Abstract
Type II polyketide synthase (PKS) systems are a rich source of structurally diverse polycyclic aromatic compounds with clinically relevant antibiotic and chemotherapeutic properties. The enzymes responsible for synthesizing the polyketide core, known collectively as the minimal cassette, hold potential for applications in synthetic biology. The minimal cassette provides polyketides of different chain lengths, which interact with other enzymes that are responsible for the varied cyclization patterns. Additionally, the type II PKS enzyme clusters offer a wide repertoire of tailoring enzymes for oxidations, glycosylations, cyclizations, and rearrangements. This review begins with the variety of chemical space accessible with type II PKS systems including the recently discovered highly reducing variants that produce polyalkenes instead of the archetypical polyketide motif. The main discussion analyzes the previous approaches with an emphasis on further research that is needed to characterize the minimal cassette enzymes in vitro. Finally, the potential type II PKS systems hold the potential to offer new tools in biocatalysis and synthetic biology, particularly in the production of novel antibiotics and biofuels.
1. Introduction
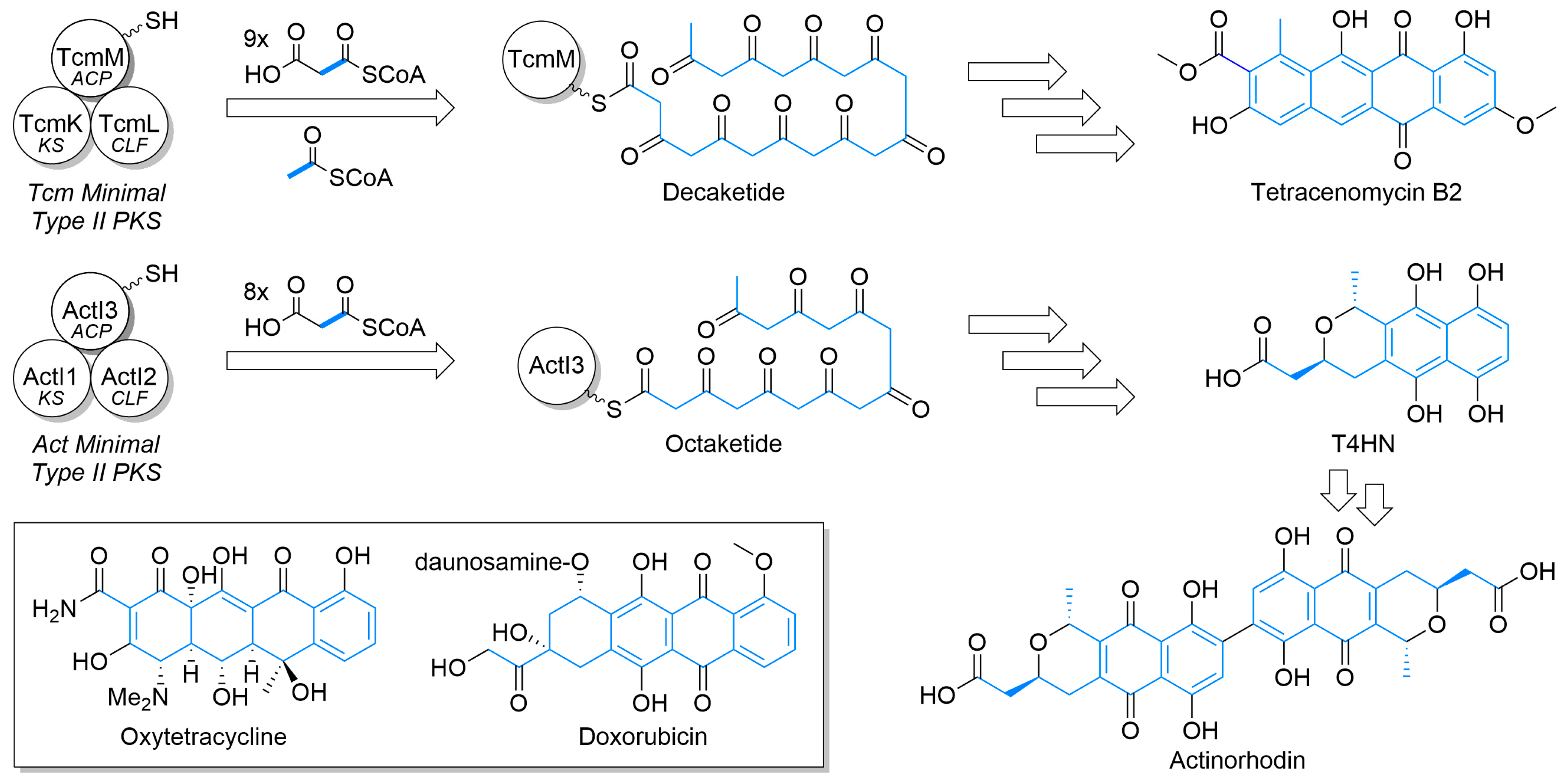
Microbes are a historically rich source of medicinal natural products with clinical applications ranging from antibiotics to anticancer drugs [1]. Within this rich mélange, type II polyketide synthase (PKS) systems of bacteria produce a number of aromatic polyketides, including potent medicinal compounds, such as tetracycline antibiotics and doxorubicin chemotherapeutics (Figure 1, inset) [2]. While the compounds produced by type II PKSs are incredibly diverse, they share common structural features. The unifying functional group is a polycyclic aromatic motif derived from the cyclization of a polyketide chain (highlighted in blue throughout the manuscript), such as those shown for tetracenomycin and actinorhodin. The initial polyketide chains of various lengths are formed through the action of a minimal type II PKS cassette, hence the eponymous name of this class of compound. The aromatic rings form the core structure of these natural products as driven by variations in polyketide chain length and tailoring enzymes that create a diverse range of ring systems and pendant functionality [3].
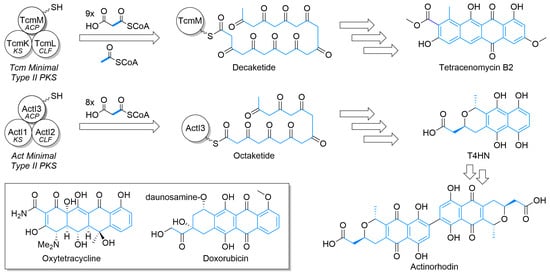
Figure 1.
The basic construction of the polyketides of tetracenomycin and actinorhodin by type II polyketide synthases and elaboration to aromatic polycycle. The minimal type II polyketide synthase (PKS) cassette, composed of an acyl carrier protein (ACP), ketosynthase (KS), and chain length factor (CLF), work in concert to produce various length polyketides that are subsequently elaborated by other enzymes to the natural product. Carbon atoms from the starter unit or malonyl extender units are highlighted in blue. Inset—Structures of oxytetracycline and doxorubicin.
The vast number and diversity of type II PKS compounds present rich opportunities to leverage these biosynthetic enzymes for synthetic biology. Despite their commonalities, type II PKSs that produce polyaromatic- or polyalkene-type structures remain understudied [4] with regard to the in vitro interrogation of individual enzymes due to difficulties affiliated with handling these systems. This gap is especially pronounced relative to the two other major PKS families, Type I [5] and Type III [6].
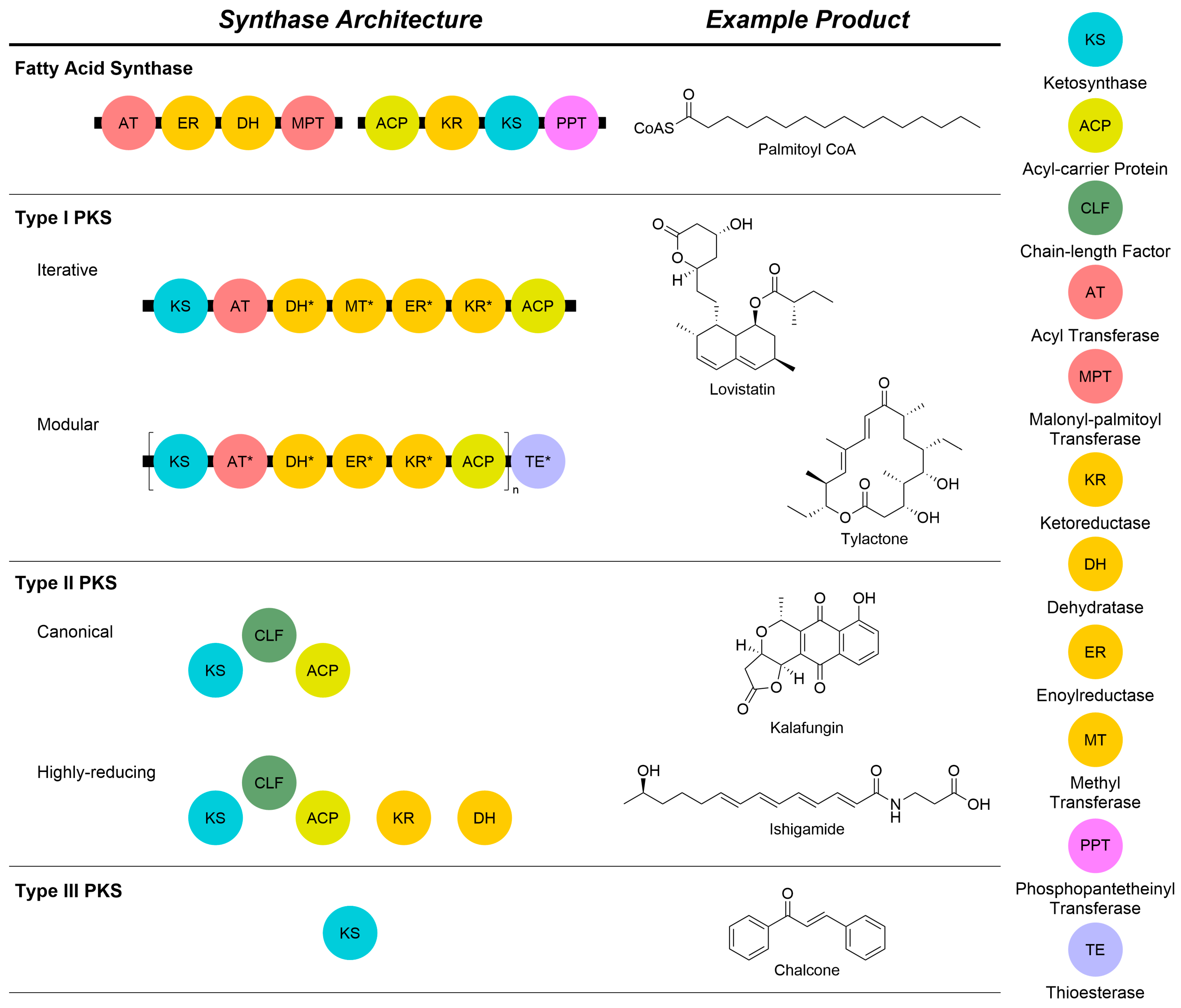
Like fatty acid synthases (FAS, Figure 2), polyketide synthases (PKS) conduct the organized extension of malonyls-CoA to make more complex compounds [7]. Type I PKSs, primarily found in bacteria and fungi, exist as massive multi-domain megasynthases [5]. These megasynthases can be subdivided into assembly line and iterative types [8,9]. Assembly line type I PKS [8] are composed of individual modules that work to extend the polyketide chain. Each module contains an acyl carrier protein (ACP), ketosynthase (KS), and acyltransferase (AT). The ACP from a previous module transfers the growing polyketide chain to the KS. The AT then trans-esterifies an acyl-CoA-derived extender unit which is loaded on to the ACP. The KS then adds the extender unit to its chain by catalyzing a Claisen-like condensation before passing it along to the ACP of the next module. The oxidation state is controlled by the presence or absence of tailoring domains. Iterative type I PKS [9], like the assembly line types, are still multi-domain megasynthases. However, the extension of the polyketide chain is accomplished by extending the chain with a single ACP, KS, and AT. Instead of passing the chain to the next module, the enzymes will continuously catalyze Claisen-like condensation reactions with the polyketide chain until it reaches the appropriate length. After chain extension is completed, the chain is passed to the next module for down-stream modifications. While it is a single megasynthase, different oxidation states and methyl substitutions are selectively accomplished by the additional domains.
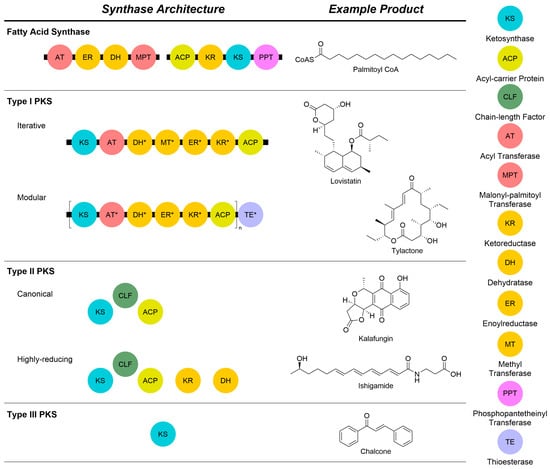
Figure 2.
Comparison of the core architectures and components of fatty acid and polyketide synthase proteins with example natural products from each class. All types rely on a core KS to conduct a Claisen reaction to extend the growing chain by an acetate unit. Different oxidation states can be achieved by the incorporation of additional domains, either within a megasynthase or as discrete proteins. A legend of abbreviations is shown on the right of the Figure. Domains marked with an asterisk are not always present.
Type III PKSs make small aromatic molecules, such as chalcones [6]. While primarily found in plants, they have also been found in bacteria and fungi. Unlike the multi-domain type I and discrete proteins of type II PKSs, the enzymes in type III PKSs exist as a pair of homodimers that fulfill the roles played by an ACP, KS, and AT. Within the dimer, the polyketide chain is iteratively extended by the active sites in each monomer. Upon completion of the chain, the dimeric pair is capable of cyclizing the chain without the need for other modules or tailoring enzymes [10]. Due to the lack of tailoring enzymes, such as those found in type I and II PKSs, the structural diversity of their products is generated by altering chain length and cyclization patterns and integrating different starter units [11]. The compounds, called starter units (SUs), are the initial molecules that serve as a base to grow a polyketide chain [12].
For further information about these PKSs, there are several excellent reviews for both type I and type III PKS systems that go further in-depth than the brief overview presented here. Herbst et al. go in-depth about the architectures of iterative type I PKSs and their relation to fatty acid synthases (FASs) [9]. For assembly line type I PKS, Nivina et al. provide a discussion about the diversity of these PKSs and their evolutionary origins [8]. An overview of type III PKS systems in both plants and bacteria is well-described by Yu et al. with their applications to metabolic engineering detailed by Palmer and Alper [10,13]. More details on the structural and mechanistic similarities between type II fatty acid synthases (FAS) and type II polyketide synthases are discussed by Chen et al., who provide an excellent comparison in their review [14]. Additionally, Smith and Tsai provide a detailed discussion regarding the comparison of type I FASs and type I PKS modular components [15]. The focus of this review is limited to type II PKSs with a focus on current in vitro work and their enzymatic potential in synthetic biology.
While there are some commonalities between all types of PKSs, such as the use of various starter units and the inclusion of malonyl-CoA as an extender unit, the differences are distinct. Type I and III PKSs rely on multi-domain megasynthases and homodimeric proteins, respectively. In contrast, type II PKSs are composed of multiple discrete enzymes that can synthesize the polyketide chain or provide later modifications such as cyclization [16]. It is the discrete nature of type II PKS enzymes that presents a potential advantage over the other systems.
While type I and III systems are more widely studied and utilized, type II PKSs offer more flexibility for designer biosynthesis. Introducing non-native modules for polyketide chain synthesis and modification in type I PKSs, while possible, presents challenges. The addition or removal of modules in type I PKSs can result in impaired function or even inactivation of the system [17]. Type III PKSs lack tailoring enzymes and are dependent on the SU and cyclization pattern from its homodimeric proteins to introduce structural diversity into the products [12]. Because their common core iteratively produces polyketides, Type II PKSs avoid these pitfalls of the Type I and III systems. The discrete proteins of type II PKSs offer more options for tailoring enzymes than type III PKSs without the concerns of module compatibility associated with type I PKSs.
2. Structural and Biosynthetic Diversity in Canonical Type II PKS
Research into type II PKS natural products began in the 1940s with the discovery of actinorhodin [18], and later in the 1970s with tetracenomycin [19], with their associated biosynthetic pathways being identified in the 1980s [20,21]. Biosynthetic pathways in bacteria usually comprise a cluster of genes encoding the proteins responsible for the biosynthesis of the target metabolite, a grouping referred to as a biosynthetic gene cluster (BGC). The BGCs responsible for the synthesis of actinorhodin and tetracenomycin are act and tcm, respectively. These two pathways laid the foundation of our understanding of type II PKS enzymes and remain the model BGCs for type II PKS studies [22]. Despite these discoveries, type II PKSs remain relatively uncharacterized and poorly utilized in comparison to type I and type III PKSs.
