Abstract
Methylation is a biochemical process involving the addition of methyl groups to proteins, lipids, and nucleic acids (both DNA and RNA). DNA methylation predominantly occurs on cytosine and adenine nucleobases, and the resulting products—most frequently 5-methylcytosine and N6-methyladenine epigenetic marks—can significantly influence gene activity at the affected genomic sites without modifying the DNA sequence called nucleotide order. Various environmental factors can alter the DNA methylation pattern. Among these, methyl donor micronutrients, such as specific amino acids, choline, and several B vitamins (including folate, pyridoxine, thiamine, riboflavin, niacin, and cobalamin), primarily regulate one-carbon metabolism. This molecular pathway stimulates glutathione synthesis and recycles intracellular methionine. Glutathione plays a pivotal role during oocyte activation by protecting against oxidative stress, whereas methionine is crucial for the production of S-adenosyl-L-methionine, which serves as the universal direct methyl donor for cellular methylation reactions. Because local DNA methylation patterns at genes regulating fertility can be inherited by progeny for multiple generations even in the absence of the original disrupting factors to which the parent was exposed, and DNA methylation levels at specific genomic sites highly correlate with age and can also be passed to offspring, nutrition can influence reproduction and life span in a transgenerational manner.
1. Introduction
“You are what you eat.”—said Ludwig Feuerbach, a German anthropologist and philosopher, in the early 19th century. Similarly, “Tell me what you eat, and I will tell who you are” was stated by Jean Anthelme Brillat-Savarin, a French lawyer and politician, in 1825. These statements clearly emphasize the importance of nutrition, which has a significant impact on both physical and mental health. However, nutrition influences not only our bodies but also several biological traits of our progeny, even when they follow completely different dietary regimens. Accumulating evidence indicates that this impact primarily manifests at the epigenetic level. Epigenetics refers to heritable changes in traits which occur independently of the DNA sequence, with DNA methylation being a key epigenetic mechanism. Genes that regulate physiology, metabolism, fertility, and aging can undergo methylation and demethylation processes that determine their expression (activity). The DNA methylation pattern of these genes in the gamete involved in fertilization can be inherited by progeny for multiple generations. Erasure of the original epigenetic pattern is likely to occur only in the great-grandchildren or great-great-grandchildren. Studies in worms have demonstrated that the removal of the parental epigenetic landscape occurs in the fifth generation. It is now well-established that DNA methylation is strongly influenced by nutrients, primarily dietary methyl donors. This connection suggests that adverse nutritional habits can severely disrupt normal physiology, interfering with fertility and the rate at which cells age in a transgenerational manner. In light of this, we can modify the above quotations as follows: “Our descendants are partly shaped by what we ate and how we lived before conception.”
2. S-Adenosyl-L-Methionine as a Universal Methyl Donor for Biochemical Methylation Reactions
During the methylation process, a methyl group (–CH3) is enzymatically added to an acceptor biomolecule. Such acceptors include lysine and arginine amino acid residues in the case of histone protein methylation [1,2,3] (histones, together with the DNA they pack, constitute the chromatin structure that forms chromosomes within the nucleus of eukaryotic cells), phosphatidylethanolamine in the case of (phospho) lipid methylation [4], and specific nucleobases, cytosine and adenine, in the case of nucleic acid (DNA and RNA) methylation [5]. Each of these enzymatic processes utilizes S-adenosyl-L-methionine (SAM) as a direct methyl donor.
SAM is generated from L-methionine and adenosine triphosphate (ATP), catalyzed by the enzyme methionine adenosyltransferase (Figure 1) [6,7]. This biochemical reaction requires H2O and releases phosphate groups. Methionine is primarily utilized in protein synthesis as an essential amino acid that cannot be synthesized by the human body and must be obtained through the diet. It also provides a sulfur atom for cysteine synthesis and serves as a principal methyl donor for cellular methylation reactions (indicated by red letters and orange highlighting in Figure 1). ATP is a major energy-carrying biomolecule, and functions as an important substrate for many biochemical reactions. When SAM donates a methyl group to an acceptor biomolecule, it is converted into S-adenosyl-L-homocysteine (SAH) by a specific methyltransferase enzyme (Figure 1 and Figure 2). SAH is then hydrolyzed to homocysteine and adenosine, catalyzed by the enzyme 5-methyltetrahydrofolate-homocysteine methyltransferase (Figure 2). Thus, homocysteine is an amino acid derivative formed by methionine demethylation. The conversion of homocysteine to methionine completes the methionine cycle. Methionine intake through diet significantly affects intracellular homocysteine levels [8].
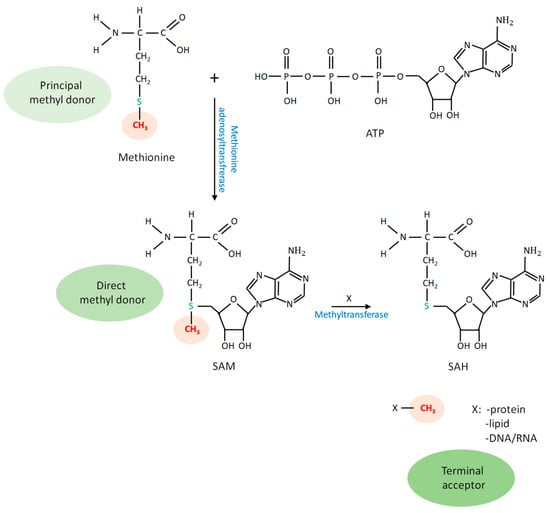
Figure 1.
Methionine as the principal methyl donor for all cellular methylation reactions. The methionine residue contains a methyl group (shown in red letters and orange highlighting) that is covalently bound to a sulfur atom. Methionine is converted into S-adenosyl-L-methionine (SAM) using an adenosine triphosphate (ATP) substrate, and SAM serves as a universal, direct methyl donor for cellular methylation processes. Specific methyltransferase enzymes use SAM as a substrate to transfer a methyl group to an acceptor molecule. As a result, SAM is converted into S-adenosyl-L-homocysteine (SAH), while the methyl group is covalently linked to the acceptor, including DNA and RNA (nucleic acids), (histone) proteins, and phospholipids. SAH is then hydrolyzed to homocysteine (the demethylated form of methionine) and adenosine.
