Notes in Networks and Systems; Springer: Cham, Switzerland, 2022; Volume 507. [Google Scholar] [CrossRef]
- Fei, Y.; Rong, G.; Wang, B.; Wang, W. Parallel L-BFGS-B algorithm on GPU. Comput. Graph. 2014, 40, 1–9. [Google Scholar] [CrossRef]
- D’Amore, L.; Laccetti, G.; Romano, D.; Scotti, G.; Murli, A. Towards a parallel component in a GPU–CUDA environment: A case study with the L-BFGS Harwell routine. Int. J. Comput. Math. 2015, 92, 59–76. [Google Scholar] [CrossRef]
- Najafabadi, M.M.; Khoshgoftaar, T.M.; Villanustre, F.; Holt, J. Large-scale distributed L-BFGS. J. Big Data 2017, 4, 22. [Google Scholar] [CrossRef]
- Powell, M.J.D. A Tolerant Algorithm for Linearly Constrained Optimization Calculations. Math. Program. 1989, 45, 547–566. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J.L. ADAM: A method for stochastic optimization. In Proceedings of the 3rd International Conference on Learning Representations (ICLR 2015), San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Amari, S.I. Backpropagation and stochastic gradient descent method. Neurocomputing 1993, 5, 185–196. [Google Scholar] [CrossRef]
- Wolberg, W.H.; Mangasarian, O.L. Multisurface method of pattern separation for medical diagnosis applied to breast cytology. Proc. Natl. Acad. Sci. USA 1990, 87, 9193–9196. [Google Scholar] [CrossRef]
- Kelly, M.; Longjohn, R.; Nottingham, K. The UCI Machine Learning Repository. Available online: https://archive.ics.uci.edu (accessed on 8 December 2024).
- Alcalá-Fdez, J.; Fernandez, A.; Luengo, J.; Derrac, J.; García, S.; Sánchez, L.; Herrera, F. KEEL Data-Mining Software Tool: Data Set Repository, Integration of Algorithms and Experimental Analysis Framework. J. Mult. Valued Log. Soft Comput. 2011, 17, 255–287. [Google Scholar]
- Weiss, S.M.; Kulikowski, C.A. Methods from Statistics, Neural Nets, Machine Learning, and Expert Systems; Morgan Kaufmann Publishers Inc.: Cambridge, MA, USA, 1991. [Google Scholar]
- Wang, M.; Zhang, Y.Y.; Min, F. Active learning through multi-standard optimization. IEEE Access 2019, 7, 56772–56784. [Google Scholar] [CrossRef]
- Tzimourta, K.D.; Tsoulos, I.; Bilero, I.T.; Tzallas, A.T.; Tsipouras, M.G.; Giannakeas, N. Direct Assessment of Alcohol Consumption in Mental State Using Brain Computer Interfaces and Grammatical Evolution. Inventions 2018, 3, 51. [Google Scholar] [CrossRef]
- Quinlan, J.R. Simplifying Decision Trees. Int. J. Man-Mach. Stud. 1987, 27, 221–234. [Google Scholar] [CrossRef]
- Evans, B.; Fisher, D. Overcoming process delays with decision tree induction. IEEE Expert 1994, 9, 60–66. [Google Scholar] [CrossRef]
- Zhou, Z.H.; Jiang, Y. NeC4.5: Neural ensemble based C4.5. IEEE Trans. Knowl. Data Eng. 2004, 16, 770–773. [Google Scholar] [CrossRef]
- Setiono, R.; Leow, W.K. FERNN: An Algorithm for Fast Extraction of Rules from Neural Networks. Appl. Intell. 2000, 12, 15–25. [Google Scholar] [CrossRef]
- Demiroz, G.; Govenir, H.A.; Ilter, N. Learning Differential Diagnosis of Eryhemato-Squamous Diseases using Voting Feature Intervals. Artif. Intell. Med. 1998, 13, 147–165. [Google Scholar]
- Horton, P.; Nakai, K. A Probabilistic Classification System for Predicting the Cellular Localization Sites of Proteins. In Proceedings of the International Conference on Intelligent Systems for Molecular Biology, St. Louis, MO, USA, 12–15 June 1996; Volume 4, pp. 109–115. [Google Scholar]
- Hayes-Roth, B.; Hayes-Roth, B.F. Concept learning and the recognition and classification of exemplars. J. Verbal Learn. Verbal Behav. 1977, 16, 321–338. [Google Scholar] [CrossRef]