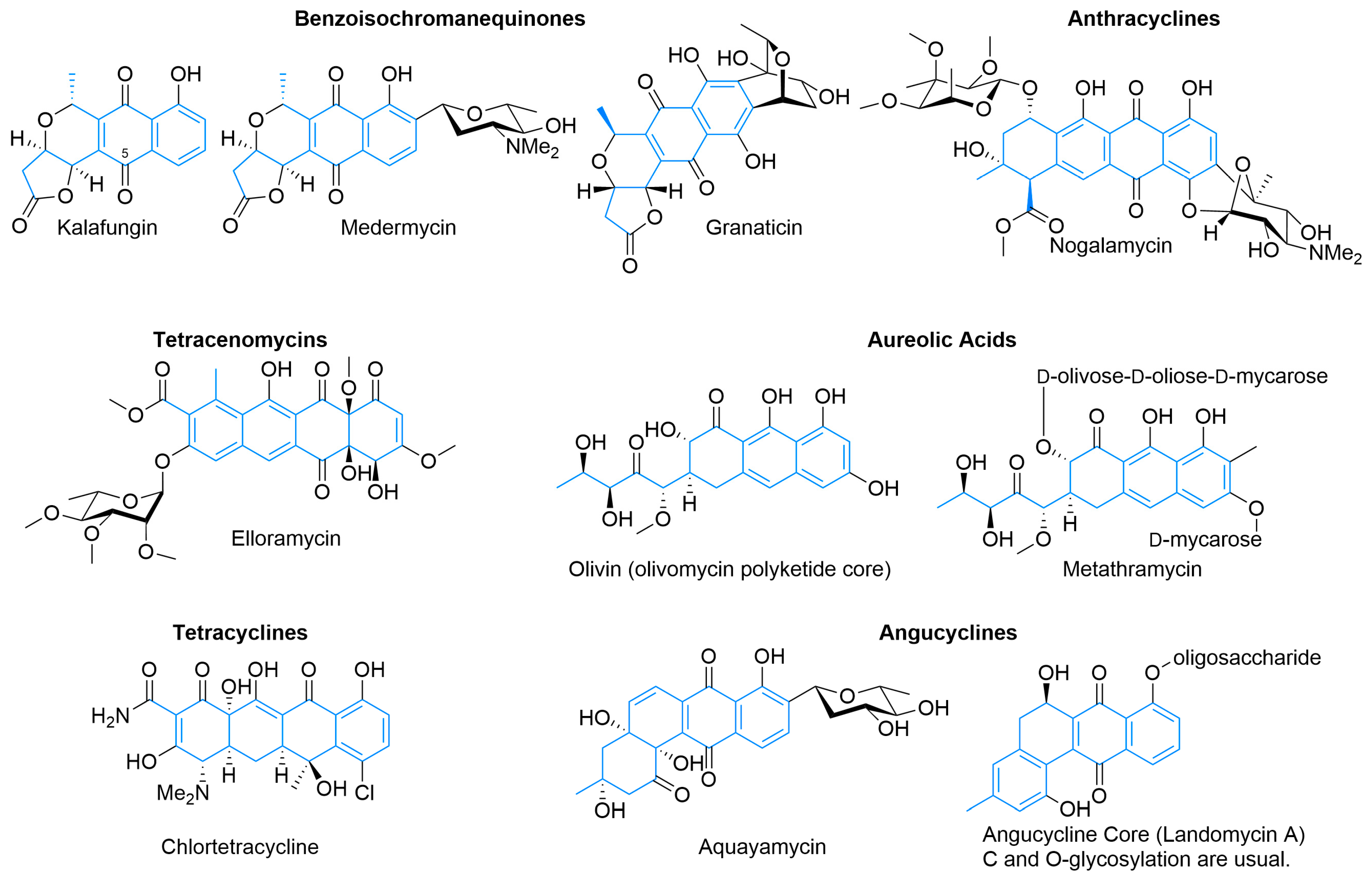
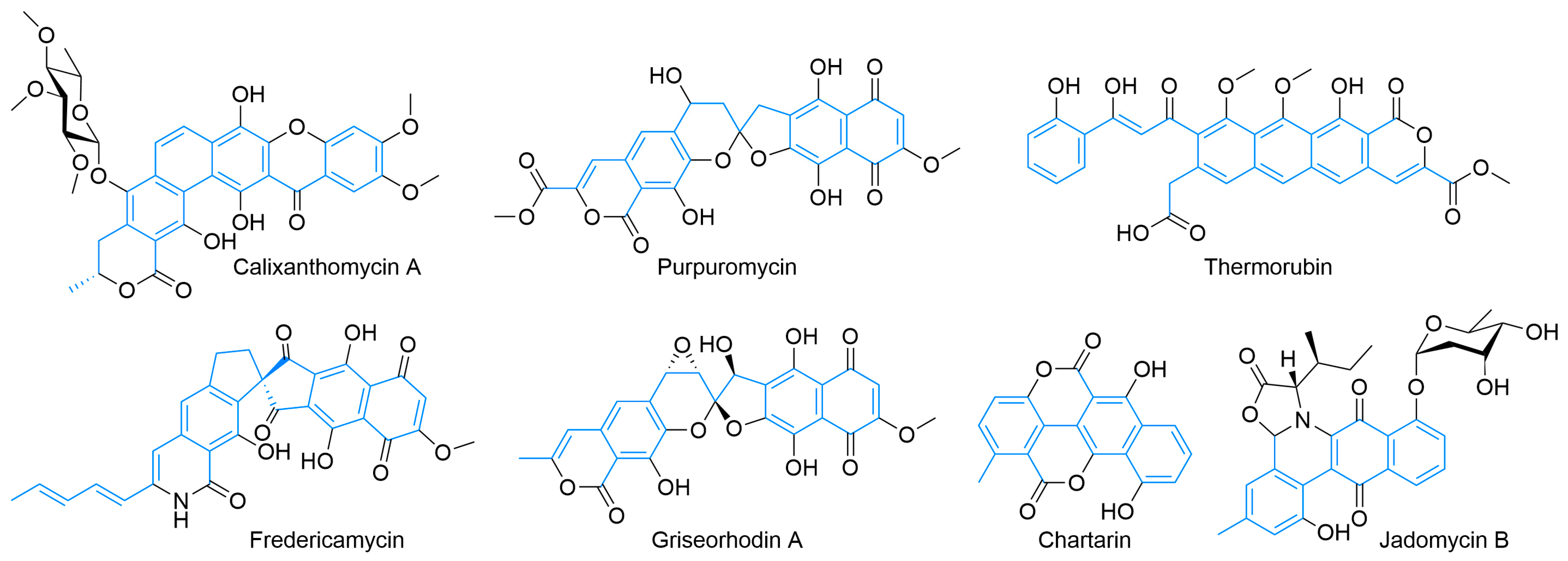
Beyond the octaketide-derived anthraquinones and smaller skeletons commonly produced by plants, fungi, and some bacteria [23,24,25], there is a wide diversity of type II PKS products. The structurally smaller, well-defined major classes (Figure 3) include benzoisochromanequinones, anthracyclines, tetracenomycins, aureolic acids, tetracyclines, and angucyclines. All structures are drawn by highlighting the polyketide-derived core in blue and proceeding clockwise from the starter unit closest to the top left corner to the ACP attachment point closest to the bottom left corner whenever possible, as exemplified by the polyketides for tetracenomycin B2 and T4HN (Figure 1).
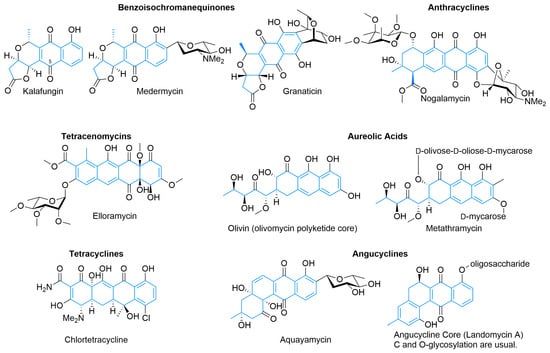
Benzoisochromanequinones (Figure 3), such as kalafungin, medermycin, and granaticin, show a variety of activities including antibiotic and potential beta-lactamase inhibitory activity [26,27,28,29,30]. These natural products share a common octaketide core exemplified by kalafungin, which has no additional oxidative or sugar decoration except C5 oxidation, as is common in members of this family [26,27]. Medermycin bears an angolosamine C-glycoside, while granaticin has a doubly attached sugar derived from 4-keto-2,6-dideoxyglucose [28,31,32]. Actinorhodin [18] (Figure 1) is another example of this class of type II PKS, but without the closure of the C3 acetate into a five-membered ring as is present in the other examples, as well as its existence as a biaryl dimer, the biosynthesis of which has only recently been elucidated [33].
The anthracyclines, for example, nogalamycin (Figure 3) and doxorubicin (Figure 1) have activity against cancer and share a tetracene core derived from a decaketide with various oxidation states and pendant sugar functionalities [34,35,36]. Doxorubicin uses propionyl-CoA as a starting unit rather than acetate, is O-glycosylated with daunosamine, and has found strong use as a chemotherapeutic. In addition to oxidation state adjustment of the tetracyclic core and side chains, it also loses the carboxylate of the final acetate unit after glycosylation [37]. Nogalamycin does not have an unusual starter unit or late-stage decarboxylation, but is more structurally interesting with the dual C- and O-linked nogalamine sugar in addition to O-glycosylated nogalose [38,39]. Despite promising initial results, neither it nor its semisynthetic analogs have found use because of high cardiotoxicity, although interest remains high [40,41,42].
Tetracenomycins, such as tetracenomycin B2 (Figure 1) and elloramycin (Figure 3), are a smaller family of type II PKS products that derive from a linear cyclization of decaketide consisting of nine malonyls-CoA and one acetyl-CoA and show antibiotic and cancer inhibition properties [19,43]. Earlier natural products from the pathway, such as tetracenomycin B2, retain more aromaticity, while more mature ones have additional tailoring to partially saturate an aromatic ring via oxidation to a diol [37]. Elloramycin is O-glycosylated with trimethylated rhamnose, with a production of the sugar encoded outside of the aglycone cluster [44].
Sharing a similar linear arrangement, the tetracyclines, such as oxytetracycline (Figure 1) and chlortetracycline (Figure 3) have potent antimicrobial activity that has been improved upon semisynthetically to create this clinically important class of antibiotics, including tetracycline, doxycycline, minocycline, and tigecycline [45,46]. Both start with the malonyl-CoA-derived malonamoyl-CoA and, after extension with eight additional malonyl-CoA units, the polyketide cyclizes into the linear tetracycle. They are highly tailored post-cyclization, with enzymes partially unsaturating most of the tetracycle, to give it a significant three-dimensional shape and adding additional carbon-, oxygen-, nitrogen-, and chlorine-based functionality. While not present in the commercially relevant compounds, C- and O-glycosylation can occur on this scaffold enhancing activity against cancer cell lines and resistant bacteria [47,48,49]. Similar scaffolds arise from fungi as well, although these originate from a different biosynthetic pathway [50,51]. A less strict interpretation of the family, where acetates from two malonyls can replace the initial malonamoyl-CoA starter unit, includes chelocardin, polyketomycin, and dutomycin, which, along with cervimycin, have spurred chemical investigations into the glycosylation of tetracycline [52,53,54,55,56]. The structure of these tetracycles, however, resembles that of premithramycinone, suggesting that these may be more closely related to aureolic acids [57,58].
Aureolic acids, such as olivin and metathramycin, are derived from a decaketide that initially cyclizes into a tetracyclic core [59,60]. Careful experimentation and hypothesis on the mithramycin biosynthetic pathway [61], which was later confirmed experimentally [62], enabled a proposed mechanism of formation that involves the action of a Baeyer–Villiger-type oxidase, followed by hydrolysis and decarboxylation to give the highly oxidized side chain off the tricyclic core, the hallmark feature of aureolic acids. This opening of the core ring system emphasizes that diversity in type II PKS products not only includes the many potential ring numbers, configurations, and oxidation states but also includes ring modification after cyclization. While the core of aureolic acids is derived exclusively from acetate and malonate, this family tends to be highly glycosylated and has potent antitumor activity [61].
Angucyclines, such as aquayamycin [63] and landomycins [64], are a large family of type II PKS products and have been thoroughly reviewed [65]. They also derive from a decaketide, but unlike the previous families where the initial cyclization results in a linear tetracycle, most (though not all) [66,67] instead cyclize directly in an angular fashion. Also introduced with this family is a decarboxylative cleavage of the tetracycle from the ACP, removing an acetate-derived carbon from the isolated natural product, hypothesized to be catalyzed by LanM2 [68]. As shown by aquayamycin and landomycin, the PKS-derived core has various oxidation patterns and can be both C- and O-glycosylated [69]. Angucyclines possess both anticancer and antimicrobial activity [65].
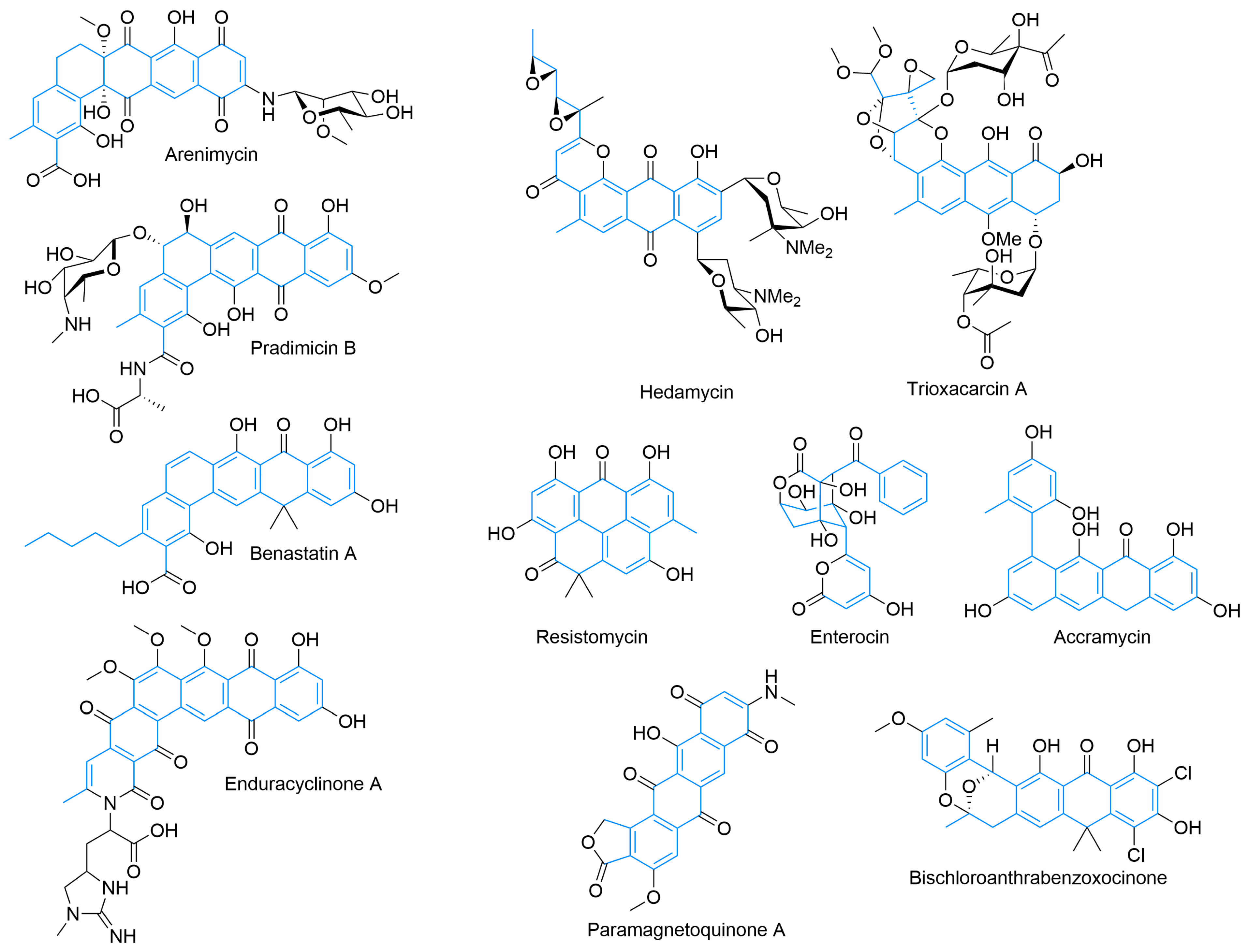
Beyond these delimited systems, there are a large number of type II PKS products with more complex features that are still the result of regular condensation-based cyclization (Figure 4). Extended angular polyphenols, such as arenimycin [70] and pradimicin B [71,72], are derived from dodecaketides and have five rings arranged in an angular fashion [73]. A variety of oxidation states and glycosylation are controlled by tailoring enzymes [74,75,76]. Activity includes antimicrobial and antifungal activity, with pradimicins behaving as lectin mimics and possessing potent antifungal activity [70,72,77]. Others, such as hedamycin [78,79] and trioxacarcin [80], have larger type-II-PKS-derived cores because of larger starter units [81,82]. These natural products also contain C- and O-glycosylation and a diverse variety of oxidations, epoxides for hedamycin, and the complicated polyketal of trioxacarcin, which contribute to their potent anti-tumor activity [83,84]. Benastatin combines features of the previous molecules as an example of an extended angular polyketide with a larger starter unit [85,86]. It is an inhibitor of glutathione S-transferase and has proven amenable to a variety of starter units [85,87].
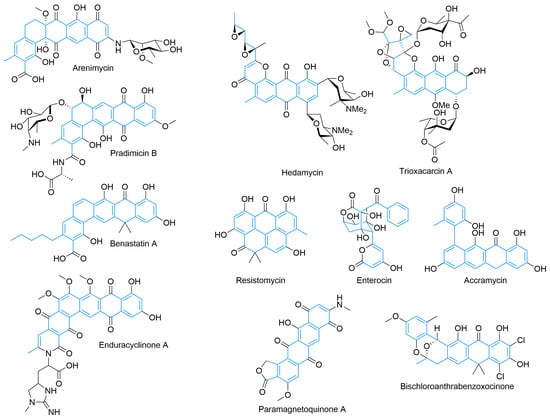
Figure 4.
Examples of type II PKS-derived natural products including extended angular polyphenols, polycycles with different starter units, unusual cyclization patterns, and structures with other atom incorporation beyond C, H, and O.
Apart from the more regular construction of linear and angular polycycles from type II PKS pathways, multiply fused rings, non-aromatized products, and multiple ring systems are also possible. Resistomycin displays a range of activities, and unlike the previous examples, it folds back on itself during cyclization, resulting in its unusual discoid ring fusion [88,89]. Other significantly complex examples [90] suggest a myriad of folding possibilities. The formation of enterocin, which contains an unusual polyhydroxy bridged tricycle at the core of the molecule, is enabled biosynthetically by the pre-cyclization rearrangement of the polyketide chain by the Favorskiiase EncM and subsequent aldol condensations [91,92,93,94]. Its structure highlights the potential diversity of type II PKS products. Accramycin, and other phenylnaphthacenoid polyketides [95], are hypothesized to arise from a tridecaketide via cyclizations to form two separate ring systems in a biaryl configuration [96]. The polycycle is offloaded in a decarboxylative fashion. Accramycin and its derivatives, some of which are halogenated [95], show promising activity against drug-resistant Gram-positive pathogens [97].
Beyond the usual H, C, and O composition of most of the previous type II PKS polyaromatic products, other atoms, such as nitrogen and halogens can be included. Enduracyclinone A is another tridecaketide and incorporates the unusual amino acid enduracididine, likely as part of the ACP-offloading and cyclization process [98]. Pradimicin contains an amino sugar and has its ACP displaced by an alanine [75]. The polycyclic core of arenimycin is aminated and glycosylated in an unknown manner as is the core of the paramagnetoquinones [99,100]. The paramagnetoquinones also contain a five-membered lactone ring, which is hypothesized to be the result of a hydroxyacetyl-CoA starter unit that can displace the ACP or cyclize with the free acid that results from offloading [99,100]. Besides the halogenated derivatives of accramycin, bischloroanthrabenzoxocinone is halogenated and shows promising activity against a new potential antimicrobial target: fatty acid synthase [100,101,102]. Bischloroanthrabenzoxocinone also contains an unusual bridging ketal, which may be formed from the polyketide during the cyclization process or as part of a ring opening and reclosure process as was elucidated for certain angucyclinones [103,104].
The previous examples of type II PKS products cyclized directly from the polyketide to the final polycycle. The exception is the oxidation/ring-opening/decarboxylation sequence that results in the aureolic acid family and possibly bischloroanthrabenzoxocinone. The rearrangement needed to produce enterocin occurs on the polyketide chain prior to cyclization. Figure 5 shows examples of type II PKS products that first cyclize into linear or angular carbocycles, but then subsequently rearrange after this process to form heterocycles with an enormously diverse level of functionality.