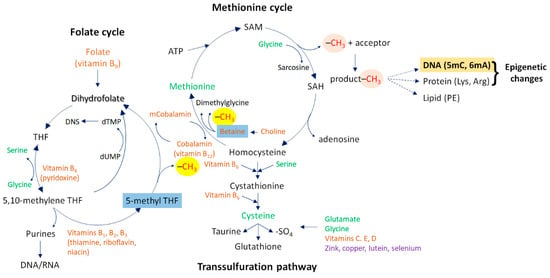
Figure 2.
One-carbon cycle metabolism. This metabolic system consists of three interconnected molecular pathways: the methionine cycle, the transsulfuration pathway, and the folate cycle. During the methionine cycle, methionine donates a methyl group for a methylation reaction. Methionine is first transferred to ATP, thereby generating SAM. This reaction is catalyzed by the enzyme methionine adenosyltransferase. When nucleobases (cytosine or adenine—highlighted in bold) or histone proteins (their lysine or arginine amino acid residues) act as final methyl acceptors, the resulting epigenetic marks (predominantly 5mC and 6mA, as well as mono-, di-, and tri-methylated lysine and arginine amino acids, respectively) can significantly alter gene activity at the affected genomic sites (highlighted in claret and bold). Upon losing the methyl group, SAM is converted to SAH and subsequently to homocysteine, the demethylated form of methionine. The conversion of homocysteine to methionine is mediated by the methyl group donor betaine and the co-factor vitamin B12 (cobalamin) or, alternatively, by the folate cycle (where 5-methyl THF provides the methyl group for the homocysteine-to-methionine conversion step). Betaine and 5-methyl THF are highlighted in blue. The folate cycle also provides essential components (purine and pyrimidine bases) for nucleic acid metabolism. It relies on several B vitamins, including vitamin B9 (folate/folic acid), B6 (pyridoxine), B1 (thiamine), B2 (riboflavin), and B3 (niacin). Alternatively, homocysteine can be converted into cysteine via the transsulfuration pathway, whose end product is glutathione. The methyl group provided by methionine through SAM for a methylation reaction is highlighted in orange, while the methyl group used for methionine synthesis is highlighted in yellow. Abbreviations: ATP, adenosine triphosphate; SAM, S-adenosyl-L-methionine; SAH, S-adenosyl-L-homocysteine; 5mC, 5-methylcytosine; 6mA, N6-methyladenine; –SO4, sulfate group; THF, tetrahydrofolate; dTMP, deoxythymidine monophosphate; dUMP, deoxyuridine monophosphate; PE, phosphatidylethanolamine.
This biochemical cycle (methionine–SAM–SAH–homocysteine–methionine), also termed the methionine demethylation–remethylation pathway, represents a part of one-carbon metabolism (Figure 2) [1]. “One-carbon” refers to the methyl group that participates in the methylation process. In this cycle, the conversion of methionine from homocysteine requires a methyl group, which is donated by betaine, a metabolite of choline (Figure 2). Another component of one-carbon metabolism is the transsulfuration pathway, which is initiated by the conversion of homocysteine into cystathionine by the enzyme cystathionine-β-synthase (Figure 2). Cystathionine is then converted into the non-essential amino acid cysteine in a vitamin B6-dependent manner, and cysteine is further metabolized into taurine, glutathione, or sulfate [9]. Glutathione is an important antioxidant compound capable of preventing cells from the damaging effects of reactive oxygen species (ROS) [10].
The third pathway constituting one-carbon metabolism is the folate cycle. This cycle contributes to the remethylation of homocysteine to methionine in a folate/folic acid- (vitamin B9) and cobalamin- (vitamin B12) dependent manner. Folate is first converted into 5-methyltetrahydrofolate (5-methyl-THF), which ultimately donates a methyl group for homocysteine-to-methionine conversion. The folate cycle also supplies essential components for DNA and RNA metabolism, including purine and pyrimidine nucleobases, as well as for the synthesis of neurotransmitters such as serotonin, dopamine, noradrenaline, and adrenaline (Figure 2).
3. DNA Methylation
The term epigenetics refers to changes in inheritance patterns which are independent of the nucleotide order (alterations in the nucleotide order are called mutations and DNA polymorphisms). Epigenetics involves chromatin modifications, comprising DNA methylation and various chemical modifications of histone proteins, such as mono-, di-, and tri-methylation at specific lysine and arginine residues of histone 2A (H2A), H3, and H4 proteins. These methylation processes rely on the availability of SAM as a direct methyl donor and specific methyltransferase enzymes (Figure 1 and Figure 2). DNA methylation generally occurs at cytosine and adenine nucleobases and is mediated by specific DNA methyltransferases. Cytosine is primarily methylated at the 5C position, generating the 5-methylcytosine (5mC) epigenetic mark (Figure 3A), which represses gene transcription. Adenine is mostly methylated at the N6 position, resulting in N6-methyladenine (6mA), which promotes gene transcription (Figure 3B). Thus, DNA methylation, together with histone modifications, can alter the transcriptional activity of genes located at the affected genomic sites, although the original DNA sequence remains intact in these loci [11].
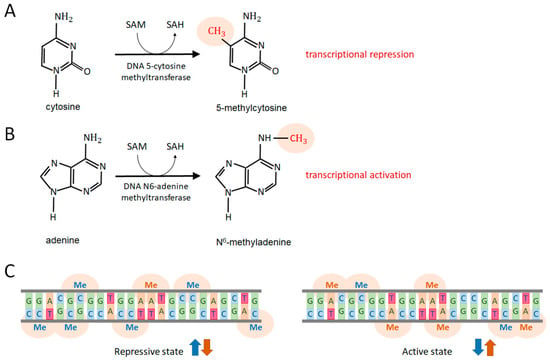
Figure 3.