- Kononenko, I.; Šimec, E.; Robnik-Šikonja, M. Overcoming the Myopia of Inductive Learning Algorithms with RELIEFF. Appl. Intell. 1997, 7, 39–55. [Google Scholar] [CrossRef]
- French, R.M.; Chater, N. Using noise to compute error surfaces in connectionist networks: A novel means of reducing catastrophic forgetting. Neural Comput. 2002, 14, 1755–1769. [Google Scholar] [CrossRef]
- Dy, J.G.; Brodley, C.E. Feature Selection for Unsupervised Learning. J. Mach. Learn. Res. 2004, 5, 845–889. [Google Scholar]
- Perantonis, S.J.; Virvilis, V. Input Feature Extraction for Multilayered Perceptrons Using Supervised Principal Component Analysis. Neural Process. Lett. 1999, 10, 243–252. [Google Scholar] [CrossRef]
- Garcke, J.; Griebel, M. Classification with sparse grids using simplicial basis functions. Intell. Data Anal. 2002, 6, 483–502. [Google Scholar] [CrossRef]
- Mcdermott, J.; Forsyth, R.S. Diagnosing a disorder in a classification benchmark. Pattern Recognit. Lett. 2016, 73, 41–43. [Google Scholar] [CrossRef]
- Cestnik, G.; Konenenko, I.; Bratko, I. Assistant-86: A Knowledge-Elicitation Tool for Sophisticated Users. In Progress in Machine Learning; Bratko, I., Lavrac, N., Eds.; Sigma Press: Wilmslow, UK, 1987; pp. 31–45. [Google Scholar]
- Heck, D.; Knapp, J.; Capdevielle, J.N.; Schatz, G.; Thouw, T. CORSIKA: A Monte Carlo Code to Simulate Extensive Air Showers. 1998. Available online: https://digbib.bibliothek.kit.edu/volltexte/fzk/6019/6019.pdf (accessed on 8 December 2024).
- Elter, M.; Schulz-Wendtland, R.; Wittenberg, T. The prediction of breast cancer biopsy outcomes using two CAD approaches that both emphasize an intelligible decision process. Med. Phys. 2007, 34, 4164–4172. [Google Scholar] [CrossRef]
- Little, M.; Mcsharry, P.; Roberts, S.; Costello, D.; Moroz, I. Exploiting Nonlinear Recurrence and Fractal Scaling Properties for Voice Disorder Detection. BioMed Eng OnLine 2007, 6, 23. [Google Scholar] [CrossRef]
- Little, M.A.; McSharry, P.E.; Hunter, E.J.; Spielman, J.; Ramig, L.O. Suitability of dysphonia measurements for telemonitoring of Parkinson’s disease. IEEE Trans. Biomed. Eng. 2009, 56, 1015–1022. [Google Scholar] [CrossRef]
- Smith, J.W.; Everhart, J.E.; Dickson, W.C.; Knowler, W.C.; Johannes, R.S. Using the ADAP learning algorithm to forecast the onset of diabetes mellitus. In Proceedings of the Symposium on Computer Applications and Medical Care IEEE Computer Society Press, Minneapolis, MN, USA, 8–10 June 1988; pp. 261–265. [Google Scholar]
- Lucas, D.D.; Klein, R.; Tannahill, J.; Ivanova, D.; Brandon, S.; Domyancic, D.; Zhang, Y. Failure analysis of parameter-induced simulation crashes in climate models. Geosci. Dev. 2013, 6, 1157–1171. [Google Scholar] [CrossRef]
- Giannakeas, N.; Tsipouras, M.G.; Tzallas, A.T.; Kyriakidi, K.; Tsianou, Z.E.; Manousou, P.; Hall, A.; Karvounis, E.C.; Tsianos, V.; Tsianos, E. A clustering based method for collagen proportional area extraction in liver biopsy images. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Milano, Italy, 25–29 August 2015; pp. 3097–3100. [Google Scholar]
- Hastie, T.; Tibshirani, R. Non-parametric logistic and proportional odds regression. JRSS (Appl. Stat.) 1987, 36, 260–276. [Google Scholar] [CrossRef]
- Cortez, P.; Silva, A.M.G. Using data mining to predict secondary school student performance. In Proceedings of the 5th FUture BUsiness TEChnology Conference (FUBUTEC 2008), Porto, Portugal, 9–11 April 2008; pp. 5–12. [Google Scholar]
- Yeh, I.; Yang, K.; Ting, T.-M. Knowledge discovery on RFM model using Bernoulli sequence. Expert. Appl. 2009, 36, 5866–5871. [Google Scholar] [CrossRef]