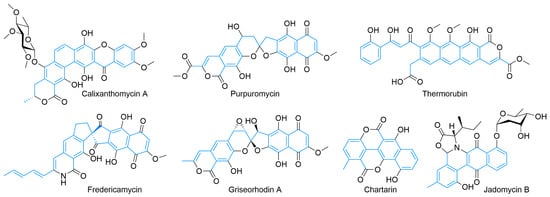
Figure 5.
Examples of type II PKS products with post-cyclization rearrangement or atom excision.
The calixanthomycins and other xanthones are predicted to derive from a tridecaketide [99,105]. Oxidation and Baeyer–Villiger rearrangement is hypothesized to create an epoxy ester that hydrolyzes, epoxide opens, and subsequently decarboxylates to form the xanthone core and reestablish aromaticity [106]. A lactone (or lactam) [107] can also be formed from the terminal carboxylate, and glycosylation can be prolific [108]. Other highly modified cores include fredericamycin [109,110,111], the rubromycin-type spirocycles purpuromycin [112], and griseorhodin [113], all of which show a variety of activity against cancer. Formally a pentadecaketide [114] (this type II PKS is primed by a non-acetate starter unit) [87,115], fredericamycin undergoes rearrangement and decarboxylation to achieve its unique quaternary carbon-based spirocyclic ring fusion [116]. Although apparently inactive in the genomes of the rubromycin family, an amide synthetase is responsible for lactam formation [117] at the ACP attachment point. Griseorhodins and other rubromycins are tridecaketides [118] and undergo substantial post-PKS modification to achieve the highly oxidized spiroketal core [119,120,121]. The antibiotic thermorubin [122,123] begins with a singular salicyclic acid starter unit; after the inclusion of eleven acetates, it is hypothesized to cyclize to an all-carbon tetracycle [124]. Tailoring enzymes are predicted to oxidatively open one polyphenolic ring and reclose it as a lactone [125], which is the first potential example of the formation of this functional group not formed from the ACP-bound acetate. Chartreusin [126,127] and its related structures [128] show antibiotic and chemotherapeutic activity. Its type-II-PKS-derived aglycone core, chartarin, is derived from a tetracenomycin-type decaketide through a series of redox reactions and decarboxylation that enables the rearrangement [129,130,131]. Derivatives of chartreusin have already been the subject of some biosynthetic pathway engineering [132]. Like chartreusin, jadomycins [133], such as jadomycin B [134], are ring-opened and decarboxylated natural products, which, in this case, are derived from an angucycline-type decaketide [135]. The biosynthetic pathway is well-investigated [134,135] with the formation of the amino-acid-containing E ring spontaneous [136], with this process being leveraged to create a variety of derivatives [137].
This sampling of type II PKS structures gives a sense of the diverse chemical space that can be accessed once the details of the function of the critically important minimal cassette are elucidated. We would also direct the reader’s attention to a number of excellent reviews detailing particular aspects of type II PKS natural products. Bililign et al. provide a detailed description of the biosynthetic pathways of benzoisochromanequinones and angucyclines along with their modifications with aryl-C-glycosides [138]. Tetracycline biosynthesis and metabolic engineering, particularly that of oxytetracycline, is well-described by Pickens and Tang [49]. A detailed review of anthracycline biosynthetic pathways and the clinical relevance of anthracycline compounds is provided by Hulst et al. [42]. A comprehensive review by Rohr and Hertweck breaks down the classes by their unifying themes [139]. Alternatively, a recent review by Xie and Zhang goes in-detail regarding the bioinformatic classification of both the minimal cassette and tailoring enzymes of each major family of compounds in type II PKSs [140], with recent tools aiding in this method of classification for type II PKS products [141].
With few exceptions [142,143], the vast majority of work for interrogating biosynthesis is a deconstructive in vivo approach, where genes are inactivated and natural product precursors are isolated. These knockouts are complemented by recombination, where combinations of mutants bearing the missing gene(s) restore natural product production. While this approach has successfully elucidated most biosynthetic pathways, it relies heavily on mass spectrometry to detect trace metabolites, a process that can misidentify isomers, and by its wholistic, in-species nature, does not usually enable assessment and in-depth study of the abilities of individual proteins [144]. In this review, we focus on in vitro work that has been conducted specifically on the minimal type II PKS cassette. By overcoming gaps in our knowledge of how this system works, both its use and more precise investigations and use of downstream cyclization and tailoring enzymes can enable their applications in synthetic biology.
3. Canonical Type II PKS
All type II PKSs follow the same general reaction scheme to create their polyketide products. A set of three enzymes, known as a minimal cassette, is composed of a ketosynthase (KS or KSα), a chain-length factor (CLF or KSβ), and an acyl carrier protein (ACP) that work together to synthesize a polyketide chain (Figure 6). Decarboxylation of the ACP-bound malonyl by the CLF and its subsequent transfer to the KS [145] is a typical method for starter unit preparation, although other methods exist [3]. Once a starter unit is established on the KS and the ACP is reloaded with malonyl, decarboxylation activates the ACP-bound malonyl and the resulting enolate attaches to the nascent polyketide held by the KS. Collapse of the tetrahedral intermediate releases the KS resulting in an ACP-bound polyketide that has been extended by one unit. Transthioesterification passes the nascent polyketide, extended by one acetate unit, back to the KS, and extension can then continue. The CLF dimerized with the KS is believed to stabilize and control the length of the growing chain [16]. Once the length dictated by the CLF is reached, additional transfer back to the KS is not possible and the full-length, ACP-bound polyketide dissociates from the KS-CLF dimer [146]. Downstream cyclases generate the characteristic aromatic core while other tailoring enzymes add functional groups [140].
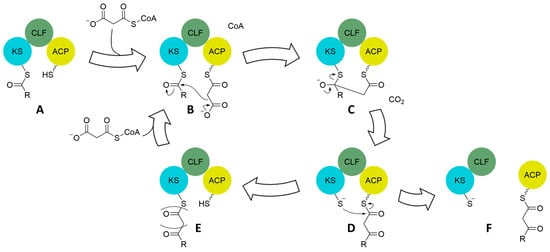
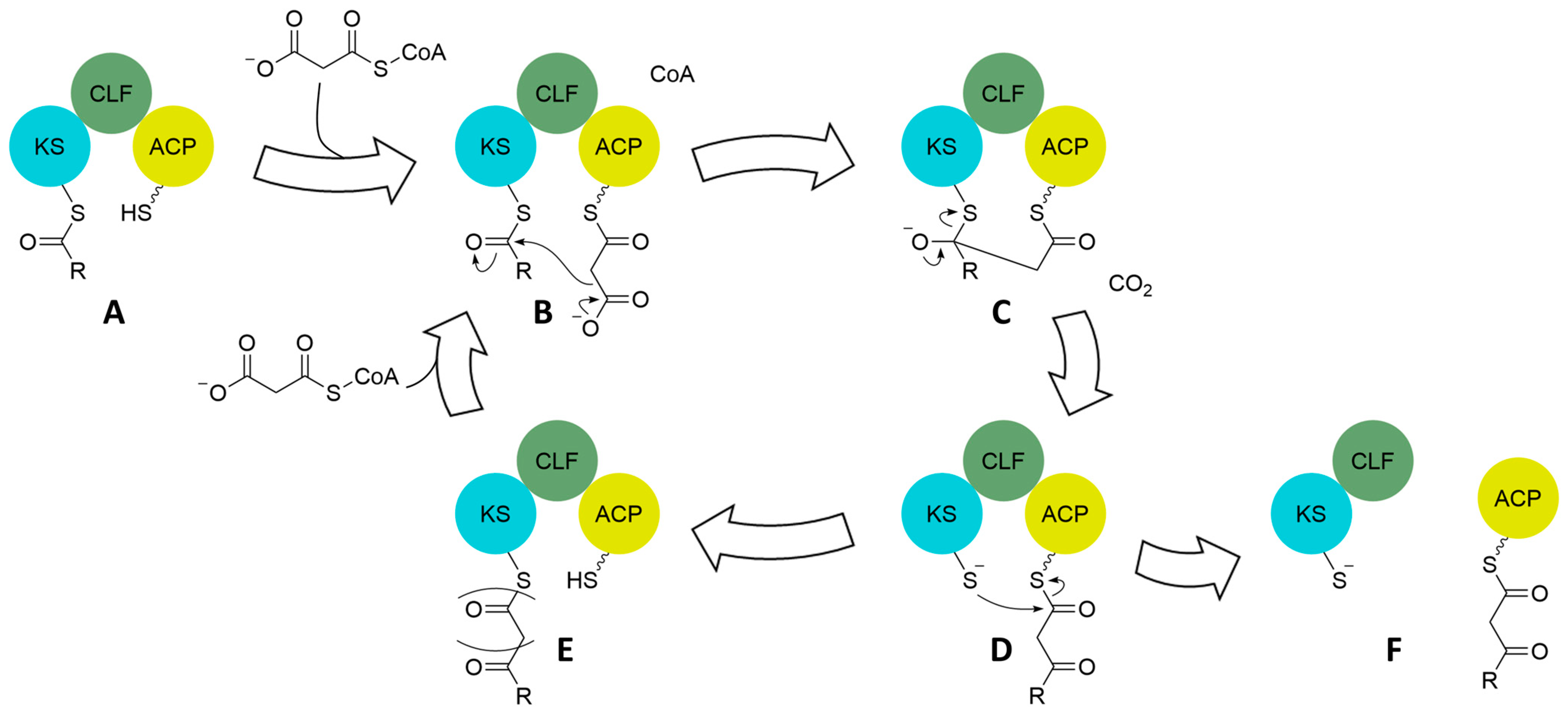
Figure 6.
Claisen-like condensation mechanism of the minimal cassette in type II PKS to grow the polyketide chain. Once the KS is loaded with a starter unit (A) via one of several mechanisms, the ACP interacts with malonyl-CoA or transacylases from primary metabolism to obtain a malonyl for chain extension (B). Decarboxylation activates the malonyl as an enolate on the ACP, which reacts with the thioester on the KS to form a tetrahedral intermediate (C). Collapse of the tetrahedral intermediate results in a polyketide extended by one acetate unit attached to the ACP (D). Continued extension involves the transfer of the extended polyketide back to the KS (E) and the acquisition of another malonyl by the ACP to reenter the extension cycle (E to B). Once the ACP-bound polyketide chain is at the appropriate length, it can no longer be accepted back onto the KS (see later Figure) causing dissociation of the ACP from the KS-CLF dimer (F) and enabling delivery of the polyketide chain to cyclases for tailoring to various ring systems.
Despite the expansive array of structures demonstrating that type II PKSs are a valuable source of natural products, individual enzymes are often poorly characterized. The KS and CLF are usually insoluble when expressed individually, resulting in the formation of inclusion bodies and precipitation in vitro [3,4,147]. As a result of their insolubility, detailed in vitro mechanistic studies involving the minimal cassette enzymes have been limited, leaving major gaps regarding the details of polyketide chain length determination, substrate recognition, and mechanisms of polyketide synthesis from non-acetate starter units. While recent structural work has shed some light on protein–protein interactions from the anthraquinone producer Photorhabdus luminescens [146], a more in-depth understanding of the specific functions and limitations of the enzymes of the type II PKS minimal cassette would enable the development of new enzymes for use in synthetic biology.
Much of the knowledge of in vitro interactions of the minimal type II PKS cassette, as well as the limitations of how they can be purified and used, arose from a series of articles describing the expression of core proteins from the act and tcm gene clusters. Initial work by Shen and Hutchinson demonstrated that the minimal type II PKS cassette (tcmKLM) could be successfully expressed from recombinant strains of the native Streptomyces glaucescens designed to overexpress the PKS [148]. After preparation as a cell-free extract, the authors showed that in the presence of acetyl-CoA and malonyl-CoA, precursors in the biosynthesis of tetracenomycin were produced. While production occurred only with the co-expression of the intact cyclase (TcmN), subsequent work enabled the expression of individual proteins, except for the KS-CLF dimer (TcmKL), which had to be co-expressed [149].
Mirroring these findings, Khosla and coworkers successfully expressed the actinorhodin minimal type II PKS (actI1-3) from recombinant Streptomyces coelicolor and showed that its use as a cell-free extract with acetyl-CoA and radio-labeled malonyl-CoA resulted in the production of spontaneously cyclized octaketide products [150]. With the inclusion of the actinorhodin ketoreductase (actIII), aromatase (actVII), and cyclase (actIV) in the cell-free preparation, they were able to generate actinorhodin pathway metabolites that coeluted with authentic standards. Separately, they were able to individually express and purify the KS-CLF dimer from S. coelicolor and demonstrate its ability to produce spontaneously cyclized octaketides in the presence of a variety of holo-ACPs, malonyl CoA/ACP transacylase, and malonyl-CoA [151].
While these seminal works unambiguously demonstrated the function of the minimal type II PKS cassette, they also revealed some of the continuing challenges with studying the function of these enzymes in vitro. Specifically, the KS and CLF could not be purified separately and needed to be co-expressed and purified as a dimer. Further, while certain ACPs could be successfully phosphopantetheinylated in vitro, many were only isolated in their holo form after being expressed from a Streptomyces spp., rather than E. coli. Subsequent in vitro work on the key minimal type II PKS cluster was thus limited to certain systems and workarounds [4,152]. There have been conflicting reports about the ability of rat type I FAS ACP to successfully substitute the native actinorhodin PKS ACP in S. coelicolor [153,154]. Earlier work by Tropf et al. reported pigment production in vivo by expressing rat type 1 FAS ACP in place of the native ACP in the act minimal type II PKS system [153]. A later study by Reed et al. recreated the experiment in vivo with rat type I FAS ACP showing that it can be converted into its holo form by E.coli acyl carrier protein synthase (ACPS), but pigment production was not observed [154]. A recent study [155] has made some advances in producing type II PKS products in E. coli. The transcriptional coupling of the two proteins resulted in the successful purification of the soluble heterodimer with co-expression, with a four nucleotide (ATGA) transcriptional overlap with KS and CLF of the alpomycin (alp) type II PKS. Likewise, breakthroughs are forthcoming with respect to ACP phosphopantetheinylation [156].
3.1. Acyl Carrier Proteins
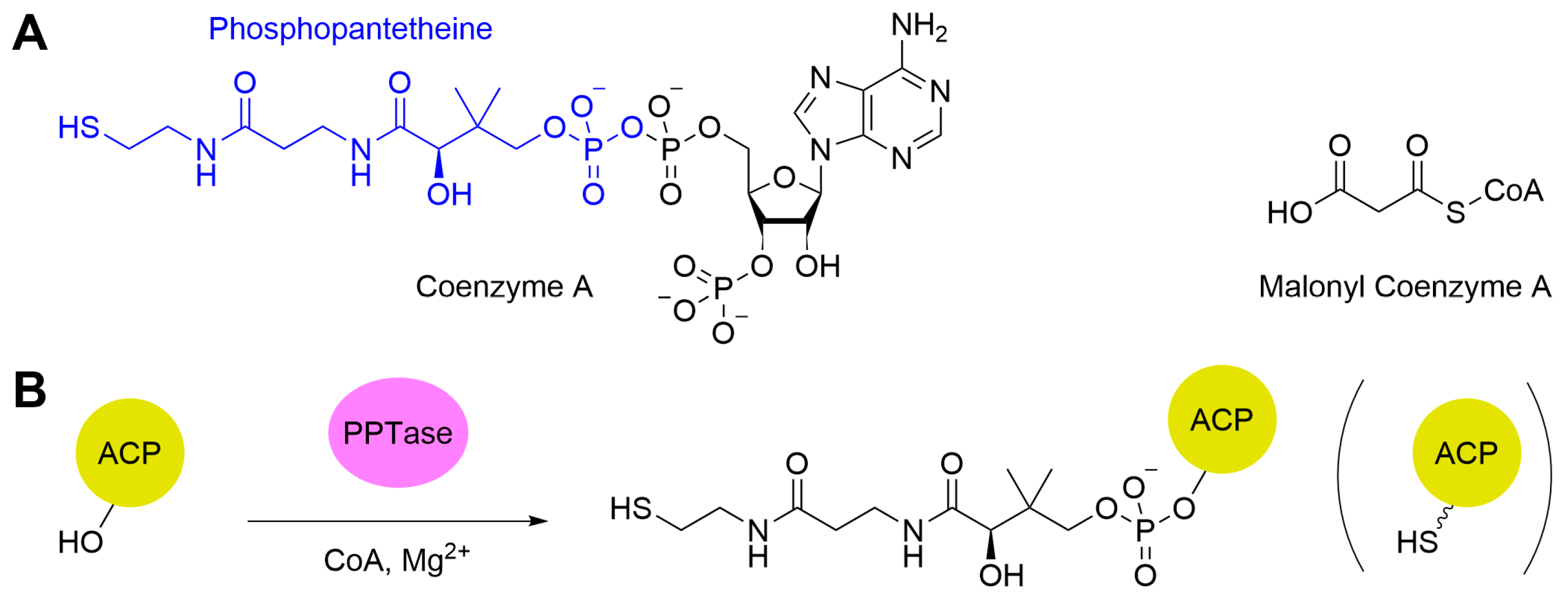
ACPs are an essential part of polyketide synthesis [157]. These enzymes serve three functions: loading the starter unit, loading the malonyl-CoA extender unit (Figure 7A), and exchanging the growing polyketide chain with the KS. In their native form, ACPs are inactive (apo-ACP). To gain catalytic activity, ACPs must undergo post-translational modification by a phosphopantetheine transferase (PPTase), which accepts coenzyme A (CoA) as a substrate and source of phosphopantetheine (PPant). The PPant is attached to a conserved serine residue on the ACP via a phosphoester bond (Figure 7B), converting the ACP to its active form (holo-ACP). The PPant prosthetic group with its reactive thiol site at the end acts as a flexible arm which allows the enzyme to be loaded with both the starter unit and the malonyl-CoA extender unit and to interact with the other proteins.
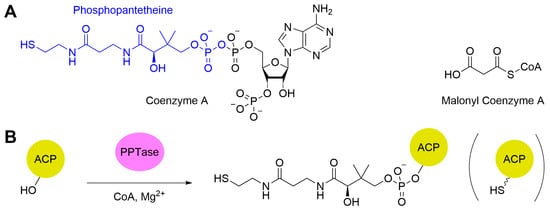
Figure 7.
(A) The structures of coenzyme A (the phosphopantetheine is highlighted in blue) and the abbreviated form of the malonyl-CoA unit. (B) Conversion of apo-ACP to holo-ACP by the action of PPTase. The squiggly line terminating in a thiol in the cartoon to the right represents the flexible phosphopantetheine arm.
PAn early experiment by Hutchinson and coworkers [158,159] revealed that the malonyl-CoA:holo-acyl carrier protein transacylase (MCAT) native to a host’s fatty acid biosynthesis pathway was capable of loading malonyls onto type II PKS holo-ACPs. However, the addition of malonyl-CoA to the ACP has also been demonstrated to be a spontaneous process, an intrinsic property to at least some ACPs [159]. To determine the origin of malonylation, Arthur et al. synthesized the Act ACP in vitro to ensure it was free from any potential MCAT contamination [159]. After refolding it with the GroEL/ES chaperone system [160] from E. coli and phosphopantetheinylation by acyl carrier protein synthase from S. coelicolor, incubation with malonyl-CoA and analysis via electrospray mass spectrometry showed loading of the malonyl extender unit onto the ACP [159]. This result confirmed that the Act ACP does not innately require MCAT for modification. This resulted in a revision of the original hypothesis with the new one being that ACPs can self-load, at least with malonyl-CoA extender units [159].