DNA methylation processes. (A) Cytosine methylation primarily occurs through the addition of a methyl group to a cytosine nucleobase at the 5C position (5mC) by the enzyme DNA 5-cytosine methyltransferase. This methylation reaction utilizes SAM as a methyl donor, converting it into SAH. 5mC represses nearby genes. (B) Adenine methylation occurs through the transfer of a methyl group to an adenine nucleobase at the N6 position (6mA), mediated by the enzyme DNA N6-adenine methyltransferase. 6mA activates the transcription of affected genes. These methyltransferase enzymes are responsible for de novo DNA methylation. Specific demethylases can remove the methyl group from 5mC and 6mA. (C) At a given genomic site, 5mC levels (blue arrows) negatively correlate with 6mA levels (brown arrows); a loss of 5mC is associated with an accumulation of 6mA. In general, 5mC represses gene activity (repressive epigenetic state), whereas 6mA activates gene transcription (activating epigenetic state). The grey lines represent the two glucose-phosphate chains of the DNA region. Nucleobases pair according to Chargaff’s rule (G pairs with C and vice versa, while A pairs with T and vice versa). Both DNA fragments (left and right) have the same nucleotide sequence, but their methylation patterns differ. Abbreviations: 5mC, 5-methylcytosine; 6mA, N6-methyladenine; SAM, S-adenosyl-L-methionine; SAH, S-adenosyl-L-homocysteine; G, guanine; C, cytosine; A, adenine; T, thymine; Me, methyl group.
DNA methylation can be characterized by several key features. First, the process is sufficient to cause significant differences in the global gene expression pattern even between identical (monozygotic) twins, whose primary genetic information (DNA sequence) is nearly indistinguishable. Second, various environmental factors (e.g., cigarette smoke, high temperatures, and dietary methyl donors such as choline and folate) and endogenous factors (e.g., ROS, produced by mitochondrial respiration, and stress hormones) can induce extensive DNA methylation and demethylation changes. Third, altered DNA methylation patterns can be inherited by the progeny of affected parents for several generations, even in the absence of the original disruptive factor to which the parent was subjected. This transgenerational inheritance has been demonstrated to affect offspring for up to four generations in the nematode Caenorhabditis elegans, a tractable genetic model for studying the mechanisms and functions of epigenetic processes [12,13,14]. In this system, the erasure of epigenetic memory occurs only in the fifth generation. Fourth, besides bacteria and plants, DNA N6-adenine methylation is a ubiquitous process among divergent animal taxa, ranging from worms to mammals [15,16,17,18,19,20]. Fifth, DNA methylation predominantly occurs at the sites of transposable elements (TEs), also called mobile genetic elements or “jumping genes”, which are highly repetitive intragenomic parasites causing, when mobilized, significant levels of genomic instability (accumulation of insertional mutations and DNA damage in functional coding and regulatory sequences) at advanced ages. However, other genomic regions can also undergo methylation and demethylation processes but at much lower levels as compared with TEs [16,18,21,22,23]. Sixth, 5-cytosine methylation and N6-adenine methylation are coupled processes as 5mC loss is associated with elevated 6mA levels at the same genomic site, and vice versa (Figure 3C) [24].
4. Nutrition and DNA Methylation
The ultimate methyl donor in methylation reactions is methionine, an essential sulfur-containing amino acid obtained through dietary intake. Methionine can be converted to cysteine, a non-essential sulfur-containing amino acid, and taurine, which is a naturally occurring aminosulfonic acid. It is also utilized in protein synthesis and the production of the crucial antioxidant glutathione. Additionally, methionine serves as the primary precursor for SAM, which in turn serves as the direct methyl donor for cellular methylation reactions. As a result, methionine metabolism (the methionine cycle; Figure 2) and, consequently, DNA and histone methylation are highly dependent on dietary intake. The efficiency of the methionine cycle is primarily influenced by dietary methionine intake. A diet low in methionine (dietary methionine restriction) reduces the rate at which the methionine cycle proceeds, thereby limiting SAM production and the availability of methyl groups for methylation reactions (Figure 2). For example, vegan diets are typically low in methionine, as plant proteins generally contain less methionine compared to animal proteins. This dietary factor has been associated with increased life span [25]. This may be a consequence of lowered 6mA levels at TE sites (6mA promotes TE transcription). Conversely, high methionine intake can lead to locus-specific DNA hypermethylation in certain genomic regions, leading to shortened life span [26].
Beyond methionine, the intracellular concentrations of other amino acids including glycine, serine, and cysteine also influence one-carbon metabolism (Figure 2). The proteogenic amino acid glycine facilitates SAM-to-SAH conversion, serine is essential for cystathionine synthesis, and cysteine and glycine contribute to glutathione production. Additionally, the serine-to-glycine transformation affects folate cycle functionality. Therefore, dietary intake of these amino acids influences the rate of one-carbon metabolism and in turn the rate of DNA and histone methylation. By affecting multiple steps in this metabolic pathway, glycine deficiency or a high-glycine diet can significantly modify DNA methylation patterns at functional genomic sites.
In addition to amino acids, several vitamins play a critical role in regulating one-carbon metabolism and SAM production (Figure 2). Among these essential organic molecules, certain B vitamins are particularly important. For example, vitamin B6 (pyridoxine), along with vitamins C, E, and D, is required for glutathione synthesis from homocysteine. Vitamin B12 (cobalamin) acts as a cofactor for homocysteine-to-methionine conversion, while vitamin B9 (folate/folic acid) is the primary substrate for the folate cycle. Additionally, vitamins B6, B1 (thiamine), B2 (riboflavin), and B3 (niacin) participate in the folate cycle by facilitating the conversion of 5,10-methylene-tetrahydrofolate (THF) to 5-methyl-THF, which serves as a methyl donor substrate for methionine synthesis (Figure 2). Notably, long-term supplementation with folic acid and vitamin B12 significantly affects genome-wide DNA methylation patterns, particularly in elderly individuals [27]. Conversely, inadequate maternal levels of B vitamins can alter DNA methylation patterns in offspring [28]. Consistent with these findings, plasma folate, together with vitamins B6 and B12 levels, influences intracellular homocysteine concentration, pregnancy outcomes, and the incidence of various human pathologies [29,30,31].