- Jeyasingh, S.; Veluchamy, M. Modified bat algorithm for feature selection with the wisconsin diagnosis breast cancer (WDBC) dataset. Asian Pac. J. Cancer Prev. APJCP 2017, 18, 1257. [Google Scholar]
- Alshayeji, M.H.; Ellethy, H.; Gupta, R. Computer-aided detection of breast cancer on the Wisconsin dataset: An artificial neural networks approach. Biomed. Signal Process. Control. 2022, 71, 103141. [Google Scholar] [CrossRef]
- Raymer, M.; Doom, T.E.; Kuhn, L.A.; Punch, W.F. Knowledge discovery in medical and biological datasets using a hybrid Bayes classifier/evolutionary algorithm. IEEE transactions on systems, man, and cybernetics. Part B Cybern. Publ. IEEE Systems Man Cybern. Soc. 2003, 33, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Zhong, P.; Fukushima, M. Regularized nonsmooth Newton method for multi-class support vector machines. Optim. Methods Softw. 2007, 22, 225–236. [Google Scholar] [CrossRef]
- Andrzejak, R.G.; Lehnertz, K.; Mormann, F.; Rieke, C.; David, P.; Elger, C.E. Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state. Phys. Rev. E 2001, 64, 061907. [Google Scholar] [CrossRef]
- Tzallas, A.T.; Tsipouras, M.G.; Fotiadis, D.I. Automatic Seizure Detection Based on Time-Frequency Analysis and Artificial Neural Networks. Comput. Intell. Neurosci. 2007, 2007, 80510. [Google Scholar] [CrossRef]
- Koivisto, M.; Sood, K. Exact Bayesian Structure Discovery in Bayesian Networks. J. Mach. Learn. Res. 2004, 5, 549–573. [Google Scholar]
- Brooks, T.F.; Pope, D.S.; Marcolini, A.M. Airfoil Self-Noise and Prediction; Technical Report; NASA: Washington, DC, USA, 1989. Available online: https://ntrs.nasa.gov/citations/19890016302 (accessed on 14 November 2024).
- Yeh, I.C. Modeling of strength of high performance concrete using artificial neural networks. Cem. Concr. Res. 1998, 28, 1797–1808. [Google Scholar] [CrossRef]
- Harrison, D.; Rubinfeld, D.L. Hedonic prices and the demand for clean Ai. Environ. Econ. Manag. 1978, 5, 81–102. [Google Scholar] [CrossRef]
- Simonoff, J.S. Smooting Methods in Statistics; Springer: Berlin/Heidelberg, Germany, 1996. [Google Scholar]
- Mackowiak, P.A.; Wasserman, S.S.; Levine, M.M. A critical appraisal of 98.6 degrees f, the upper limit of the normal body temperature, and other legacies of Carl Reinhold August Wunderlich. J. Am. Med. Assoc. 1992, 268, 1578–1580. [Google Scholar] [CrossRef]
- Gropp, W.; Lusk, E.; Doss, N.; Skjellum, A. A high-performance, portable implementation of the MPI message passing interface standard. Parallel Comput. 1996, 22, 789–828. [Google Scholar] [CrossRef]
- Chandra, R. Parallel Programming in OpenMP; Morgan Kaufmann: Cambridge, MA, USA, 2001. [Google Scholar]




| Parameter | Value |
|---|---|
| chromosome_count | 500 |
| chromosome_size | 100 |
| selection_rate | 0.1 |
| mutation_rate | 0.05 |
| generations | 500 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsoulos, I.G.; Varvaras, I.; Charilogis, V. RbfCon: Construct Radial Basis Function Neural Networks with Grammatical Evolution. Software 2024, 3, 549-568. https://doi.org/10.3390/software3040027
Tsoulos IG, Varvaras I, Charilogis V. RbfCon: Construct Radial Basis Function Neural Networks with Grammatical Evolution. Software. 2024; 3(4):549-568. https://doi.org/10.3390/software3040027
Chicago/Turabian StyleTsoulos, Ioannis G., Ioannis Varvaras, and Vasileios Charilogis. 2024. "RbfCon: Construct Radial Basis Function Neural Networks with Grammatical Evolution" Software 3, no. 4: 549-568. https://doi.org/10.3390/software3040027
APA StyleTsoulos, I. G., Varvaras, I., & Charilogis, V. (2024). RbfCon: Construct Radial Basis Function Neural Networks with Grammatical Evolution. Software, 3(4), 549-568. https://doi.org/10.3390/software3040027