The ACP can also be involved in starter unit synthesis. A study by Bisang et al. revealed that decarboxylation of the malonyl-bound ACP to form the acetate starter unit is catalyzed by the KS-CLF dimer [145]. Residues Cys169 and Gln161 on the KS and CLF, respectively, were hypothesized to have catalytic activity towards malonyl-ACP. Three sets of mutants were created through site-direct mutagenesis to test the catalytic activity, C169A, Q161A, and the double mutant. The incubation of malonyl-ACP with the double mutant showed no change in the malonyl-ACP concentrations compared to a control reaction lacking the native KS-CLF dimer. Incubation with the native CLF and KS C169A mutant resulted in approximately 40% decarboxylation of malonyl-ACP in 60 min, which showed that the CLF contributes to the decarboxylative activity of the KS-CLF dimer towards malonyl-ACP. Incubation with CLF Q161A and the native KS also showed catalytic activity with 20% substrate decarboxylation after 60 min. This experiment provided evidence that the decarboxylative ability of the KS alone was insufficient normal rates of polyketide production. Beyond the ability of KS-CLF dimer mutants to decarboxylate, the ability of the dimer to produce polyketides as part of a minimal cassette with ACP and malonyl-CoA substrate was monitored. In the absence of pre-acetylated ACP, polyketide production by the KS-CLF mutant occurred, but only after a notable lag phase.
Despite these notable successes, as well as critical breakthroughs by Burkart and colleagues enabling the analysis of protein–protein interactions with the ACP [14], the phosphopantetheinylation of most type II PKS ACPs is not straightforward, limiting the ability to study the individual enzymes of the minimal cluster. However, a recent study by Charkoudian and coworkers [156] has provided insight into the interactions between PPTase and ACP that enable recognition and phosphopantetheinylation. The promiscuous B. subtilis PPTase Sfp [161], widely used for phosphopantetheinylation and genome incorporated for some E. coli strains [162], has been known to fail when modifying type II PKS ACP both in vitro and in vivo. NMR spectroscopy of an ACP from Gloeocapsa sp. PCC 7428 and Sfp revealed that enzymes interact to form a complex, but that the expected post-translational modification does not occur. Multiple sequence analysis of GloACP with ACPs known to be modified by Sfp revealed that GloACP carried polar residues in the −3 and +1 positions relative to the conserved serine attachment point of the ppant in contrast to nonpolar residues in modifiable ACPs. To test the significance of these residues, they completed site-directed mutagenesis to modify these positions. The MS analysis of the resulting Q31G, T35L, and Q31G/T35L mutants after incubation with Sfp revealed that only the T35L and Q31G/T35L mutants were modified to their holo form, leading to the conclusion that T35L was essential residue for phosphopantetheinylation. The double mutant Q31G/T35L showed the greatest levels of phosphopantetheinylation, suggesting that the Q31G mutation improves efficiency, but is not essential for successful recognition by Sfp. Furthermore, these ACP mutations were shown to be compatible with its cognate KS-CLF; upon incubation with the other required protein components, substrates, and cofactors, a m/z signal consistent with the expected salicylyl undecyl ketide was observed.
3.2. KS-CLF Heterodimer
The ketosynthases (KS) and chain length factor (CLF), also known as KSα and KSβ, respectively, compose the other two-thirds of the minimal cassette [3]. The KS and CLF form a heterodimer responsible for catalyzing and sequestering the growing polyketide chain. The KS catalyzes the chain-lengthening Claisen reaction and serves as an anchor for the polyketide chain, which allows the ACP to leave the complex and reload with malonyl-CoA. The CLF fulfills two separate roles in the minimal cassette: polyketide chain length determination and decarboxylation of malonyl-CoA [16,22].
Investigations into the roles played by the KS and CLF in polyketide chain length began in 2000 [22]. The sequences of act and tcm KSs and CLFs were aligned and divided into six sections. A series of chimeric proteins were constructed with each section of the act KS and CLF being systematically exchanged with a corresponding respective section of the tcm enzymes [22]. While not all combinations produced were functional, some hybrids produced polyketides of lengths primarily corresponding to the native CLF. This experiment resulted in three important conclusions: (1) the influence of the KS on chain length is limited to the D region; (2) the CLF primarily determines chain length; and (3) it is possible to produce functional recombinant KS-CLF dimers [22].
In 2004, the crystal structure of the act KS-CLF heterodimer was resolved, providing insight into the function of the KS-CLF complex [16]. The interface between the two proteins formed an amphipathic tunnel which differed from the hydrophobic tunnels found in fatty acid synthases (FASs). Computational modeling provided strong evidence that the polyketide chain grows into, and is subsequently stabilized, by the tunnel. The resulting structure formed by the tunnel led to three hypotheses about the KS-CLF function: (1) the chain length is influenced by “gates” in the tunnel (Figure 8); (2) the first cyclization of the chain is performed inside the dimer; and (3) the buried chain cannot be accessed by tailoring enzymes until after it dissociates from the complex.
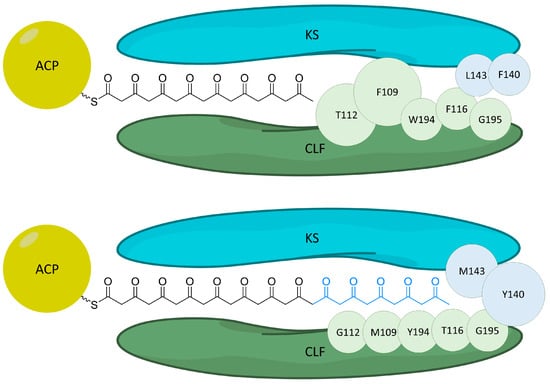
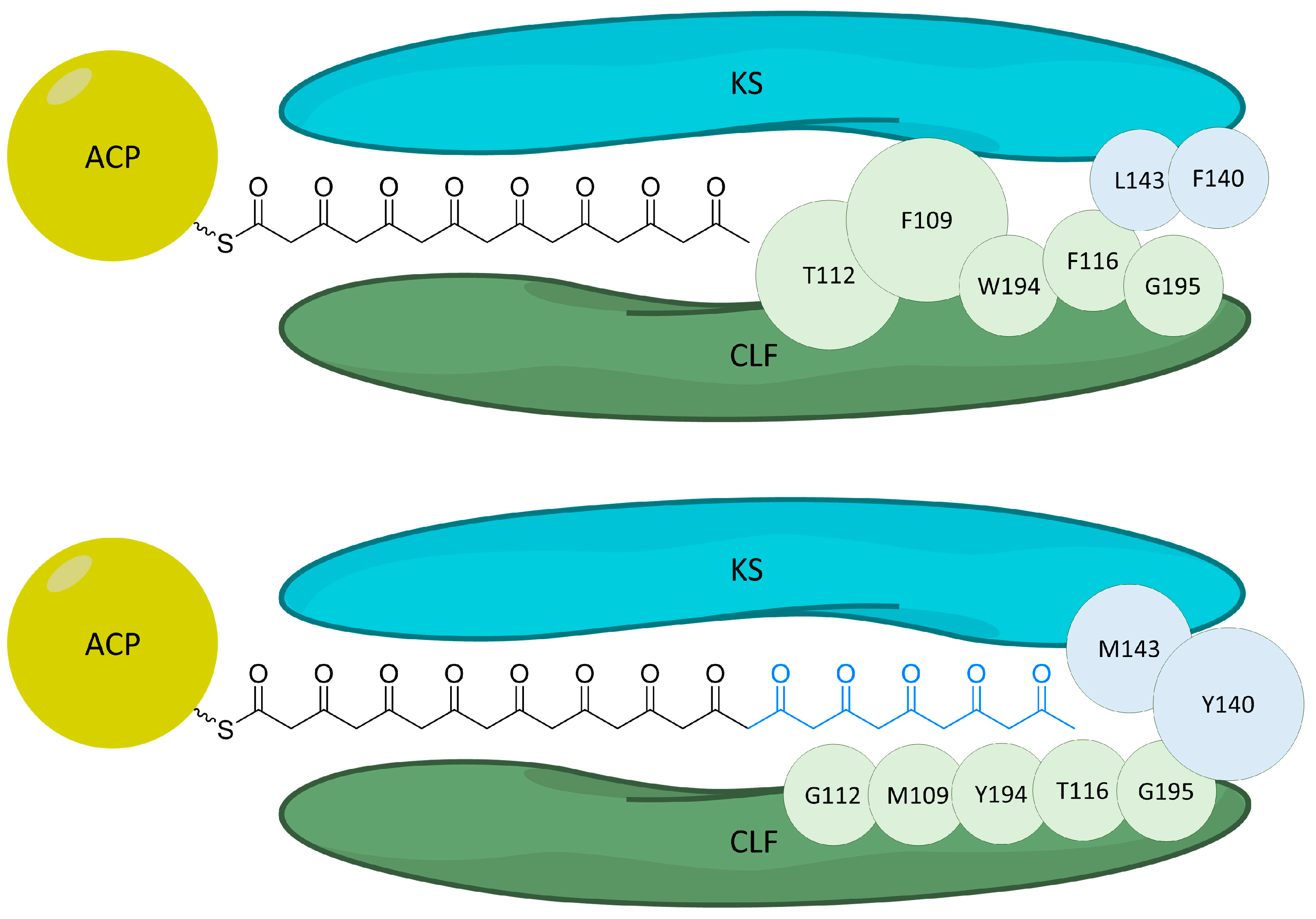
Figure 8.
Cartoon depicting the gating residues [16] for octaketide (top) production in the actinorhodin system and (bottom) the corresponding residues for tridecaketide production in the griseorhodin system. Residues from the CLF are shown in green while residues from the KS are shown in blue. The larger threonine112 and phenylalanine109 (top) cause growth to stop at the octaketide because the octaketide cannot be accommodated in the binding tunnel and thus cannot be transferred back to the KS for a continued extension. These are replaced by smaller residues (bottom) for the tridecaketide, which terminate at methionine143 and tyrosine140.
Evidence for the first hypothesis is supported by prior work by Tang, Tsai, and Kholsa [163], where mutations to the act KS-CLF resulted in the production of a decaketide instead of the original octaketide. Site-direct mutagenesis of the act chain length factor was used to substitute smaller amino acids in key residues in the protein with the goal of altering polyketide chain length. Two CLF mutants were created (F116A and F109A/F116A) and tested both in vivo and in vitro. As a result of the mutations, polyketide chain length shifted from an octaketide to a decaketide. The double mutant resulted in a ratio that was 96% decaketide in vitro, which clearly established that the altering amino acid size in the tunnel resulted in a changed polyketide length.
The second hypothesis, that the first cyclization of the chain is performed inside the dimer, is primarily supported by the presence of C7-C12 or C9-C14 cyclization patterns in the aromatic products [16]. While many other patterns of cyclization are possible, they are rarely seen. Evidence opposing this hypothesis is the existence of cyclases that catalyze C7-C12 and C9-C14 cyclization patterns [140,164], although these may simply accelerate the process. Further research is needed to support or disprove this hypothesis.
The third hypothesis is supported by the existence of the buried tunnel used to stabilize polyketide chain. The authors believe that tailoring enzymes would be unable to reach the chain until after dissociation from the complex [16].
A more recent crystallographic study has elucidated the interactions of the KS with the ACP during the extension process [146]. Bode, Groll, and coworkers succeeded in expressing type II PKS enzymes from the ant pathway [165] of Photorhabdus luminescens in E. coli, including the KS-CLF dimer complex along with the ACP and its phosphopantetheinylation components. The KS-CLF complex emerged from the E. coli with a hexaketide bound to the KS, a fact the authors attribute to the action of an endogenous E. coli ACP interacting with the dimer, despite the dimer’s inability to produce polyketides with other closely related type II PKS ACPs. Co-crystallization with the holo-ACP AntF along with the KS-CLF dimer AntDE (PDB ID: 6SMP) clearly captured a static image of the tetrahedral intermediate that is generated as the ACP hands off the growing polyketide chain back to the KS. This study directly supports the hypothesis that a host’s native MCAT is a predominate pathway for malonylation of ACP, reveals details about protein–protein interactions, and highlights contacts and length restrictions for nascent polyketides growing in the tunnel formed by the KS-CLF interface.
Despite these key advances, continued investigations into the functional manipulation of the KS-CLF complex, and to a lesser extend type II PKS tailoring enzymes, have been limited by handling issues. While E. coli has been a successful host for the production of a range of natural products, including those by type I PKS systems, its use for the expression of type II PKS proteins and secondary metabolites has been largely unsuccessful. The primary bottleneck in type II PKS expression in E. coli is the insolubility of the KS-CLF heterodimer [147].
Several different methods to improve solubility and expression in E. coli have been previously explored with limited success. Fusing KS-CLF proteins with solubility tags still resulted in the formation of inclusion bodies [4]. The overexpression of σ54, a transcription initiation factor, in E. coli successfully increased the production of oxytetracycline to 2.0 mg/L due to upregulating oxyB (CLF). However, it still did not resolve the issue of KS-CLF insolubility [166]. The method of overexpressing σ54 in E. coli has not been tried with other type II PKS gene clusters. Transcriptional overlap between the KS and CLF in combination with pGro7 chaperonin plasmids has also been used with some success to improve solubility with the angucyclinone (Alp) KS-CLF [155,167]. The hypothesis behind transcriptional overlap was developed by analysising KS-CLF gene clusters in GenBank. The authors observed that the majority of KS-CLF genes shared a transcription overlap of four nucleotides and hypothesized that recreating the overlap would improve solubility. The alp KS-CLF were given a four-nucleotide overlap (ATGA) and produced a stable heterodimer when purified. Other nucleotide overlaps of 10, 18, and 41 were tested, with only the 41 nucleotide overlap showing limited improvements in solubility [155].
Alternative hosts for the heterologous expression of type II PKS systems are available, though they lack the versatility and facility of protein expression in E.coli. Streptomyces coelicolor and Streptomyces lividans are two common alternatives for type II PKS expression [168]. Unlike E. coli, the physiology of Streptomyces spp. is closer to the native actinomycete producers. While tools for genetic manipulation of Streptomyces spp. are well established, they do not have the same range and versatility as those for E.coli [142]. In addition, Streptomyces hosts present other issues not found in E. coli. Streptomyces spp. are a rich source of natural products, and genome sequencing has revealed many cryptic biosynthetic gene clusters. The presence of these gene clusters poses issues such as cross-talk between biosynthetic pathways, the production of unintended natural products, and decreased production due to resources being diverted to other metabolic pathways. Additionally, streptomyces spp. have a slower growth rate than E. coli [4].
3.3. Starter Units
In all classes of polyketide synthases, starter units (SUs) are the first units to be loaded onto the KS and incorporated into the growing polyketide chain. From seminal research with the act and tcm clusters [18,19], acetate is regarded as the canonical starter unit for type II PKSs [22]. Canonical acetate starter units are sourced from malonyl groups and, based on analogies to other systems, were proposed to be decarboxylated either during their addition to the PKS (such as KSQ, a ketosynthase-like decarboxylase domain, in pikromycin PKS) or via separate machinery (such as for the loading of Ery PKS for erythromycin) [7,169]. The leading hypothesis is that malonyl-CoA ACP is decarboxylated to form the acetate starter unit that can then join the KS-CLF through a condensation reaction [145,146]. For non-canonical starter units, several processes may be used [3]. A KSIII, such as the one proposed in the trioxacarcin pathway [81], or another specialized KS [115] may deliver the substituted acetate to the type II PKS ACP. Alternatively, a priming ACP can deliver starter units to the KS, as proposed for hedamycin [82]. Recent work has demonstrated that aryl carboxylates can be loaded directly onto the holo-ACP [156]. This flexibility of the starter unit (examples are shown in Table 1) contributes to the vast structural diversity of type II PKS products and, with complete deciphering of the minimal cluster, lends itself to synthetic biology applications.

Table 1.
Structures and names of starter units with associated organism, natural product, and method of identification in type II PKSs.
3.4. Aromatases and Cyclases
Aromatases and cyclases in type II PKSs are tailoring enzymes that control the formation of ring structures from the polyketide chain by catalyzing the formation of C-C bonds. Since cyclases can produce aromatic rings upon cyclization, aromatase and cyclases are often grouped under the sole category of “cyclases” [140]. The polyketide chain produced by the minimal cassette is unstable, and without tailoring enzymes, it will spontaneously cyclize. In the presence of cyclases, spontaneous cyclization of the chain is suppressed and deliberate cyclization can be directed [140]. Due to the unstable nature of the polyketide chain, the mechanism of cyclization by cyclases are difficult to study. However, gene knockout studies and in vitro work have enabled the elucidation of their cyclization patterns [140,149,164,176].
Cyclases can be categorized based on their protein class [140]. Didomain TcmN-like cyclases, based on the tetracenomycin BGC, catalyze the first formation of a ring structure following a C7-C12 pattern [140,164]. Metallo-β-lactamase and OxyN-like cyclases catalyze a C5-C14 cyclization pattern for the formation of a second ring structure [140,176]. TcmJ-like cyclases catalyze the formation of second and third ring structures in tricyclic compounds by forming C7-C16 and C5-C18 bonds, respectively [140,149].
Cyclases in type II PKS systems normally group well within established classes, such as for the creation of linear tetracycles, tetracenomycins, and tetracyclines (Figure 3). The tetracenomycin family relies on the following archetypes: TcmN, which cyclizes the first two rings [177,178,179]; TcmJ, believed to complete the third ring [44]; and TcmI, which closes the fourth [180]. Oxytetracycline biosynthesis also relies on three cyclases, but they are not homologous to the tcm series: OxyK, which cyclizes the first ring [181]; OxyN, which cyclizes the second (and possibly the third ring, though this step may be spontaneous) [182]; and OxyH, an unusual CoA-ligase-like protein which forms the fourth ring [183]. Intriguingly, of the three predicted cyclases in the thermorubin cluster [125], two resemble the tcm/elm family while one resembles the oxy family, and the ordering is blended. This finding suggests that after establishing the controlling features of starter unit and length with minimal type II PKS cassettes, the mixing of engineered cyclases from various systems may enable the controlled creation of novel ring architectures.