Finally, certain micronutrients, such as choline and its derivative betaine, as well as trace elements including zinc, copper, and selenium, also modulate one-carbon metabolism (Figure 2). Among these factors, choline and betaine are particularly important. Choline is essential for the synthesis of the neurotransmitter acetylcholine, thereby playing a role in several neuronal functions, and it also influences gene expression. Additionally, through its derivative betaine, choline contributes to methionine synthesis from homocysteine (Figure 2). Numerous studies have demonstrated that dietary choline deficiency affects both global and locus-specific DNA methylation patterns [32,33,34].
The Dutch Famine, which was a severe food crisis occurring in the Netherlands during the last months of World War II, represents a real-life example of how under-nutrition/starvation affects DNA methylation patterns (epigenome) in a transgenerational manner. This event caused various metabolic and cardiovascular diseases still in the offspring although they were exposed to normal nutritional regimens [35].
5. Epigenetic Regulation of Fertility and Embryo Viability
Fertility and embryo viability, and consequently overall reproduction, are regulated by numerous genetic factors in both sexes (Table 1) [36,37,38,39]. These genes encode proteins with diverse functions, including transcription factors that influence gene activity (e.g., WT1, FOXO1A, FOXO3A, SOX8, TCF7L2, and SOX3); components involved in cellular maintenance processes such as autophagy (cellular self-digestion) and DNA repair (e.g., ATG7, ATG9A, and WDR11, as well as HMF1, BUB1B, MCM8/9, ERCC6, MSH5, REC8, SPIDR, MEI1, TOP6BL, RECIL4, and FANCA); factors essential for development and pattern formation (e.g., NOBOX, BMP15, GDF9, NANOS3, SOHLH1/2, HOXA13, various members of the HOXB protein family, and LHX1); multiple growth hormones and hormone receptors (e.g., AR, FSHR, ESR1/2, LHCGR, INS, INSR, FGF8, FGFR1, FSHB, GNRH1/2, LHB, SHBG, AMH, AMHR2, and FGFR1); signaling components (e.g., WNT4 and CYP19A1); and cell cycle regulatory proteins (e.g., CDKN1B, RB1CC1, RBBP8, and KISS1) (for gene function, see Table 1). These gene products determine the maturation of gametes, the efficiency of fertilization, the implantation and normal development of the embryo, and ultimately the proper function of the offspring’s reproductive system. In women, early dysfunction of the reproductive system, such as premature ovarian insufficiency (POI), can result from the malfunction of multiple genes [40].

Table 1.
DNA methylation at genes implicated in human fertility. Representative, rather than full, references are shown. Gene list largely relies on data published by Refs. [35,36,37,38]. “-” indicates the lack of information rather than evidence for not. The number of fertility-related genes affected by methylation changes is expected to increase as further research is conducted.
Unsurprisingly, decreased expression of genes involved in reproduction [36,37,38,39] has frequently been associated with changes in DNA methylation. Of the 216 genes listed in Table 1, 133 (approximately 62%) have documented evidence linking altered gene activity to changes in DNA methylation patterns, primarily through 5mC demethylation, which predominantly affects regulatory regions of implicated genes [41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173]. For the majority of genes without observed methylation changes, it is possible that these factors have not yet been investigated from this perspective (indicated by “-” in Table 1). Consequently, the number of fertility-related genes affected by methylation changes is expected to increase as further research is conducted.
Infertility is a global and increasingly prevalent health issue in developed societies. Oxidative stress, primarily caused by ROS, affects the entire reproductive life span of both men and women. In women, for example, ROS significantly impact oocyte maturation and ovulation. Glutathione, an antioxidant derived from homocysteine, plays a crucial role in maintaining female fertility [174]. The availability of glutathione ultimately depends on the activity of the transsulfuration pathway, which is regulated by the methylation cycle. Consequently, reproduction (including fertility and embryo viability) is linked to two distinct, methylation cycle-dependent mechanisms: first, through the methylation of genes involved in reproduction (Table 1), and, second, via the transsulfuration pathway associated with the cycle, which produces the antioxidant glutathione (Figure 2).
6. Nutrition and Reproduction
Human reproduction, fertility, and the development of healthy offspring are closely linked to dietary habits and micronutrient intake, particularly through epigenetic mechanisms. Proper nutrition plays a dual role in optimizing fertility and maintaining epigenetic stability. Choline, several B vitamins—including B1 (thiamine), B2 (riboflavin), B3 (niacin), B6 (pyridoxine), B9 (folate), and B12 (cobalamin)—and methionine are critical components of one-carbon metabolism, a pathway essential for DNA methylation of genes implicated in fertility (Figure 1 and Figure 2, Table 1) [36,37,38,39,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173].
Vitamin B9, derived from leafy greens and fruits, contributes to the production of methionine from homocysteine, which serves as the ultimate source of methyl groups for DNA methylation processes. Its deficiency can lead to hypomethylation and epigenetic instability, thereby increasing risk of several diseases. Folate absorption differs between natural sources (~50% bioavailability) and synthetic folic acid (~85% bioavailability). Pregnant women require 600 µg of folate per day to support normal embryonic development and reduce the risk of neural tube defects. Choline, which is converted to betaine and also influences methionine homeostasis (Figure 2), supports neurogenesis and plays a role in phospholipid synthesis, lipid metabolism, and acetylcholine production. The recommended daily intake is 400 mg of choline for adults and 480 mg for pregnant women [174,175,176,177,178,179,180,181].
Vitamins B6 and B12 act as essential cofactors in the folate cycle, thereby facilitating DNA methylation processes (Figure 2). These nutrients directly influence fertility. Omega-3 fatty acids improve oocyte and sperm quality by reducing oxidative stress, while antioxidants (such as vitamins A, C, and E) protect DNA and enhance oocyte quality by generating glutathione (Figure 2). Personalized nutrition, tailored to genetic and sex-specific needs, may optimize reproductive health [182].
Both obesity and underweight can disrupt endocrine homeostasis and impair ovulatory function, thereby adversely affecting fertility. Extremes in body mass index (BMI)—particularly values above 25 or below 20—have been associated with subfertility in both sexes [183]. Nutritional interventions, whether aimed at weight loss or healthy weight gain, should be guided by qualified healthcare professionals.
In addition to impairing fertility, obesity induces epigenetic alterations—most notably changes in DNA methylation—in metabolically active tissues, such as adipose tissue, skeletal muscle, and blood. These modifications affect gene expression and may contribute to the development of obesity. Specifically, altered methylation patterns in genes such as HIF3A, CPT1A and ABCG1 have been associated with increased BMI and waist circumference, implicating DNA methylation in the regulation of metabolic pathways [183].
Maternal obesity and excessive gestational weight gain have also been linked to an increased risk of obesity and type 2 diabetes (T2D) in offspring. Epigenetic mechanisms—including aberrant DNA methylation and histone modifications—appear to mediate these intergenerational effects by altering fetal gene expression and metabolic programming. For example, changes in DNA methylation can impair the development and function of pancreatic β-cells and reduce insulin sensitivity, thereby contributing to T2D susceptibility in later life [184].
Maternal obesity can thus epigenetically reprogram key biological systems involved in energy balance, appetite regulation, and inflammatory responses in the offspring. Persistent alterations, such as reduced leptin sensitivity in the hypothalamus, may result in long-term disruptions to energy homeostasis. Moreover, impaired β-cell development may compromise insulin production and glucose regulation. Importantly, if these epigenetic modifications occur in the germ-line, they may be transmitted to subsequent generations. This transgenerational epigenetic inheritance implies that the metabolic consequences of maternal obesity may extend beyond the immediate offspring, influencing disease risk in descendants across multiple generations [185,186].
The Western diet, characterized by high fat and sugar content, processed meats, and sugary beverages, adversely affects reproductive health [187]. In women, it is associated with polycystic ovary syndrome (PCOS) and lower progesterone levels, while in men, it results in decreased sperm concentration and motility. Conversely, the Mediterranean diet—rich in olive oil, vegetables, fruits, nuts, and legumes, with moderate fish intake and low sweets consumption—positively impacts fertility, presumably by maintaining epigenetic balance, which is essential for the optimal expression of genetic factors implicated in male and female fertility (Table 1) [41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173]. High adherence to this diet reduces conception difficulties and improves in vitro fertilization (IVF) success by enhancing embryo quality. In men, it improves sperm parameters, particularly motility and concentration, due to its antioxidant and essential fatty acid content, which reduce oxidative stress [187,188].
The widespread adoption of unhealthy dietary patterns and neglect of seasonality distort macro- and micro-nutrient intake, negatively affecting male and female fertility. Deficiencies in folate, vitamin B12, and iron impair female reproductive function, while supplementation can improve fertility outcomes by maintaining epigenetic homeostasis (DNA methylation, predominantly 5-cytosine methylation). Both obesity and low body weight cause hormonal imbalances and ovulatory dysfunction, with extreme BMI values (<20 or >25) linked to fertility issues in both sexes. Folate and B12 supplementation support DNA methylation (Figure 2), cell division, and reproductive function. Optimal nutrition should prioritize nutrient-dense natural foods, such as vegetables, healthy fats, and high-quality proteins while avoiding processed foods and refined sugars. Dietary and lifestyle modifications significantly contribute to fertility and reproductive health [183,184,185,186,187,188,189].
Because N6-adenine methylation predominantly occurs in TE loci, genes involved in fertility control and regulated epigenetically predominantly underlay 5-cytosine methylation (Table 1). This implies that dietary methyl donors promote 5-cytosine methylation at these sites, thereby repressing the transcriptional activity of these genetic factors.
7. DNA Methylation and Aging
Aging is driven by the lifelong, progressive accumulation of damaged and superfluous macromolecules and organelles (collectively called cellular damage) that can interfere with cellular processes [190]. As a consequence, affected cells undergo a functional decline over time (senescence) and, eventually, die. Massive levels of cell death can then lead to the development of various age-associated diseases, such as cancer, neurodegeneration, diabetes, tissue atrophy, fibrosis, and immune deficiency. Ultimately, these diseases contribute to organismal death. The regulation of the aging process (that is, factors that influence the rate at which cells age) has been the subject of intense research for several decades [191,192,193]. Diverse regulatory proteins (e.g., the kinase target of rapamycin (TOR kinase), forkhead box A (FOXA1), p53, heat shock factor-1 (HSF1), and sirtuin 1 (Sirt1)), signaling pathways (e.g., insulin/insulin-like growth factor (IGF1), receptor tyrosine kinase (RTK)/Ras/mitogen-activated protein kinase (MAPK), and transforming growth factor (TGF) beta signaling), metabolic systems (e.g., the translation machinery and mitochondrial respiratory chain), cellular processes (e.g., autophagy), and germ-line activity have been implicated in life-span determination across evolutionarily divergent eukaryotic organisms, ranging from yeast to mammals [194,195,196,197,198,199]. However, the primary genetic mechanisms underlying aging remain unresolved [191].
Accumulating evidence indicates that the activity of TEs (“jumping genes”) is predominantly responsible for genome instability (the accumulation of mutations and DNA damage), a hallmark of essentially all aging cells [18,200,201]. Notably, non-aging cells (e.g., germ-line and cancer stem cells, which maintain unlimited proliferation capacity—a phenomenon known as replicative immortality), but not aging cells (somatic cells that become terminally differentiated, undergo senescence, and eventually die), exclusively exhibit the activity of the Piwi-piRNA pathway (P element-induced wimpy testis in Drosophila—Piwi-interacting RNA), whose fundamental biological function is to inhibit TE activity. TEs are repetitive (as they exist in high copy numbers within the genome) genetic factors, and function as intragenomic parasites of retroviral origin, capable of relocating to new genomic sites. This mobility causes insertional mutations and DNA damage, primarily in the form of double-strand DNA breaks, in functional regulatory and coding sequences. In early adulthood, somatic cells exhibit negligible levels of TE activity. However, as the organism ages, TEs become progressively mobilized and increasingly mutagenic. Recent genetic data have shown that age-related TE mobilization is coupled with changes in a specific epigenetic process, DNA methylation [20,22,23].