4. Highly Reducing Type II PKS Systems and Other New Findings
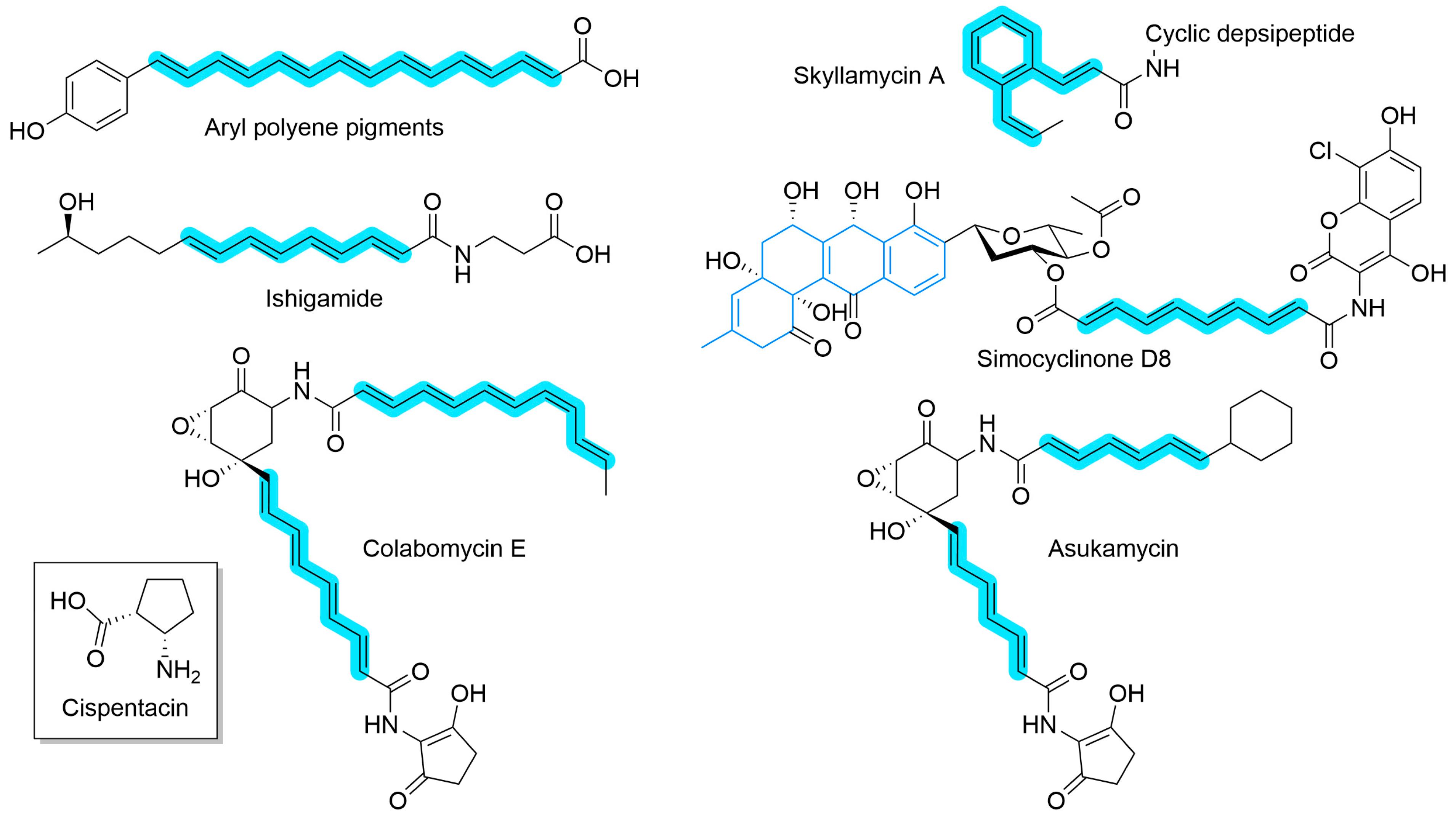
While canonical type II PKS systems have been known for decades, the recent discovery of highly reducing (HR) variants has both challenged the type II PKS paradigm and expanded their known biosynthetic repertoire. The HR type II PKSs share the same core features as the canonical type II PKS; a minimal cassette composed of an ACP, KS, and CLF which work in combination to produce a polyketide chain. What distinguishes HR from the canonical type is the reduction in the polyketide chain to a polyene chain. Some polyene natural products (Figure 9), such as aryl polyene pigments [184] and ishigamide [185], are predominately linear and, after loading with a starter unit, rely predominately on HR type II PKSs for their structure. Others, such as skyllamycin A [186], are depsipeptides where the HR type II PKS is responsible for side chain formation and can include tailoring enzymes that are hypothesized to catalyze cyclization. HR type II PKS can also be used alongside canonical type II PKS, for example in the construction of simocyclinone D8 [187], where a glycosylated anguidine core is linked to a substituted coumarin by a polyene. Multiple HR type II PKSs may also produce varied and complex structures, such as the colabomycins [188] and asukamycin [189].
The first HR type II PKS system described as such arose from Streptomyces sp. MSC090213JE08 [185]. Bioinformatic analysis of its genome with antiSMASH revealed a proposed cryptic biosynthetic gene cluster corresponding to a type II PKS system. To express the proposed cluster in vivo, genes from the Streptomyces antibiotic regulatory protein (SARP) transcriptional activator family were inserted into the genome to force the cryptic clusters to express. Metabolic profiling of the recombinant strain controlled by the SARP7 gene using LC-MS indicated the production of a new metabolite.
The structure of the unknown compound, later named ishigamide (Figure 9), was identified through a combination of single-dimensional and two-dimensional NMR as 3-((2E,4E,6E,8E)-13-hydroxytetradeca-2,4,6,8-tetraenamido)propanoic acid [185]. The presence of β-alanine and unsaturated fatty acid moieties suggested the product might be synthesized and reduced by a PKS pathway with the peptide bond catalyzed by nonribosomal peptide synthetase (NRPS), a peptide ligase, or an ATP-grasp enzyme. Three potential biosynthetic clusters (type-1-PKS-like, type-II-PKS-like, and type-III-PKS-like) in the genome were identified as possible sources of ishigamide. To determine which cluster was responsible, mutants were generated with key enzymes in each cluster disrupted. Only in the cluster containing the type II PKS was the production of ishigamide abolished [185].
Later studies were able to successfully reconstitute ishigamide PKS (IgaPKS) in vitro to investigate the genes behind the minimal cassette [190]. The incubation of the holo-ACP (Iga10), KS (Iga11), and CLF (Iga12) with hexanonyl-CoA, malonyl-CoA, and NADPH did not yield any polyene product by LC-MS. Instead, the analysis revealed the presence of 3-oxooctanoyl, indicating the ACP was loaded with the hexanoyl-CoA starter unit. This led to the hypothesis that the minimal cassette could only extend the chain after the reduction from a ketide to an alkene.
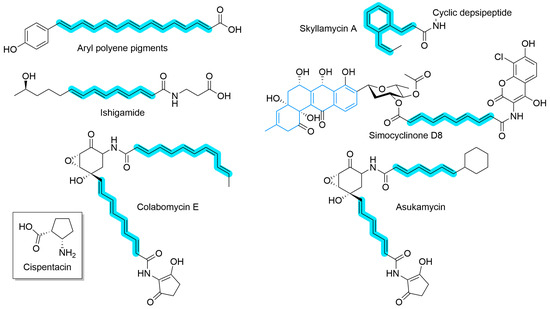
Figure 9.
Natural products that utilize HR type II PKS in their biosynthesis. The polyene(s) derived from the HR type II PKS are highlighted in blue. Simocyclinone D8 also includes a canonical type II PKS core (blue bonds). Inset: The structure of cispentacin, a non-canonical amino acid formed through the action of a type-II-PKS-like minimal cassette [191].
Figure 9.
Natural products that utilize HR type II PKS in their biosynthesis. The polyene(s) derived from the HR type II PKS are highlighted in blue. Simocyclinone D8 also includes a canonical type II PKS core (blue bonds). Inset: The structure of cispentacin, a non-canonical amino acid formed through the action of a type-II-PKS-like minimal cassette [191].
To test this hypothesis, the ketoreductase (Iga13) and dehydrogenase (Iga16) were incubated with the minimal cassette under the same conditions. LC-MS analysis revealed four unsaturated carboxylic acid products that were detected: (2E)-octenoic acid, (2E,4E)-deca-2,4-dienoic acid, (2E,4E,6E)-dodeca-2,4,6-trienoic acid, and (2E,4E,6E,8E)-tetradeca-2,4,6,8-tetraenoic acid. Each product formed corresponds to a different step in the chain extension process. The formation of these products only after the addition of ketoreductase (KR) and dehydratase (DH) proteins strongly suggests that reduction in the keto group is necessary before the chain can be extended [190].
The unusual nature of the IgaPKS, and its requirement of KS and DH enzymes outside of the minimal cassette for chain elongation, led to computational and experimental investigation into the structural basis of the KS-CLF complex [192]. X-ray crystallography of the Iga KS-CLF confirmed that the two enzymes form the expected heterodimer. However, an analysis of Iga11, the ketosynthase, revealed that it is more similar to KSs from FAS systems than other type II PKSs. Loading and cross-linking experiments revealed protein–protein interactions and demonstrated the flexibility of the hexanoyl starter unit in the active site.
Analysis of the KS-CLF substrate pocket revealed the presence of additional polar amino acids, differing from the ambiphilic pocket of most canonical type II PKSs. Site-directed mutagenesis was used to systematically substitute each polar amino acid in the substrate pocket with alanine. The testing of the KS-CLF mutants in combination with the ACP, KR, and DH revealed that they disfavor transfer of the β-ketoacyl group from the ACP to the KS, effectively favoring the formation of the polyene prior to additional extension by the PKS. As with other type II PKS systems, the chain length was controlled by a residue in the CLF [192].
In addition to the canonical and HR type II PKSs, a novel type-II-PKS-like system has been discovered that is responsible for the biosynthesis of cispentacin, a nonproteinogenic amino acid with a five-membered non-aromatic skeleton. Bioinformatic analysis of the amipurimycin (Amc) BGC identified AmcB and AmcF as putative ACP and KS, respectively. The MS analysis of AmcB confirmed that it can undergo phosphopantetheinylation, which identifies its function as an ACP. An in vitro analysis of AmcH and AmcB confirmed the role of AmcH as a 2-oxoglutaryl-ACP ligase with AmcB as its substrate. The coexpression of AmcF and AmcG resulted in the formation of a heterodimer characteristic of KS-CLF heterodimers found in typical type II PKS systems. However, upon the incubation of the heterodimer AmcF-AmcG with AmcB (ACP) and AmcH (2-oxoglutaryl-ACP ligase), the 2-oxoglutaryl starter unit was extended only by one malonyl unit. The heterodimeric AmcF-AmcG also appears to catalyze an intramolecular aldol condensation, resulting in a five-member ring that is subsequently tailored by other enzymes into cispentacin [191].
With the addition of HR to canonical type II PKSs, not only can a variety of length polyketides be formed to furnish intricate polycycles, but polyenes that might link ring systems [187] or form additional rings [190,191] become available as well. These intriguing new systems highlight the continuing diversity unlocked from natural product biosynthesis, even among enzyme families that share many similarities. Further investigations into HR type II PKSs will likely reveal additional structural diversity and how this fascinating new class of type II PKS can be adapted to create designed compounds.
5. Future Directions
Previous studies into type II PKS systems have yielded fundamental knowledge about functions of the ACP, KS, and CLF along with tailoring enzymes. Additionally, the natural products derived from type II PKS systems have provided clinically relevant drugs, both in their unmodified and derivatized forms. Given the potential for novel natural and unnatural products, pursuing further research is critical to learn more about these remarkable enzymes and unlock their potential for use in chemoenzymatic catalysis and synthetic biology. For this potential of type II PKS systems to be fully realized, however, gaps in understanding the properties of the enzymes, which can best be elucidated with in vitro approaches, must be addressed.
5.1. Acyl Carrier Proteins
Initial studies into the ACPs have revealed insight into their structure and mechanism. However, our understanding of ACP-protein interactions remained incomplete. For ACPs to be consistently utilized, further work is needed to understand how type II PKS ACPs interact with both the native and non-native PPTase for conversion into their functional holo forms. Research by Charkoudian et al. [156] has shown that interactions between Sfp and GloACP are heavily influenced by key residues. Confirming the applicability of these mutations to other non-actinobacterial ACPs would enable the critical breakthrough of establishing how ACPs interact with the minimal cassette, enabling their consistent modification and routine use. Beyond this essential extension function, details of how they can be effectively loaded, both with malonyl and starter units, as well as how they interact with other enzymes, be it cyclases to form ring systems or KR/DH proteins to create polyenes, must also be broadly addressed to begin working toward rationally designed molecules.
5.2. Ketosynthase and Chain-Length Factors
The KS and CLF are fundamental for the production of the polyketide chain that cyclizes to form the diverse structures found in type II PKS natural products [146], or in the case of HR type II PKSs, to form a polyene [192]. The most significant challenge to address with these enzymes is the successful production of soluble proteins both in vivo and in vitro.
While previous works have shown success with the expression of these proteins in Streptomyces, expression in E. coli remains challenging [4]. By identifying the methods of soluble expression in E. coli, natural product production levels could be improved due to the wide range of genetic engineering tools readily available for E. coli hosts. The reliance on the co-expression of the KS-CLF is a major hurdle in dissecting the interactions between the two and how each might be selectively adapted. For in vitro work, soluble KSs and CLFs would enable further characterization of type II PKS BGCs and their intermediate products in addition to creating combinatorial biosynthetic pathways. Furthermore, the ability to create polyketide chains in vitro expands options for discrete testing of the function of downstream tailoring enzymes, such as cyclases, for the production of novel natural products.
Once technical concerns are addressed, there is great potential for creating novel products by investigating factors that alter polyketide chain length and acceptance of alternative starter units. Altering the chain length of the polyketide could result in novel cyclization patterns and products. Incorporating novel starter units into established biosynthetic pathways would further enhance the diversity and functionality of their natural products. Different polyene chain lengths and their cyclization to aromatic compounds, as well as the possibility of polymer-type compounds, are a possibility as well with the recent identification of HR type II PKSs.
5.3. Cyclases
The function of cyclases has been primarily limited to in vivo gene knockout studies due to issues when handling the minimal cassette in vitro and the instability of the polyketide products it produces. Once the handling properties of the minimal cassette are optimized, the incorporation and in vitro characterization of cyclase would provide foundational knowledge pertaining to their mechanisms in addition to identifying their promiscuity towards non-native chain lengths. By identifying their promiscuity and factors related to chain length recognition, novel cyclization patterns and products can be synthesized, setting predetermined scaffolds for further modification by tailoring enzymes, such as oxidases and glycosylases.
5.4. Derivatized Scaffolds
The polycyclic aromatic cores of the type II PKS have more synthetic potential to offer than simply the ring structures. Incredibly complex rings can be produced through oxidations and rearrangements as well as the incorporation of other atoms to produce heterocycles. The type II PKS systems also offer a range of tailoring enzymes that can act after cyclization including oxygenases, methyltransferases, and glycosylases [3]. The benefit of these enzymes can be seen from both a synthetic biology and synthetic organic chemistry standpoint. For synthetic biology, additional functional groups such as alcohols, ketones, and epoxides can be stereoselectivity added to the polyaromatic scaffolds, directly accessing desired compounds or advanced precursors that, in combination with synthetic chemistry, offer a starting point for further derivatization and modification. Indeed, there are already established reactions for the modification of biobased molecules [193]. Plentiful and varied functional groups, including ketones, alcohols, and amines, can be added, and groups can readily be further oxidized or reduced. The ability to biocatalytically create and selectively append glycosides is especially promising, given the historical difficulties of installing these groups chemically [194]. Alkenes, such as those found in the highly reducing variant of type II PKS, could be hydrogenated into alkanes.
5.5. Biofuel
The structures of highly reducing type II PKS, due to their long hydrocarbon chains, show promise for use in biofuel production. Long-chain alcohols, such as 1-octonal, have been considered a renewable alternative to petroleum diesel due to their similar fuel properties [195]. While the biosynthesis of 1-octonal has been successfully constructed in vivo, the production yields were low due to the activities of the enzymes involved in chain elongation [196]. The long hydrocarbons chains produced by the HR type II PKS present a potential alternative for the biosynthetic or semi-synthesis of 1-octanol. Additionally, modifications to the CLF in a PKS system create the potential for the synthesis of both longer and shorter hydrocarbon chains in alcohols.
6. Conclusions
In summary, a large body of meticulous work has demonstrated the impact of type II PKS natural products in medicine. This study has also significantly advanced our knowledge of the genetics of the BGCs and the functions of their associated enzymes. The in vivo production of antibiotics and anticancer therapeutics has been an undeniable boon, but the full potential of these systems has not yet been fully realized. To truly harness the biosynthetic capabilities of type II PKSs, additional work is needed to characterize the functionalities of the minimal cassette, much of which is best performed in vitro in order to elucidate the range, flexibility, and compatibility of the various components. Additionally, nascent research into the highly reducing variant of type II PKS has yet to reveal the structural diversity and enzymatic capabilities of these systems. Beyond their established importance in the production of antibiotics and chemotherapeutics, type II PKS systems can continue to impact other fields of medicine as well as expand to diverse fields, such as the production of biofuels. As a whole, type II PKS systems show promise in the field of synthetic biology once further enzymatic characterization unlocks their full potential.
Author Contributions
Conceptualization, M.A.J.R. and A.N.L.; Formal Analysis, M.A.J.R. and A.N.L.; Data Curation, M.A.J.R. and A.N.L.; Writing—Original Draft Preparation; M.A.J.R. and A.N.L.; Writing—Review and Editing; M.A.J.R. and A.N.L.; Visualization, A.N.L.; Supervision, A.N.L.; Project Administration, A.N.L.; Funding Acquisition, A.N.L. All authors have read and agreed to the published version of the manuscript.