In mammalian genomes, most of the 5mC epigenetic marks are located in TE loci [21]. A recent analysis of human blood cells demonstrated that 5mC demethylation (the loss of methylation at 5mC sites) within the genomic loci of certain TE families gradually increases with age (Figure 4) [22,23]. This phenomenon has been applied to improve the so-called epigenetic (DNA methylation) clock that reflects 5mC levels at multiple genomic loci in the context of chronological ages. Conversely, in C. elegans (worms) and the fruit fly Drosophila melanogaster (insects), 6mA levels at active TE sequences progressively rise during adulthood [16,20]. The higher is the average 6mA level in the loci of a given TE family (as determined in numerous individual genomes obtained from tissue samples), the older is the organism. Thus, DNA methylation levels at specific genomic sites serve as accurate markers for determining biological age (the DNA methylation clock). It is possible that similar age-associated changes in 6mA levels also exist in the human genome [23].
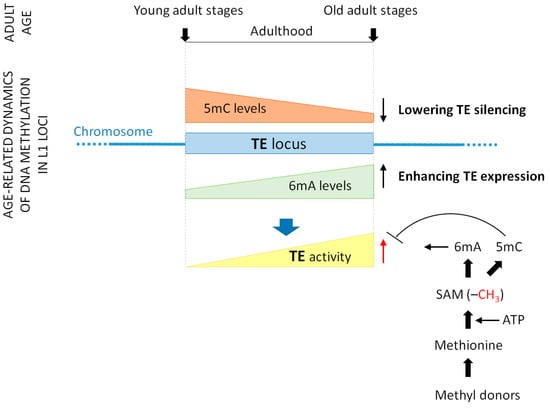
Figure 4.
DNA methylation changes at transposable element sites during aging. Transposable elements (TEs), which are repetitive DNA sequences capable of relocating to new genomic sites, play a pivotal role in the genetic mechanisms underlying aging. Their mutagenic activity and ability to induce DNA damage appear to be primary contributors to genome instability, a hallmark of nearly all aging cells. During aging, TEs become progressively more mobile, and this increasing activity is regulated by specific DNA methylation (epigenetic) processes. 5-cytosine methylation represses TE transcription, while N6-adenine methylation promotes it. As aging progresses, 5-methylcytosine (5mC) levels gradually decrease (represented by the brown triangle), whereas N6-methyladenine (6mA) levels steadily increase (represented by the green triangle) at TE sites, leading to decreased repression and increased expression of TEs. As a cumulative result, TE activity increases over time (the yellow triangle). N6-adenine methylation relies on the availability of SAM, the direct methyl donor. Methyl donor nutrients, such as vitamins B2, B6, B9, B12, and choline, as well as betaine and amino acid methionine, elevate intracellular concentrations of methionine and SAM. Dietary methyl donors promote N6-adenine (hyper) methylation, but inhibit 5mC demethylation. Because N6-adenine methylation is an active process, nutrition may influence it more significantly than 5mC demethylation. For simplicity, a single TE locus is illustrated (represented by the blue rectangle). Abbreviations: TE, transposable element; 5mC, 5-methylcytosine; 6mA, N6-methyladenine; SAM, S-adenosyl-L-methionine; ATP, adenosine triphosphate; G, guanine; C, cytosine; A, adenine; T, thymine; Me, methyl group.
These findings suggest that TE mobility is largely regulated by DNA methylation and demethylation processes. Lowering 5mC levels (that is, 5mC demethylation) in TE loci decreases their repression (inactivity), whereas increasing 6mA levels at these sites elevates their expression (activity). Consequently, TE sequences become increasingly mobile throughout adulthood (Figure 4). TE activity has been shown to be sensitive to exogenous factors (e.g., temperature, oxygen levels, and food availability) and endogenous factors (e.g., regulatory proteins, signal transduction systems, and ROS) that influence life span. Collectively, these findings indicate that TEs may play a fundamental role in the genetic mechanisms underlying aging [201], with their activity being regulated by DNA methylation and demethylation processes.
8. Reproductive Aging
Aging significantly affects reproductive capacity in both males and females [202,203]. The female reproductive system ages at a faster rate compared to other organ systems. Female fertility peaks in the third decade of life but declines drastically by the late thirties and forties. This decline is primarily attributed to the deterioration in both the quality and quantity of oocytes, driven by a reduction in ovarian follicular reserve, increased chromosomal abnormalities, and cumulative environmental damage.
In males, aging leads to a decline in both sperm production and sperm quality. Increased oxidative stress damages sperm DNA, inducing mutations that elevate the risk of spontaneous abortion and disease in offspring. A study involving 2.678 men reported a significant reduction in semen volume (OR: 2.2), sperm concentration (OR: 2.09), and motility (OR: 11.91) in individuals over 50 years of age, along with an increased risk of DNA fragmentation (OR: 4.58). Antioxidant therapies have been shown to improve sperm quality, while decreasing DNA fragmentation not only improves spontaneous fertility, but also has a positive effect on the success of assisted reproductive treatments.
These age-dependent changes may be linked to alterations in the methylation levels of TE sites, leading to increased TE activity, which in turn can be significantly influenced by nutritional factors. In oocytes, the re-establishment of cytosine methylation (5mC) occurs only upon ovulation and fertilization, making maternal age a critical determinant of the genetic stability of offspring [200]. In contrast, TE activity in sperm-producing cells and spermatozoa remains largely repressed, with only moderate activation via passive demethylation. Consequently, while male fertility persists into advanced age, it exhibits a gradual decline over time.
9. Nutrition and Aging
It is well established that dietary energy intake significantly influences life span. Calorie restriction and intermittent fasting have been demonstrated to extend life span in various animal phyla including humans [204]. The molecular mechanisms underlying the influence of calorie intake on the aging process, commonly referred to as the nutritional signaling axis, have been extensively studied. These mechanisms involve TOR kinase, the autophagic-lysosomal degradation system, insulin/IGF1 signaling, and specific sirtuin proteins [191,192,193,194,195,205,206,207].