Funding
M.A.J.R. and A.N.L. thank the Virginia Tech Department of Chemistry for supporting this research.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Pham, J.V.; Yilma, M.A.; Feliz, A.; Majid, M.T.; Maffetone, N.; Walker, J.R.; Kim, E.; Cho, H.J.; Reynolds, J.M.; Song, M.C.; et al. A Review of the Microbial Production of Bioactive Natural Products and Biologics. Front. Microbiol. 2019, 10, 1404. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, R.; Chen, X.; Sun, X.; Yan, Y.; Shen, X.; Yuan, Q. Biosynthesis of aromatic polyketides in microorganisms using type II polyketide synthases. Microb. Cell Fact 2020, 19, 110. [Google Scholar] [CrossRef]
- Hertweck, C.; Luzhetskyy, A.; Rebets, Y.; Bechthold, A. Type II polyketide synthases: Gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 2007, 24, 162–190. [Google Scholar] [CrossRef]
- Gao, X.; Wang, P.; Tang, Y. Engineered polyketide biosynthesis and biocatalysis in Escherichia coli. Appl. Microbiol. Biotechnol. 2010, 88, 1233–1242. [Google Scholar] [CrossRef]
- Keatinge-Clay, A.T. The structures of type I polyketide synthases. Nat. Prod. Rep. 2012, 29, 1050. [Google Scholar] [CrossRef]
- Austin, M.B.; Noel, J.P. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 2003, 20, 79–110. [Google Scholar] [CrossRef]
- Hertweck, C. The Biosynthetic Logic of Polyketide Diversity. Angew. Chem. Int. Ed. 2009, 48, 4688–4716. [Google Scholar] [CrossRef]
- Nivina, A.; Yuet, K.P.; Hsu, J.; Khosla, C. Evolution and Diversity of Assembly-Line Polyketide Synthases. Chem. Rev. 2019, 119, 12524–12547. [Google Scholar] [CrossRef]
- Herbst, D.A.; Townsend, C.A.; Maier, T. The architectures of iterative type I PKS and FAS. Nat. Prod. Rep. 2018, 35, 1046–1069. [Google Scholar] [CrossRef]
- Yu, D.; Xu, F.; Zeng, J.; Zhan, J. Type III polyketide synthases in natural product biosynthesis. IUBMB Life 2012, 64, 285–295. [Google Scholar] [CrossRef]
- Katsuyama, Y.; Ohnishi, Y. Chapter Sixteen—Type III Polyketide Synthases in Microorganisms. In Methods in Enzymology; Hopwood, D.A., Ed.; Academic Press: Cambridge, MA, USA, 2012; Volume 515, pp. 359–377. [Google Scholar]
- Shimizu, Y.; Ogata, H.; Goto, S. Type III Polyketide Synthases: Functional Classification and Phylogenomics. ChemBioChem 2017, 18, 50–65. [Google Scholar] [CrossRef]
- Palmer, C.M.; Alper, H.S. Expanding the Chemical Palette of Industrial Microbes: Metabolic Engineering for Type III PKS-Derived Polyketides. Biotechnol. J. 2019, 14, 1700463. [Google Scholar] [CrossRef]
- Chen, A.; Re, R.N.; Burkart, M.D. Type II fatty acid and polyketide synthases: Deciphering protein–protein and protein–substrate interactions. Nat. Prod. Rep. 2018, 35, 1029–1045. [Google Scholar] [CrossRef]
- Smith, S.; Tsai, S.-C. The type I fatty acid and polyketide synthases: A tale of two megasynthases. Nat. Prod. Rep. 2007, 24, 1041. [Google Scholar] [CrossRef]
- Keatinge-Clay, A.T.; Maltby, D.A.; Medzihradszky, K.F.; Khosla, C.; Stroud, R.M. An antibiotic factory caught in action. Nat. Struct. Mol. Biol. 2004, 11, 888–893. [Google Scholar] [CrossRef]
- Malico, A.A.; Nichols, L.; Williams, G.J. Synthetic biology enabling access to designer polyketides. Curr. Opin. Chem. Biol. 2020, 58, 45–53. [Google Scholar] [CrossRef]
- Brockmann, H.; Pini, H. Actinorhodine, a red pigment from Actinomycetes. Sci. Nat. 1947, 34, 190. [Google Scholar] [CrossRef]
- Weber, W.; Zähner, H.; Siebers, J.; Schröder, K.; Zeeck, A. Stoffwechselprodukte von Mikroorganismen. 175. Mitteilung. Tetracenomycin C. Arch. Microbiol. 1979, 121, 111–116. [Google Scholar] [CrossRef]
- Motamedi, H.; Hutchinson, C.R. Cloning and heterologous expression of a gene cluster for the biosynthesis of tetracenomycin C, the anthracycline antitumor antibiotic of Streptomyces glaucescens. Proc. Natl. Acad. Sci. USA 1987, 84, 4445–4449. [Google Scholar] [CrossRef]
- Hallam, S.E.; Malpartida, F.; Hopwood, D.A. Nucleotide sequence, transcription and deduced function of a gene involved in polyketide antibiotic synthesis in Streptomyces coelicolor. Gene 1988, 74, 305–320. [Google Scholar] [CrossRef]
- Burson, K.K.; Khosla, C. Dissecting the Chain Length Specificity in Bacterial Aromatic Polyketide Synthases using Chimeric Genes. Tetrahedron 2000, 56, 9401–9408. [Google Scholar] [CrossRef]
- Qun, T.; Zhou, T.; Hao, J.; Wang, C.; Zhang, K.; Xu, J.; Wang, X.; Zhou, W. Antibacterial activities of anthraquinones: Structure–activity relationships and action mechanisms. RSC Med. Chem. 2023, 14, 1446–1471. [Google Scholar] [CrossRef]
- Bringmann, G.; Irmer, A.; Feineis, D.; Gulder, T.A.M.; Fiedler, H.-P. Convergence in the biosynthesis of acetogenic natural products from plants, fungi, and bacteria. Phytochemistry 2009, 70, 1776–1786. [Google Scholar] [CrossRef] [PubMed]
- Cheemalamarri, C.; Batchu, U.R.; Thallamapuram, N.P.; Katragadda, S.B.; Reddy Shetty, P. A review on hydroxy anthraquinones from bacteria: Crosstalk’s of structures and biological activities. Nat. Prod. Res. 2022, 36, 6186–6205. [Google Scholar] [CrossRef] [PubMed]
- Kakinuma, S.; Ikeda, H.; Omura, S.; Hopwood, D.A. Biosynthesis of kalafungin in Streptomyces tanashiensis. J. Antibiot. 1990, 43, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Bergy, M.E. Kalafungin, A New Broad Spectrum Antibiotic. J. Antibiot. 1968, 21, 454–457. [Google Scholar] [CrossRef] [PubMed]
- Takano, S.; Hasuda, K.; Ito, A.; Koide, Y.; Ishii, F.; Haneda, I.; Chihara, S.; Koyama, Y. A new antibiotic, medermycin. J. Antibiot. 1976, 29, 765–768. [Google Scholar] [CrossRef] [PubMed]
- Corbaz, R.; Ettlinger, L.; Gäumann, E.; Kalvoda, J.; Keller-Schierlein, W.; Kradolfer, F.; Manukian, B.K.; Neipp, L.; Prelog, V.; Reusser, P.; et al. Stoffwechselprodukte von Actinomyceten. 9. Mitteilung. Granaticin. Helv. Chim. Acta 1957, 40, 1262–1269. [Google Scholar] [CrossRef]
- Jabila Mary, T.R.; Kannan, R.R.; Muthamil Iniyan, A.; Carlton Ranjith, W.A.; Nandhagopal, S.; Vishwakarma, V.; Prakash Vincent, S.G. β-lactamase inhibitory potential of kalafungin from marine Streptomyces in Staphylococcus aureus infected zebrafish. Microbiol. Res. 2021, 244, 126666. [Google Scholar] [CrossRef]
- Okabe, T.; Nomoto, K.; Funabashi, H.; Okuda, S.; Suzuki, H.; Tanaka, N. Lactoquinomycin, a novel anticancer antibiotic. II. Physico-chemical properties and structure assignment. J. Antibiot. 1985, 38, 1333–1336. [Google Scholar] [CrossRef]
- Ichinose, K.; Bedford, D.J.; Tornus, D.; Bechthold, A.; Bibb, M.J.; Peter Revill, W.; Floss, H.G.; Hopwood, D.A. The granaticin biosynthetic gene cluster of Streptomyces violaceoruber Tü22: Sequence analysis and expression in a heterologous host. Chem. Biol. 1998, 5, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Watari, S.; Taguchi, T.; Ishikawa, K.; Kumamoto, T.; Okamoto, S.; Ichinose, K. Actinorhodin Biosynthesis Terminates with an Unprecedented Biaryl Coupling Reaction. Angew. Chem. Int. Ed. 2023, 62, e202214400. [Google Scholar] [CrossRef] [PubMed]
- Bhuyan, B.K.; Dietz, A. Fermentation, taxonomic, and biological studies on nogalamycin. Antimicrob. Agents Chemother. 1965, 5, 836–844. [Google Scholar] [PubMed]
- Arora, S.K. Molecular structure, absolute stereochemistry, and interactions of nogalamycin, a DNA-binding anthracycline antitumor antibiotic. J. Am. Chem. Soc. 1983, 105, 1328–1332. [Google Scholar] [CrossRef]
- Arena, E.; D’Alessandro, N.; Dusonchet, L.; Gebbia, N.; Gerbasi, F.; Palazzoadriano, M.; Raineri, A.; Rausa, L.; Tubaro, E. Analysis of the pharmacokinetic characteristics, pharmacological and chemotherapeutic activity of 14-Hydroxy-daunomycin (Adriamycin), a new drug endowed with an antitumour activity. Arzneimittelforschung 1971, 21, 1258–1263. [Google Scholar] [PubMed]
- Hutchinson, C.R. Biosynthetic Studies of Daunorubicin and Tetracenomycin C. Chem. Rev. 1997, 97, 2525–2536. [Google Scholar] [CrossRef]
- Wiley, P.F.; Elrod, D.W.; Marshall, V.P. Biosynthesis of the anthracycline antibiotics nogalamycin and steffimycin B. J. Org. Chem. 1978, 43, 3457–3461. [Google Scholar] [CrossRef]
- Siitonen, V.; Nji Wandi, B.; Törmänen, A.-P.; Metsä-Ketelä, M. Enzymatic Synthesis of the C-Glycosidic Moiety of Nogalamycin R. ACS Chem. Biol. 2018, 13, 2433–2437. [Google Scholar] [CrossRef]
- Wiley, P.F. Improved Antitumor Activity by Modification of Nogalamycin. J. Nat. Prod. 1979, 42, 569–582. [Google Scholar] [CrossRef]
- McGovren, J.P.; Neil, G.L.; Denlinger, R.H.; Hall, T.L.; Crampton, S.L.; Swenberg, J.A. Chronic Cardiotoxicity Studies in Rabbits with 7-con-O-Methylnogarol, a New Anthracycline Antitumor Agent1. Cancer Res. 1979, 39, 4849–4855. [Google Scholar] [PubMed]
- Hulst, M.B.; Grocholski, T.; Neefjes, J.J.C.; Van Wezel, G.P.; Metsä-Ketelä, M. Anthracyclines: Biosynthesis, engineering and clinical applications. Nat. Prod. Rep. 2022, 39, 814–841. [Google Scholar] [CrossRef] [PubMed]
- Drautz, H.; Reuschenbach, P.; Zähner, H.; Rohr, J.; Zeeck, A. Metabolic products of microorganisms. 225. Elloramycin, a new anthracycline-like antibiotic from Streptomyces olivaceus. Isolation, characterization, structure and biological properties. J. Antibiot. 1985, 38, 1291–1301. [Google Scholar] [CrossRef]
- Ramos, A.; Lombó, F.; Braña, A.F.; Rohr, J.; Méndez, C.; Salas, J.A. Biosynthesis of elloramycin in Streptomyces olivaceus requires glycosylation by enzymes encoded outside the aglycon cluster. Microbiology 2008, 154, 781–788. [Google Scholar] [CrossRef]
- Finlay, A.C.; Hobby, G.L.; P’an, S.Y.; Regna, P.P.; Routien, J.B.; Seeley, D.B.; Shull, G.M.; Sobin, B.A.; Solomons, I.A.; Vinson, J.W.; et al. Terramycin, a new antibiotic. Science 1950, 111, 85. [Google Scholar] [CrossRef]
- Duggar, B.M. Aureomycin: A Product of the Continuing Search for New Antibiotics. Ann. N. Y. Acad. Sci. 1948, 51, 177–181. [Google Scholar] [CrossRef]
- Tymiak, A.A.; Aklonis, C.; Bolgar, M.S.; Kahle, A.D.; Kirsch, D.R.; O’Sullivan, J.; Porubcan, M.A.; Principe, P.; Trejo, W.H. Dactylocyclines: Novel tetracycline glycosides active against tetracycline-resistant bacteria. J. Org. Chem. 1993, 58, 535–537. [Google Scholar] [CrossRef]
- Hatsu, M.; Sasaki, T.; Gomi, S.; Kodama, Y.; Sezaki, M.; Inouye, S.; Kondo, S. A new tetracycline antibiotic with antitumor activity. II. The structural elucidation of SF2575. J. Antibiot. 1992, 45, 325–330. [Google Scholar] [CrossRef]
- Pickens, L.B.; Tang, Y. Decoding and engineering tetracycline biosynthesis. Metab. Eng. 2009, 11, 69–75. [Google Scholar] [CrossRef]
- Kabuto, C.; Silverton, J.V.; Akiyama, T.; Sankawa, U.; Hutchison, R.D.; Steyn, P.S.; Vleggaar, R. X-ray structure of viridicatumtoxin: A new class of mycotoxin from Penicillium viridicatum Westling. J. Chem. Soc. Chem. Commun. 1976, 18, 728–729. [Google Scholar] [CrossRef]
- Thomas, R. A Biosynthetic Classification of Fungal and Streptomycete Fused-Ring Aromatic Polyketides. ChemBioChem 2001, 2, 612–627. [Google Scholar] [CrossRef]
- Momose, I.; Chen, W.; Nakamura, H.; Naganawa, H.; Iinuma, H.; Takeuchi, T. Polyketomycin, a New Antibiotic from Streptomyces sp. MK277-AF1. II. Structure Determination. J. Antibiot. 1998, 51, 26–32. [Google Scholar] [CrossRef]
- Mitscher, L.A.; Juvarkar, J.V.; Rosenbrook, W.; Andres, W.W.; Schenck, J.R.; Egan, R.S. Structure of chelocardin, a novel tetracycline antibiotic. J. Am. Chem. Soc. 1970, 92, 6070–6071. [Google Scholar] [CrossRef]
- Xuan, L.-J.; Xu, S.-H.; Zhang, H.-L.; Xu, Y.-M.; Chen, M.-Q. Dutomycin, a new anthracycline antibiotic from Streptomyces. J. Antibiot. 1992, 45, 1974–1976. [Google Scholar] [CrossRef]
- Herold, K.; Gollmick, F.A.; Groth, I.; Roth, M.; Menzel, K.D.; Möllmann, U.; Gräfe, U.; Hertweck, C. Cervimycin A–D: A Polyketide Glycoside Complex from a Cave Bacterium Can Defeat Vancomycin Resistance. Chem. Eur. J. 2005, 11, 5523–5530. [Google Scholar] [CrossRef]
- Wu, H.; Selvaraj, K.; Yang, G.; Wang, Y.; Chen, G. Study for C4-O-Glycosylation of Tetracycline. Tetrahedron Lett. 2023, 133, 154829. [Google Scholar] [CrossRef]
- Rohr, J.; Weißbach, U.; Beninga, C.; Künzel, E.; Rohr, J.; Siems, K.; Bindseil, K.U.; Lombó, F.; Prado, L.; Braña, A.F.; et al. The structures of premithramycinone and demethylpremithramycinone, plausible early intermediates of the aureolic acid group antibiotic mithramycin. Chem. Commun. 1998, 437–438. [Google Scholar] [CrossRef]
- Lombó, F.; Siems, K.; Braña, A.F.; Méndez, C.; Bindseil, K.; Salas, J.A. Cloning and insertional inactivation of Streptomyces argillaceus genes involved in the earliest steps of biosynthesis of the sugar moieties of the antitumor polyketide mithramycin. J. Bacteriol. 1997, 179, 3354–3357. [Google Scholar] [CrossRef] [PubMed]
- Berlin, Y.A.; Chuprunova, O.A.; Klyashchitskii, B.A.; Kolosov, M.N.; Peck, G.Y.; Piotrovich, L.A.; Shemyakin, M.M.; Vasina, I.V. Olivomycin. III. The structure of olivin. Tetrahedron Lett. 1966, 7, 1425–1430. [Google Scholar] [CrossRef]
- Stevenson, L.J.; Bracegirdle, J.; Liu, L.; Sharrock, A.V.; Ackerley, D.F.; Keyzers, R.A.; Owen, J.G. Metathramycin, a new bioactive aureolic acid discovered by heterologous expression of a metagenome derived biosynthetic pathway. RSC Chem. Biol. 2021, 2, 556–567. [Google Scholar] [CrossRef]
- Rohr, J.; Méndez, C.; Salas, J.A. The Biosynthesis of Aureolic Acid Group Antibiotics. Bioorg. Chem. 1999, 27, 41–54. [Google Scholar] [CrossRef]
- Wheeler, R.; Yu, X.; Hou, C.; Mitra, P.; Chen, J.M.; Herkules, F.; Ivanov, D.N.; Tsodikov, O.V.; Rohr, J. Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis. Angew. Chem. Int. Ed. 2020, 59, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Sezaki, M.; Kondo, S.; Maeda, K.; Umezawa, H.; Ohno, M. The structure of aquayamycin. Tetrahedron 1970, 26, 5171–5190. [Google Scholar] [CrossRef] [PubMed]
- Yushchuk, O.; Kharel, M.; Ostash, I.; Ostash, B. Landomycin biosynthesis and its regulation in Streptomyces. Appl. Microbiol. Biotechnol. 2019, 103, 1659–1665. [Google Scholar] [CrossRef] [PubMed]
- Kharel, M.K.; Pahari, P.; Shepherd, M.D.; Tibrewal, N.; Nybo, S.E.; Shaaban, K.A.; Rohr, J. Angucyclines: Biosynthesis, mode-of-action, new natural products, and synthesis. Nat. Prod. Rep. 2012, 29, 264–325. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, E.; Ogasawara, Y.; Liu, H.-W. A Biosynthetic Pathway for BE-7585A, a 2-Thiosugar-Containing Angucycline-Type Natural Product. J. Am. Chem. Soc. 2010, 132, 7405–7417. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.J.; Halley, K.A. Biosynthesis of the benz[a]anthraquinone antibiotic PD 116198: Evidence for a rearranged skeleton. J. Am. Chem. Soc. 1991, 113, 5092–5093. [Google Scholar] [CrossRef]
- Kharel, M.K.; Pahari, P.; Shaaban, K.A.; Wang, G.; Morris, C.; Rohr, J. Elucidation of post-PKS tailoring steps involved in landomycin biosynthesis. Org. Biomol. Chem. 2012, 10, 4256. [Google Scholar] [CrossRef]
- Liu, M.; Yang, Y.J.; Gong, G.; Li, Z.; Zhang, L.; Guo, L.; Xu, B.; Zhang, S.M.; Xie, Z.P. Angucycline and angucyclinone derivatives from the marine-derived Streptomyces sp. Chirality 2022, 34, 421–427. [Google Scholar] [CrossRef]