Beyond calorie intake, the quality of nutrition, particularly the intake of essential micronutrients, antioxidants, and amino acids, also plays a crucial role in determining life span. As previously discussed, genomic instability, a major driver of aging, is primarily caused by the increasing mobilization of TEs over the life span, which exert a mutagenic effect and are regulated through DNA methylation. The epigenetic marker 5mC inhibits TE mobilization, whereas the 6mA marker promotes it (Figure 4). Given that 5mC demethylation along TE sequences appears to be a passive process [22], modifications in 6mA levels seem to represent more specific epigenetic regulatory mechanisms for TE activity (Figure 4). In C. elegans, 6mA signals are predominantly associated with TE loci, and age-dependent fluctuations in 6mA levels are almost exclusively detected at TE sites, with no such modifications observed at TE-independent loci [20].
Consequently, the one-carbon metabolic cycle, which influences DNA methylation, plays a pivotal role in shaping the global 6mA pattern of the genome, thereby regulating TE activity. This regulation ultimately determines the rate of aging and, consequently, life span. Nutritional sources of methyl donors that increase intracellular SAM concentrations (Figure 2) promote 6mA formation and, in turn, accelerate the aging process [208]. Overconsumption of certain B vitamins may induce metabolic disorders and degenerative diseases through this molecular pathway [209].
10. Transgenerational Effects of Nutrition on Reproduction and Aging
In C. elegans, phenotypic effects resulting from modifications in epigenetic patterns (including global DNA methylation levels) can be inherited up to four generations, even in the absence of the original upsetting factors by which the parental generation was influenced [12,13,14]. The reversal of these altered epigenetic patterns typically occurs only in the fifth generation, though it remains unclear whether this process is complete or partial. A similar phenomenon—transgenerational inheritance, whereby epigenetic patterns persist across generations through germ-line transmission—may also exist in humans [210,211,212]. In humans, epigenetic disruptors can include genetic mutations as well as various endogenous and environmental factors, such as endocrine imbalances, high-fat diets, obesity, diabetes, undernutrition, and trauma/shock.
Methyl donor micronutrients directly influence intracellular SAM levels, which, in turn, affect the rate of DNA methylation (Figure 1 and Figure 2). Most genes involved in fertility (Table 1), as well as active TE loci associated with genomic instability (characterized by mutation accumulation and DNA damage contributing to aging), are regulated by DNA methylation and demethylation. If an altered local DNA methylation pattern emerges prior to reproduction, it can be transmitted to subsequent generations. Thus, dietary habits have the potential to significantly impact reproductive outcomes and life span across multiple generations, though the magnitude and persistence of these effects in humans, compared to model organisms like C. elegans, remain an area of ongoing research [211]. This transgenerational effect can occur even if the descendants adhere to optimal dietary practices. Foods rich in methionine—such as egg powder, soybeans, parmesan cheese, caviar, and mackerel—or those with low methionine content—such as eggplant, bananas, and onions—can therefore transgenerationally influence reproduction and aging through epigenetic inheritance.
11. Conclusions
Nutrition plays a fundamental role in maintaining the balance of DNA and histone protein methylation processes. Several amino acids—most notably methionine, the primary methyl group donor—along with essential vitamins (predominantly B9 and B6) and other micronutrients (primarily choline) contribute to the availability of methionine, the key substrate for cellular methylation reactions. Disruptions in one-carbon cycle metabolism, and consequently in DNA and histone methylation, can lead to gene inactivation or hyperactivation, affecting gene function and regulatory networks. Reproduction and aging are biological processes that are highly influenced by DNA methylation, as the majority of genes involved in these processes undergo epigenetic modifications throughout the life cycle. Consequently, diet has a profound impact on reproductive quality and the rate of aging. Notably, nutritional influences on epigenetic patterns can persist across multiple generations (transgenerational inheritance). Thus, dietary choices not only shape an individual’s reproductive capacity and life span but also have long-term consequences for offspring, potentially affecting fertility and longevity from children to (great-)great-grandchildren.
Author Contributions
Conceptualization, F.C.D. and T.V.; methodology, F.C.D., T.V. and M.S.; formal analysis, F.C.D., T.V. and M.S.; data curation, F.C.D. and T.V.; writing—original draft preparation, F.C.D., T.V. and M.S.; writing—review and editing, F.C.D., T.V. and M.S.; visualization, F.C.D. and T.V.; supervision, T.V. and M.S.; funding acquisition, T.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was founded by the grants OTKA (Hungarian Scientific Research Fund; K132439) and GYORSÍTÓSÁV (2023-1.1.2-GYORSÍTÓSÁV-2024-00005) to T.V.
Acknowledgments
T.V. was supported by HUN-REN-ELTE (Hungarian Research Network-Eötvös Loránd University) Genetics Research Group (01062). T.V. is particularly grateful to Annamária Hammer for providing inspiration for the subject (one-carbon metabolism and DNA methylation).