- Asolkar, R.N.; Kirkland, T.N.; Jensen, P.R.; Fenical, W. Arenimycin, an antibiotic effective against rifampin- and methicillin-resistant Staphylococcus aureus from the marine actinomycete Salinispora arenicola. J. Antibiot. 2010, 63, 37–39. [Google Scholar] [CrossRef] [PubMed]
- Tsunakawa, M.; Nishio, M.; Ohkuma, H.; Tsuno, T.; Konishi, M.; Naito, T.; Oki, T.; Kawaguchi, H. The structure of pradimicins A, B and C: A novel family of antifungal antibiotics. J. Org. Chem. 1989, 54, 2532–2536. [Google Scholar] [CrossRef]
- Walsh, T.J.; Giri, N. Pradimicins: A novel class of broad-spectrum antifungal compounds. Eur. J. Clin. Microbiol. Infect. Dis. 1997, 16, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Kakushima, M.; Sawada, Y.; Nishio, M.; Tsuno, T.; Oki, T. Biosynthesis of pradimicin A. J. Org. Chem. 1989, 54, 2536–2539. [Google Scholar] [CrossRef]
- Kakinuma, S.; Suzuki, K.; Hatori, M.; Saitoh, K.; Hasegawa, T.; Furumai, T.; Oki, T. Biosynthesis of the pradimicin family of antibiotics. III. Biosynthetic pathway of both pradimicins and benanomicins. J. Antibiot. 1993, 46, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.K.; Paudel, S.; Dhakal, D.; Van, P.T.T.; Ghimire, G.P.; Sohng, J.K. Genetic evidence for the involvement of glycosyltransferase PdmQ and PdmS in biosynthesis of pradimicin from Actinomadura hibisca. Microbiol. Res. 2015, 174, 9–16. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, X.; Xiao, Z.; Zhou, J.; Song, X.; Wang, X.; Hu, L.; Wang, Y.; Sun, P.; Wang, W.; et al. O-methyltransferase-like enzyme catalyzed diazo installation in polyketide biosynthesis. Nat. Commun. 2023, 14, 5372. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Yukishige, I. Chapter 1—Molecular architecture and therapeutic potential of lectin mimics. In Advances in Carbohydrate Chemistry and Biochemistry; Horton, D., Ed.; Academic Press: Cambridge, MA, USA, 2012; Volume 68, pp. 1–58. [Google Scholar]
- Schmitz, H.; Crook, K.E., Jr.; Bush, J.A. Hedamycin, a new antitumor antibiotic. I. Production, isolation, and characterization. Antimicrob. Agents Chemother. 1966, 6, 606–612. [Google Scholar] [PubMed]
- Bradner, W.T.; Heinemann, B.; Gourevitch, A. Hedamycin, a new antitumor antibiotic. II. Biological properties. Antimicrob. Agents Chemother. 1966, 1966, 613–618. [Google Scholar]
- Shirahata, K.; Iida, T.; Hirayama, N. Structures of trioxacarcin A, a new antitumor antibiotic, and its related compounds. Tennen Yuki Kagobutsu Toronkai Koen Yoshishu 1981, 24, 199–206. [Google Scholar]
- Zhang, M.; Hou, X.-F.; Qi, L.-H.; Yin, Y.; Li, Q.; Pan, H.-X.; Chen, X.-Y.; Tang, G.-L. Biosynthesis of trioxacarcin revealing a different starter unit and complex tailoring steps for type II polyketide synthase. Chem. Sci. 2015, 6, 3440–3447. [Google Scholar] [CrossRef]
- Das, A.; Khosla, C. In Vivo and In Vitro Analysis of the Hedamycin Polyketide Synthase. Chem. Biol. 2009, 16, 1197–1207. [Google Scholar] [CrossRef]
- Hansen, M.; Yun, S.; Hurley, L. Hedamycin intercalates the DNA helix and, through carbohydrate-mediated recognition in the minor groove, directs N7-alkylation of guanine in the major groove in a sequence-specific manner. Chem. Biol. 1995, 2, 229–240. [Google Scholar] [CrossRef]
- Pfoh, R.; Laatsch, H.; Sheldrick, G.M. Crystal structure of trioxacarcin A covalently bound to DNA. Nucleic Acids Res. 2008, 36, 3508–3514. [Google Scholar] [CrossRef]
- Aoyama, T.; Naganawa, H.; Muraoka, Y.; Nakamura, H.; Aoyagi, T.; Takeuchi, T.; Iitaka, Y. Benastatins A and B, new inhibitors of glutathione S-transferase, produced by Streptomyces sp. MI384-DF12. II. Structure determination of benastatins A and B. J. Antibiot. 1992, 45, 1391–1396. [Google Scholar] [CrossRef][Green Version]
- Lackner, G.; Schenk, A.; Xu, Z.; Reinhardt, K.; Yunt, Z.S.; Piel, J.; Hertweck, C. Biosynthesis of Pentangular Polyphenols: Deductions from the Benastatin and Griseorhodin Pathways. J. Am. Chem. Soc. 2007, 129, 9306–9312. [Google Scholar] [CrossRef]
- Xu, Z.; Schenk, A.; Hertweck, C. Molecular Analysis of the Benastatin Biosynthetic Pathway and Genetic Engineering of Altered Fatty Acid−Polyketide Hybrids. J. Am. Chem. Soc. 2007, 129, 6022–6030. [Google Scholar] [CrossRef]
- Brockmann, H.; Schmidt-Kastner, G. Resistomycin, a new antibiotic from actinomycetes. Sci. Nat. 1951, 38, 479. [Google Scholar] [CrossRef]
- Jakobi, K.; Hertweck, C. A Gene Cluster Encoding Resistomycin Biosynthesis in Streptomyces resistomycificus; Exploring Polyketide Cyclization beyond Linear and Angucyclic Patterns. J. Am. Chem. Soc. 2004, 126, 2298–2299. [Google Scholar] [CrossRef] [PubMed]
- Zaleta-Rivera, K.; Charkoudian, L.K.; Ridley, C.P.; Khosla, C. Cloning, Sequencing, Heterologous Expression, and Mechanistic Analysis of A-74528 Biosynthesis. J. Am. Chem. Soc. 2010, 132, 9122–9128. [Google Scholar] [CrossRef] [PubMed]
- Miyairi, N.; Sakai, H.-I.; Konomi, T.; Imanaka, H. Enterocin, a new antibiotic taxonomy, isolation and characterization. J. Antibiot. 1976, 29, 227–235. [Google Scholar] [CrossRef]
- Tokuma, Y.; Miyairi, N.; Morimoto, Y. Structure of enterocin; X-ray analysis of m-bromobenzoyl enterocin dihydrate. J. Antibiot. 1976, 29, 1114–1116. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Kalaitzis, J.A.; Moore, B.S. EncM, a versatile enterocin biosynthetic enzyme involved in Favorskii oxidative rearrangement, aldol condensation, and heterocycle-forming reactions. Proc. Natl. Acad. Sci. USA 2004, 101, 15609–15614. [Google Scholar] [CrossRef] [PubMed]
- Teufel, R.; Miyanaga, A.; Michaudel, Q.; Stull, F.; Louie, G.; Noel, J.P.; Baran, P.S.; Palfey, B.; Moore, B.S. Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement. Nature 2013, 503, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Maglangit, F.; Deng, H. Cell Factory for Phenylnaphthacenoid Polyketide Production. SynBio 2023, 1, 89–102. [Google Scholar] [CrossRef]
- Maglangit, F.; Fang, Q.; Leman, V.; Soldatou, S.; Ebel, R.; Kyeremeh, K.; Deng, H. Accramycin A, a New Aromatic Polyketide, from the Soil Bacterium, Streptomyces sp. MA37. Molecules 2019, 24, 3384. [Google Scholar] [CrossRef]
- Maglangit, F.; Zhang, Y.; Kyeremeh, K.; Deng, H. Discovery of New Antibacterial Accramycins from a Genetic Variant of the Soil Bacterium, Streptomyces sp. MA37. Biomolecules 2020, 10, 1464. [Google Scholar] [CrossRef]
- Monciardini, P.; Bernasconi, A.; Iorio, M.; Brunati, C.; Sosio, M.; Campochiaro, L.; Landini, P.; Maffioli, S.I.; Donadio, S. Antibacterial Aromatic Polyketides Incorporating the Unusual Amino Acid Enduracididine. J. Nat. Prod. 2019, 82, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.-S.; Brady, S.F. Mining Soil Metagenomes to Better Understand the Evolution of Natural Product Structural Diversity: Pentangular Polyphenols as a Case Study. J. Am. Chem. Soc. 2014, 136, 18111–18119. [Google Scholar] [CrossRef]
- Iorio, M.; Cruz, J.; Simone, M.; Bernasconi, A.; Brunati, C.; Sosio, M.; Donadio, S.; Maffioli, S.I. Antibacterial Paramagnetic Quinones from Actinoallomurus. J. Nat. Prod. 2017, 80, 819–827. [Google Scholar] [CrossRef]
- Herath, K.B.; Jayasuriya, H.; Guan, Z.; Schulman, M.; Ruby, C.; Sharma, N.; Macnaul, K.; Menke, J.G.; Kodali, S.; Galgoci, A.; et al. Anthrabenzoxocinones from Streptomyces sp. as Liver X Receptor Ligands and Antibacterial Agents. J. Nat. Prod. 2005, 68, 1437–1440. [Google Scholar] [CrossRef]
- Kodali, S.; Galgoci, A.; Young, K.; Painter, R.; Silver, L.L.; Herath, K.B.; Singh, S.B.; Cully, D.; Barrett, J.F.; Schmatz, D.; et al. Determination of Selectivity and Efficacy of Fatty Acid Synthesis Inhibitors. J. Biol. Chem. 2005, 280, 1669–1677. [Google Scholar] [CrossRef]
- Ma, M.; Rateb, M.E.; Teng, Q.; Yang, D.; Rudolf, J.D.; Zhu, X.; Huang, Y.; Zhao, L.-X.; Jiang, Y.; Li, X.; et al. Angucyclines and Angucyclinones from Streptomyces sp. CB01913 Featuring C-Ring Cleavage and Expansion. J. Nat. Prod. 2015, 78, 2471–2480. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, S.S.; Van Der Heul, H.U.; Xiao, X.; Nuutila, A.; Baars, L.R.; Wu, C.; Metsä-Ketelä, M.; Van Wezel, G.P. Unravelling key enzymatic steps in C-ring cleavage during angucycline biosynthesis. Commun. Chem. 2023, 6, 281. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-Q.; Li, G.; Lou, H.-X. Isolation, Biosynthesis, and Biological Activity of Polycyclic Xanthones from Actinomycetes. Front. Microbiol. 2022, 13, 922089. [Google Scholar] [CrossRef]
- Kudo, F.; Yonezawa, T.; Komatsubara, A.; Mizoue, K.; Eguchi, T. Cloning of the biosynthetic gene cluster for naphthoxanthene antibiotic FD-594 from Streptomyces sp. TA-0256. J. Antibiot. 2011, 64, 123–132. [Google Scholar] [CrossRef]
- Kang, H.-S.; Brady, S.F. Arixanthomycins A–C: Phylogeny-Guided Discovery of Biologically Active eDNA-Derived Pentangular Polyphenols. ACS Chem. Biol. 2014, 9, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, X.; Zhu, J.; Shen, Y. Amexanthomycins A–J, pentangular polyphenols produced by Amycolatopsis mediterranei S699∆rifA. Appl. Microbiol. Biotechnol. 2018, 102, 689–702. [Google Scholar] [CrossRef]
- Warnick-Pickle, D.J.; Byrne, K.M.; Pandey, R.C.; White, R.J. Fredericamycin A a new antitumor antibiotic. II. Biological properties. J. Antibiot. 1981, 34, 1402–1407. [Google Scholar] [CrossRef]
- Misra, R.; Pandey, R.C.; Silverton, J.V. Fredericamycin A, an antitumor antibiotic of a novel skeletal type. J. Am. Chem. Soc. 1982, 104, 4478–4479. [Google Scholar] [CrossRef]
- Pandey, R.C.; Toussaint, M.W.; Stroshane, R.M.; Kalita, C.C.; Aszalos, A.A.; Garretson, A.L.; Wei, T.T.; Byrne, K.M.; Stroshane, R.M.; White, R.J. Fredericamycin A a new antitumor antibiotic. I. Production, isolation and physicochemical properties. J. Antibiot. 1981, 34, 1389–1401. [Google Scholar] [CrossRef]
- Coronelli, C.; Pagani, H.; Bardone, M.R.; Lancini, G.C. Purpuromycin, a New Antibiotic Isolated from Actinoplanes Ianthinogenes N. Sp. J. Antibiot. 1974, 27, 161–168. [Google Scholar] [CrossRef]
- Eckardt, K.; Tresselt, D.; Ihn, W. Chemical structure of antibiotic Griseorhodin A. Z. Chem. 1976, 16, 486. [Google Scholar] [CrossRef]
- Byrne, K.M.; Hilton, B.D.; White, R.J.; Misra, R.; Pandey, R.C. Biosynthesis of fredericamycin A, a new antitumor antibiotic. Biochemistry 1985, 24, 478–486. [Google Scholar] [CrossRef]
- Das, A.; Szu, P.-H.; Fitzgerald, J.T.; Khosla, C. Mechanism and Engineering of Polyketide Chain Initiation in Fredericamycin Biosynthesis. J. Am. Chem. Soc. 2010, 132, 8831–8833. [Google Scholar] [CrossRef]
- Wendt-Pienkowski, E.; Huang, Y.; Zhang, J.; Li, B.; Jiang, H.; Kwon, H.; Hutchinson, C.R.; Shen, B. Cloning, Sequencing, Analysis, and Heterologous Expression of the Fredericamycin Biosynthetic Gene Cluster from Streptomyces griseus. J. Am. Chem. Soc. 2005, 127, 16442–16452. [Google Scholar] [CrossRef]
- Chen, Y.; Wendt-Pienkowski, E.; Ju, J.; Lin, S.; Rajski, S.R.; Shen, B. Characterization of FdmV as an Amide Synthetase for Fredericamycin A Biosynthesis in Streptomyces griseus ATCC 43944. J. Biol. Chem. 2010, 285, 38853–38860. [Google Scholar] [CrossRef]
- Li, A.; Piel, J. A Gene Cluster from a Marine Streptomyces Encoding the Biosynthesis of the Aromatic Spiroketal Polyketide Griseorhodin A. Chem. Biol. 2002, 9, 1017–1026. [Google Scholar] [CrossRef]
- Toplak, M.; Saleem-Batcha, R.; Piel, J.; Teufel, R. Catalytic Control of Spiroketal Formation in Rubromycin Polyketide Biosynthesis. Angew. Chem. Int. Ed. 2021, 60, 26960–26970. [Google Scholar] [CrossRef]
- Frensch, B.; Lechtenberg, T.; Kather, M.; Yunt, Z.; Betschart, M.; Kammerer, B.; Lüdeke, S.; Müller, M.; Piel, J.; Teufel, R. Enzymatic spiroketal formation via oxidative rearrangement of pentangular polyketides. Nat. Commun. 2021, 12, 1431. [Google Scholar] [CrossRef] [PubMed]
- Yunt, Z.; Reinhardt, K.; Li, A.; Engeser, M.; Dahse, H.-M.; Gütschow, M.; Bruhn, T.; Bringmann, G.; Piel, J. Cleavage of Four Carbon−Carbon Bonds during Biosynthesis of the Griseorhodin A Spiroketal Pharmacophore. J. Am. Chem. Soc. 2009, 131, 2297–2305. [Google Scholar] [CrossRef] [PubMed]
- Craveri, R.; Coronelli, C.; Pagani, H.; Sensi, P. Thermorubin, a new antibiotic from a thermoactinomycete. Clin. Med. 1964, 71, 511. [Google Scholar] [PubMed]
- Johnson, F.; Chandra, B.; Iden, C.R.; Naiksatam, P.; Kahen, R.; Okaya, Y.; Lin, S.-Y. Thermorubin 1. Structure studies. J. Am. Chem. Soc. 1980, 102, 5580–5585. [Google Scholar] [CrossRef]
- Aragozzini, F.; Craveri, R.; Maconi, E.; Ricca, G.S.; Scolastico, C. Thermorubin biosynthesis: Evidence for the involvement of both salicylic acid and an undecaketide. J. Chem. Soc. Perkin Trans. 1 1988, 1865. [Google Scholar] [CrossRef]
- McCord, J.P.; Kohanov, Z.A.; Lowell, A.N. Thermorubin Biosynthesis Initiated by a Salicylate Synthase Suggests an Unusual Conversion of Phenols to Pyrones. ACS Chem. Biol. 2022, 17, 3169–3177. [Google Scholar] [CrossRef] [PubMed]
- Simonitsch, E.; Eisenhuth, W.; Stamm, O.A.; Schmid, H. Über die Struktur des Chartreusins (Vorläufige Mitteilung). Helv. Chim. Acta 1960, 43, 58–63. [Google Scholar] [CrossRef]
- Leach, B.E.; Calhoun, K.M.; Johnson, L.E.; Teeters, C.M.; Jackson, W.G. Chartreusin, a New Antibiotic Produced by Streptomyces chartreusis, a New Species. J. Am. Chem. Soc. 1953, 75, 4011–4012. [Google Scholar] [CrossRef]
- Konishi, M.; Sugawara, K.; Kofu, F.; Nishiyama, Y.; Tomita, K.; Miyaki, T.; Kawaguchi, H. Elsamicins, new antitumor antibiotics related to chartreusin. I. Production, isolation, characterization and antitumor activity. J. Antibiot. 1986, 39, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Jiao, F.W.; Wang, Y.S.; You, X.T.; Wei, W.; Chen, Y.; Yang, C.L.; Guo, Z.K.; Zhang, B.; Liang, Y.; Tan, R.X.; et al. An NADPH-Dependent Ketoreductase Catalyses the Tetracyclic to Pentacyclic Skeletal Rearrangement in Chartreusin Biosynthesis. Angew. Chem. Int. Ed. 2021, 60, 26378–26384. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.S.; Zhang, B.; Zhu, J.; Yang, C.L.; Guo, Y.; Liu, C.L.; Liu, F.; Huang, H.; Zhao, S.; Liang, Y.; et al. Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis. J. Am. Chem. Soc. 2018, 140, 10909–10914. [Google Scholar] [CrossRef]
- Xu, Z.; Jakobi, K.; Welzel, K.; Hertweck, C. Biosynthesis of the Antitumor Agent Chartreusin Involves the Oxidative Rearrangement of an Anthracyclic Polyketide. Chem. Biol. 2005, 12, 579–588. [Google Scholar] [CrossRef]
- Ueberschaar, N.; Xu, Z.; Scherlach, K.; Metsä-Ketelä, M.; Bretschneider, T.; Dahse, H.-M.; Görls, H.; Hertweck, C. Synthetic Remodeling of the Chartreusin Pathway to Tune Antiproliferative and Antibacterial Activities. J. Am. Chem. Soc. 2013, 135, 17408–17416. [Google Scholar] [CrossRef]
- Ayer, S.W.; McInnes, A.G.; Thibault, P.; Walter, J.A.; Doull, J.L.; Parnell, T.; Vining, L.C. Jadomycin, a novel 8H-benz[b]oxazolo[3,2-f]phenanthridine antibiotic from from streptomyces venezuelae ISP5230. Tetrahedron Lett. 1991, 32, 6301–6304. [Google Scholar] [CrossRef]
- Doull, J.L.; Ayer, S.W.; Singh, A.K.; Thibault, P. Production of a novel polyketide antibiotic, jadomycin B, by Streptomyces venezuelae following heat shock. J. Antibiot. 1993, 46, 869–871. [Google Scholar] [CrossRef]
- De Koning, C.B.; Ngwira, K.J.; Rousseau, A.L. Chapter Two—Biosynthesis, synthetic studies, and biological activities of the jadomycin alkaloids and related analogues. Alkaloids Chem. Biol. 2020, 84, 125–199. [Google Scholar]
- Rix, U.; Zheng, J.; Remsing Rix, L.L.; Greenwell, L.; Yang, K.; Rohr, J. The Dynamic Structure of Jadomycin B and the Amino Acid Incorporation Step of Its Biosynthesis. J. Am. Chem. Soc. 2004, 126, 4496–4497. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.W.; Martinez-Farina, C.F.; Smithen, D.A.; Yin, H.; Monro, S.; Thompson, A.; Mcfarland, S.A.; Syvitski, R.T.; Jakeman, D.L. Eight-Membered Ring-Containing Jadomycins: Implications for Non-enzymatic Natural Products Biosynthesis. J. Am. Chem. Soc. 2015, 137, 3271–3275. [Google Scholar] [CrossRef]
- Bililign, T.; Griffith, B.R.; Thorson, J.S. Structure, activity, synthesis and biosynthesis of aryl-C-glycosides. Nat. Prod. Rep. 2005, 22, 742. [Google Scholar] [CrossRef]
- Rohr, J.; Hertweck, C. 1.07—Type II PKS. In Comprehensive Natural Products II; Liu, H.-W., Mander, L., Eds.; Elsevier: Oxford, UK, 2010; pp. 227–303. [Google Scholar]
- Xie, S.; Zhang, L. Type II Polyketide Synthases: A Bioinformatics-Driven Approach. ChemBioChem 2023, 24, e202200775. [Google Scholar] [CrossRef]
- Villebro, R.; Shaw, S.; Blin, K.; Weber, T. Sequence-based classification of type II polyketide synthase biosynthetic gene clusters for antiSMASH. J. Ind. Microbiol. Biotechnol. 2019, 46, 469–475. [Google Scholar] [CrossRef]
- Zhang, W.; Tang, Y. Chapter 16 In Vitro Analysis of Type II Polyketide Synthase. In Methods Enzymology; Academic Press: Cambridge, MA, USA, 2009; Volume 459, pp. 367–393. [Google Scholar]
- Kharel, M.K.; Rohr, J. Delineation of gilvocarcin, jadomycin, and landomycin pathways through combinatorial biosynthetic enzymology. Curr. Opin. Chem. Biol. 2012, 16, 150–161. [Google Scholar] [CrossRef]
- Koch, A.A.; Schmidt, J.J.; Lowell, A.N.; Hansen, D.A.; Coburn, K.M.; Chemler, J.A.; Sherman, D.H. Probing Selectivity and Creating Structural Diversity through Hybrid Polyketide Synthases. Angew. Chem. Int. Ed. 2020, 59, 13575–13580. [Google Scholar] [CrossRef] [PubMed]
- Bisang, C.; Long, P.F.; Corte’S, J.; Westcott, J.; Crosby, J.; Matharu, A.-L.; Cox, R.J.; Simpson, T.J.; Staunton, J.; Leadlay, P.F. A chain initiation factor common to both modular and aromatic polyketide synthases. Nature 1999, 401, 502–505. [Google Scholar] [CrossRef]
- Bräuer, A.; Zhou, Q.; Grammbitter, G.L.C.; Schmalhofer, M.; Rühl, M.; Kaila, V.R.I.; Bode, H.B.; Groll, M. Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis. Nat. Chem. 2020, 12, 755–763. [Google Scholar] [CrossRef]
- Yang, D.; Eun, H.; Prabowo, C.P.S. Metabolic Engineering and Synthetic Biology Approaches for the Heterologous Production of Aromatic Polyketides. Int. J. Mol. Sci. 2023, 24, 8923. [Google Scholar] [CrossRef]
- Shen, B.; Hutchinson, C.R. Enzymatic Synthesis of a Bacterial Polyketide from Acetyl and Malonyl Coenzyme A. Science 1993, 262, 1535–1540. [Google Scholar] [CrossRef]
- Bao, W.; Wendt-Pienkowski, E.; Hutchinson, C.R. Reconstitution of the Iterative Type II Polyketide Synthase for Tetracenomycin F2 Biosynthesis. Biochemistry 1998, 37, 8132–8138. [Google Scholar] [CrossRef]
- Carreras, C.W.; Pieper, R.; Khosla, C. Efficient Synthesis of Aromatic Polyketides In Vitro by the Actinorhodin Polyketide Synthase. J. Am. Chem. Soc. 1996, 118, 5158–5159. [Google Scholar] [CrossRef]
- Carreras, C.W.; Khosla, C. Purification and In Vitro Reconstitution of the Essential Protein Components of an Aromatic Polyketide Synthase. Biochemistry 1998, 37, 2084–2088. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Jang, W.D.; Lee, S.Y. Production of Carminic Acid by Metabolically Engineered Escherichia coli. J. Am. Chem. Soc. 2021, 143, 5364–5377. [Google Scholar] [CrossRef] [PubMed]
- Tropf, S.; Peter Revill, W.; Bibb, M.J.; Hopwood, D.A.; Schweizer, M. Heterologously expressed acyl carrier protein domain of rat fatty acid synthase functions in Escherichia coli fatty acid synthase and Streptomyces coelicolor polyketide synthase systems. Chem. Biol. 1998, 5, 135–146. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reed, M.A.C.; Schweizer, M.; Szafranska, A.E.; Arthur, C.; Nicholson, T.P.; Cox, R.J.; Crosby, J.; Crump, M.P.; Simpson, T.J. The type I rat fatty acid synthase ACP shows structural homology and analogous biochemical properties to type II ACPs. Org. Biomol. Chem. 2003, 1, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hua, K.; Liu, D.; Wu, Z.-L.; Wang, Y.; Zhang, H.; Deng, Z.; Pfeifer, B.A.; Jiang, M. Heterologous Biosynthesis of Type II Polyketide Products Using E. coli. ACS Chem. Biol. 2020, 15, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Li, K.K.; Cho, Y.I.; Tran, M.A.; Wiedemann, C.; Koweek, R.S.; Hoàng, N.K.; Hamrick, G.S.; Bowen, M.A.; Kokona, B.; Beld, J.; et al. Strategic Engineering Unlocks In Vitro Type II Polyketide Biosynthesis; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2023. [Google Scholar] [CrossRef]
- Beld, J.; Sonnenschein, E.C.; Vickery, C.R.; Noel, J.P.; Burkart, M.D. The phosphopantetheinyl transferases: Catalysis of a post-translational modification crucial for life. Nat. Prod. Rep. 2014, 31, 61–108. [Google Scholar] [CrossRef] [PubMed]
- Summers, R.G.; Ali, A.; Shen, B.; Wessel, W.A.; Hutchinson, C.R. Malonyl-Coenzyme A:Acyl Carrier Protein Acyltransferase of Streptomyces glaucescens: A Possible Link between Fatty Acid and Polyketide Biosynthesis. Biochemistry 1995, 34, 9389–9402. [Google Scholar] [CrossRef] [PubMed]
- Arthur, C.J.; Szafranska, A.; Evans, S.E.; Findlow, S.C.; Burston, S.G.; Owen, P.; Clark-Lewis, I.; Simpson, T.J.; Crosby, J.; Crump, M.P. Self-Malonylation Is an Intrinsic Property of a Chemically Synthesized Type II Polyketide Synthase Acyl Carrier Protein. Biochemistry 2005, 44, 15414–15421. [Google Scholar] [CrossRef]
- Georgopoulos, C.P.; Hendrix, R.W.; Casjens, S.R.; Kaiser, A.D. Host participation in bacteriophage lambda head assembly. J. Mol. Biol. 1973, 76, 45–60. [Google Scholar] [CrossRef]
- Lambalot, R.H.; Gehring, A.M.; Flugel, R.S.; Zuber, P.; LaCelle, M.; Marahiel, M.A.; Reid, R.; Khosla, C.; Walsh, C.T. A new enzyme superfamily—The phosphopantetheinyl transferases. Chem. Biol. 1996, 3, 923–936. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, B.A.; Admiraal, S.J.; Gramajo, H.; Cane, D.E.; Khosla, C. Biosynthesis of Complex Polyketides in a Metabolically Engineered Strain of E. coli. Science 2001, 291, 1790–1792. [Google Scholar] [CrossRef]
- Tang, Y.; Tsai, S.-C.; Khosla, C. Polyketide Chain Length Control by Chain Length Factor. J. Am. Chem. Soc. 2003, 125, 12708–12709. [Google Scholar] [CrossRef]
- Mcdaniel, R.; Ebert-Khosla, S.; Hopwood, D.A.; Khosla, C. Engineered Biosynthesis of Novel Polyketides: ActVII and actIV Genes Encode Aromatase and Cyclase Enzymes, Respectively. J. Am. Chem. Soc. 1994, 116, 10855–10859. [Google Scholar] [CrossRef]
- Brachmann, A.O.; Joyce, S.A.; Jenke-Kodama, H.; Schwär, G.; Clarke, D.J.; Bode, H.B. A Type II Polyketide Synthase is Responsible for Anthraquinone Biosynthesis in Photorhabdus luminescens. ChemBioChem 2007, 8, 1721–1728. [Google Scholar] [CrossRef]
- Stevens, D.C.; Conway, K.R.; Pearce, N.; Villegas-Peñaranda, L.R.; Garza, A.G.; Boddy, C.N. Alternative Sigma Factor Over-Expression Enables Heterologous Expression of a Type II Polyketide Biosynthetic Pathway in Escherichia coli. PLoS ONE 2013, 8, e64858. [Google Scholar] [CrossRef] [PubMed]
- Wall, J.G.; Plückthun, A. Effects of overexpressing folding modulators on the in vivo folding of heterologous proteins in Escherichia coli. Curr. Opin. Biotechnol. 1995, 6, 507–516. [Google Scholar] [CrossRef]
- Medema, M.H.; Breitling, R.; Takano, E. Chapter twenty-one—Synthetic Biology in Streptomyces Bacteria. In Methods Enzymology; Voigt, C., Ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 497, pp. 485–502. [Google Scholar]
- Long, P.F.; Wilkinson, C.J.; Bisang, C.P.; Cortés, J.; Dunster, N.; Oliynyk, M.; Mccormick, E.; Mcarthur, H.; Mendez, C.; Salas, J.A.; et al. Engineering specificity of starter unit selection by the erythromycin-producing polyketide synthase. Mol. Microbiol. 2002, 43, 1215–1225. [Google Scholar] [CrossRef]
- Paulick, R.C.; Casey, M.L.; Whitlock, H.W. A carbon-13 nuclear magnetic resonance study of the biosynthesis of daunomycin from sodium acetate (carbon-13). J. Am. Chem. Soc. 1976, 98, 3370–3371. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.J.; Milne, G.W.A.; Minghetti, A. Propionate precursors in the biosynthesis of daunomycin and adriamycin: A 13C nuclear magnetic resonance study. Phytochemistry 1979, 18, 178–179. [Google Scholar] [CrossRef]
- Waldman, A.J.; Balskus, E.P. Lomaiviticin Biosynthesis Employs a New Strategy for Starter Unit Generation. Org. Lett. 2014, 16, 640–643. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Williams, D.J. Oxytetracycline biosynthesis: Origin of the carboxamide substituent. J. Chem. Soc. Chem. Commun. 1983, 677. [Google Scholar] [CrossRef]
- Wang, P.; Gao, X.; Chooi, Y.-H.; Deng, Z.; Tang, Y. Genetic characterization of enzymes involved in the priming steps of oxytetracycline biosynthesis in Streptomyces rimosus. Microbiology 2011, 157, 2401–2409. [Google Scholar] [CrossRef]
- Seto, H.; Sato, T.; Urano, S.; Uzawa, J.; Yonehara, H. Utilization of13C-13C coupling in structural and biosynthetic studies. VII1 the structure and biosynthesis of vulgamycin. Tetrahedron Lett. 1976, 17, 4367–4370. [Google Scholar] [CrossRef]
- Pickens, L.B.; Tang, Y. Oxytetracycline Biosynthesis. J. Biol. Chem. 2010, 285, 27509–27515. [Google Scholar] [CrossRef]
- Ames, B.D.; Korman, T.P.; Zhang, W.; Smith, P.; Vu, T.; Tang, Y.; Tsai, S.-C. Crystal structure and functional analysis of tetracenomycin ARO/CYC: Implications for cyclization specificity of aromatic polyketides. Proc. Natl. Acad. Sci. USA 2008, 105, 5349–5354. [Google Scholar] [CrossRef]
- Summers, R.G.; Wendt-Pienkowski, E.; Motamedi, H.; Hutchinson, C.R. Nucleotide sequence of the tcmII-tcmIV region of the tetracenomycin C biosynthetic gene cluster of Streptomyces glaucescens and evidence that the tcmN gene encodes a multifunctional cyclase-dehydratase-O-methyl transferase. J. Bacteriol. 1992, 174, 1810–1820. [Google Scholar] [CrossRef][Green Version]
- Meurer, G.; Gerlitz, M.; Wendt-Pienkowski, E.; Vining, L.C.; Rohr, J.; Richard Hutchinson, C. Iterative type II polyketide synthases, cyclases and ketoreductases exhibit context-dependent behavior in the biosynthesis of linear and angular decapolyketides. Chem. Biol. 1997, 4, 433–443. [Google Scholar] [CrossRef][Green Version]
- Thompson, T.B.; Katayama, K.; Watanabe, K.; Hutchinson, C.R.; Rayment, I. Structural and Functional Analysis of Tetracenomycin F2 Cyclase from Streptomyces glaucescens: A TYPE II POLYKETIDE CYCLASE. J. Biol. Chem. 2004, 279, 37956–37963. [Google Scholar] [CrossRef]
- Zhang, W.; Ames Brian, D.; Tsai, S.-C.; Tang, Y. Engineered Biosynthesis of a Novel Amidated Polyketide, Using the Malonamyl-Specific Initiation Module from the Oxytetracycline Polyketide Synthase. Appl. Environ. Microbiol. 2006, 72, 2573–2580. [Google Scholar] [CrossRef]
- Zhang, W.; Watanabe, K.; Wang, C.C.C.; Tang, Y. Investigation of Early Tailoring Reactions in the Oxytetracycline Biosynthetic Pathway. J. Biol. Chem. 2007, 282, 25717–25725. [Google Scholar] [CrossRef]
- Xu, W.; Raetz, L.B.; Wang, P.; Tang, Y. An ATP-dependent ligase catalyzes the fourth ring cyclization in tetracycline biosynthesis. Tetrahedron 2016, 72, 3599–3604. [Google Scholar] [CrossRef]
- Grammbitter, G.L.C.; Schmalhofer, M.; Karimi, K.; Shi, Y.-M.; Schöner, T.A.; Tobias, N.J.; Morgner, N.; Groll, M.; Bode, H.B. An Uncommon Type II PKS Catalyzes Biosynthesis of Aryl Polyene Pigments. J. Am. Chem. Soc. 2019, 141, 16615–16623. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Katsuyama, Y.; Onaka, H.; Fujie, M.; Satoh, N.; Shin-Ya, K.; Ohnishi, Y. Production of a Novel Amide-Containing Polyene by Activating a Cryptic Biosynthetic Gene Cluster in Streptomyces sp. MSC090213JE08. ChemBioChem 2016, 17, 1464–1471. [Google Scholar] [CrossRef] [PubMed]
- Bracegirdle, J.; Hou, P.; Nowak, V.V.; Ackerley, D.F.; Keyzers, R.A.; Owen, J.G. Skyllamycins D and E, Non-Ribosomal Cyclic Depsipeptides from Lichen-Sourced Streptomyces anulatus. J. Nat. Prod. 2021, 84, 2536–2543. [Google Scholar] [CrossRef] [PubMed]
- Bilyk, O.; Brötz, E.; Tokovenko, B.; Bechthold, A.; Paululat, T.; Luzhetskyy, A. New Simocyclinones: Surprising Evolutionary and Biosynthetic Insights. ACS Chem. Biol. 2016, 11, 241–250. [Google Scholar] [CrossRef]
- Petříčková, K.; Pospíšil, S.; Kuzma, M.; Tylová, T.; Jágr, M.; Tomek, P.; Chroňáková, A.; Brabcová, E.; Anděra, L.; Krištůfek, V.; et al. Biosynthesis of Colabomycin E, a New Manumycin-Family Metabolite, Involves an Unusual Chain-Length Factor. ChemBioChem 2014, 15, 1334–1345. [Google Scholar] [CrossRef] [PubMed]
- Rui, Z.; Petříčková, K.; Škanta, F.; Pospíšil, S.; Yang, Y.; Chen, C.-Y.; Tsai, S.-F.; Floss, H.G.; Petříček, M.; Yu, T.-W. Biochemical and Genetic Insights into Asukamycin Biosynthesis. J. Biol. Chem. 2010, 285, 24915–24924. [Google Scholar] [CrossRef]
- Zhang, J.; Yuzawa, S.; Thong, W.L.; Shinada, T.; Nishiyama, M.; Kuzuyama, T. Reconstitution of a Highly Reducing Type II PKS System Reveals 6π-Electrocyclization Is Required for o-Dialkylbenzene Biosynthesis. J. Am. Chem. Soc. 2021, 143, 2962–2969. [Google Scholar] [CrossRef]
- Hibi, G.; Shiraishi, T.; Umemura, T.; Nemoto, K.; Ogura, Y.; Nishiyama, M.; Kuzuyama, T. Discovery of type II polyketide synthase-like enzymes for the biosynthesis of cispentacin. Nat. Commun. 2023, 14, 8065. [Google Scholar] [CrossRef]
- Du, D.; Katsuyama, Y.; Horiuchi, M.; Fushinobu, S.; Chen, A.; Davis, T.D.; Burkart, M.D.; Ohnishi, Y. Structural basis for selectivity in a highly reducing type II polyketide synthase. Nat. Chem. Biol. 2020, 16, 776–782. [Google Scholar] [CrossRef]
- Schwartz, T.J.; Shanks, B.H.; Dumesic, J.A. Coupling chemical and biological catalysis: A flexible paradigm for producing biobased chemicals. Curr. Opin. Biotechnol. 2016, 38, 54–62. [Google Scholar] [CrossRef]
- Lowell, A.N.; Demars, M.D.; Slocum, S.T.; Yu, F.; Anand, K.; Chemler, J.A.; Korakavi, N.; Priessnitz, J.K.; Park, S.R.; Koch, A.A.; et al. Chemoenzymatic Total Synthesis and Structural Diversification of Tylactone-Based Macrolide Antibiotics through Late-Stage Polyketide Assembly, Tailoring, and C—H Functionalization. J. Am. Chem. Soc. 2017, 139, 7913–7920. [Google Scholar] [CrossRef]
- Cheon, S.; Kim, H.M.; Gustavsson, M.; Lee, S.Y. Recent trends in metabolic engineering of microorganisms for the production of advanced biofuels. Curr. Opin. Chem. Biol. 2016, 35, 10–21. [Google Scholar] [CrossRef]
- Dekishima, Y.; Lan, E.I.; Shen, C.R.; Cho, K.M.; Liao, J.C. Extending Carbon Chain Length of 1-Butanol Pathway for 1-Hexanol Synthesis from Glucose by Engineered Escherichia coli. J. Am. Chem. Soc. 2011, 133, 11399–11401. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).