Conflicts of Interest
Authors F.C.D. and T.V. are employed by the companies Rejuven-Eat Ltd. (Siófok, Hungary) and Vellab Biotech Ltd. (Szeged, Hungary), respectively. Authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Mentch, S.J.; Locasale, J.W. One-carbon metabolism and epigenetics: Understanding the specificity. Ann. N. Y. Acad. Sci. 2016, 1363, 91–98. [Google Scholar] [CrossRef]
- Friso, S.; Udali, S.; De Santis, D.; Choi, S.W. One-carbon metabolism and epigenetics. Mol. Asp. Med. 2017, 54, 28–36. [Google Scholar] [CrossRef]
- Creegan, R.; Hunt, W.; McManus, A.; Rainey-Smith, S.R. Diet, nutrients and metabolism: Cogs in the wheel driving Alzheimer’s disease pathology? Br. J. Nutr. 2015, 113, 1499–1517. [Google Scholar] [CrossRef]
- Smith, Z.D.; Hetzel, S.; Meissner, A. DNA methylation in mammalian development and disease. Nat. Rev. Genet. 2025, 26, 7–30. [Google Scholar] [CrossRef]
- Anderson, O.S.; Sant, K.E.; Dolinoy, D.C. Nutrition and epigenetics: An interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J. Nutr. Biochem. 2012, 23, 853–859. [Google Scholar] [CrossRef]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef]
- Heijer, M.D.; Graafsma, S.; Lee, S.Y.; van Landeghem, B.; Kluijtmans, L.; Verhoef, P.; Beaty, T.H.; Blom, H. Homocysteine levels—Before and after methionine loading—In 51 Dutch families. Eur. J. Hum. Genet. 2005, 13, 753–762. [Google Scholar] [CrossRef][Green Version]
- Pasquale, L.R.; Borrás, T.; Fingert, J.H.; Wiggs, J.L.; Ritch, R. Exfoliation syndrome: Assembling the puzzle pieces. Acta Ophthalmol. 2016, 94, e505–e512. [Google Scholar] [CrossRef]
- Pompella, A.; Visvikis, A.; Paolicchi, A.; De Tata, V.; Casini, A.F. The changing faces of glutathione, a cellular protagonist. Biochem. Pharmacol. 2003, 66, 1499–1503. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Greer, E.L.; Maures, T.J.; Ucar, D.; Hauswirth, A.G.; Mancini, E.; Lim, J.P.; Benayoun, B.A.; Shi, Y.; Brunet, A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 2011, 479, 365–371. [Google Scholar] [CrossRef]
- Villalobos, T.V.; Ghosh, B.; DeLeo, K.R.; Alam, S.; Ricaurte-Perez, C.; Wang, A.; Mercola, B.M.; Butsch, T.J.; Ramos, C.D.; Das, S.; et al. Tubular lysosome induction couples animal starvation to healthy aging. Nat. Aging 2023, 3, 1091–1106. [Google Scholar] [CrossRef]
- Sigmond, T.; Vellai, T. Lysosomal alteration links food limitation to longevity. Nat. Aging 2023, 3, 1048–1050. [Google Scholar] [CrossRef]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizábal-Corrales, D.; Hsu, C.-H.; Aravind, L.; He, C.; Shi, Y. DNA Methylation on N6-Adenine in C. elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef]
- Zhang, G.; Huang, H.; Liu, D.; Cheng, Y.; Liu, X.; Zhang, W.; Yin, R.; Zhang, D.; Zhang, P.; Liu, J.; et al. N6-methyladenine DNA modification in Drosophila. Cell 2015, 161, 893–906. [Google Scholar] [CrossRef]
- Wu, T.P.; Wang, T.; Seetin, M.G.; Lai, Y.; Zhu, S.; Lin, K.; Liu, Y.; Byrum, S.D.; Mackintosh, S.G.; Zhong, M.; et al. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature 2016, 532, 329–333. [Google Scholar] [CrossRef]
- Sturm, Á.; Saskői, É.; Hotzi, B.; Tarnóci, A.; Barna, J.; Bodnár, F.; Sharma, H.; Kovács, T.; Ari, E.; Weinhardt, N.; et al. Downregulation of transposable elements extends lifespan in Caenorhabditis elegans. Nat. Commun. 2023, 14, 5278. [Google Scholar] [CrossRef]
- Hao, Z.; Wu, T.; Cui, X.; Zhu, P.; Tan, C.; Dou, X.; Hsu, K.-W.; Lin, X.-T.; Peng, P.-H.; Zhang, L.-S.; et al. N6-Deoxyadenosine Methylation in Mammalian Mitochondrial DNA. Mol. Cell 2020, 78, 382–395.e8. [Google Scholar] [CrossRef]
- Sturm, Á.; Sharma, H.; Bodnár, F.; Aslam, M.; Kovács, T.; Németh, Á.; Hotzi, B.; Billes, V.; Sigmond, T.; Tátrai, K.; et al. N6-Methyladenine Progressively Accumulates in Mitochondrial DNA during Aging. Int. J. Mol. Sci. 2023, 24, 14858. [Google Scholar] [CrossRef]
- Yoder, J.A.; Walsh, C.P.; Bestor, T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997, 13, 335–340. [Google Scholar] [CrossRef]
- Morandini, F.; Lu, J.Y.; Rechsteiner, C.; Shadyab, A.H.; Casanova, R.; Snively, B.M.; Seluanov, A.; Gorbunova, V. Transposable element 5mC methylation state of blood cells predicts age and disease. Nat. Aging 2025, 5, 193–204. [Google Scholar] [CrossRef]
- Hotzi, B.; Vellai, T. Transposable element methylation tracks age. Nat. Aging 2025, 5, 179–181. [Google Scholar] [CrossRef]
- Luo, G.-Z.; He, C. DNA N6-methyladenine in metazoans: Functional epigenetic mark or bystander? Nat. Struct. Mol. Biol. 2017, 24, 503–506. [Google Scholar] [CrossRef]
- McCarty, M.F.; Barroso-Aranda, J.; Contreras, F. The low-methionine content of vegan diets may make methionine restriction feasible as a life extension strategy. Med. Hypotheses 2009, 72, 125–128. [Google Scholar] [CrossRef]
- Waterland, R.A. Assessing the effects of high methionine intake on DNA methylation. J. Nutr. 2006, 136 (Suppl. S6), 1706S–1710S. [Google Scholar] [CrossRef]
- Kok, D.E.G.; Dhonukshe-Rutten, R.A.M.; Lute, C.; Heil, S.G.; Uitterlinden, A.G.; van der Velde, N.; van Meurs, J.B.J.; van Schoor, N.M.; Hooiveld, G.J.E.J.; de Groot, L.C.P.G.M.; et al. The effects of long-term daily folic acid and vitamin B12 supplementation on genome-wide DNA methylation in elderly subjects. Clin. Epigenet. 2015, 7, 121. [Google Scholar] [CrossRef]
- McCullough, L.E.; Miller, E.E.; Mendez, M.A.; Murtha, A.P.; Murphy, S.K.; Hoyo, C. Maternal B vitamins: Effects on offspring weight and DNA methylation at genomically imprinted domains. Clin. Epigenet. 2016, 8, 8. [Google Scholar] [CrossRef]
- Tanaka, T.; Scheet, P.; Giusti, B.; Bandinelli, S.; Piras, M.G.; Usala, G.; Lai, S.; Mulas, A.; Corsi, A.M.; Vestrini, A.; et al. Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am. J. Hum. Genet. 2009, 84, 477–482. [Google Scholar] [CrossRef]
- Furness, D.; Fenech, M.; Dekker, G.; Khong, T.Y.; Roberts, C.; Hague, W. Folate, vitamin B12, vitamin B6 and homocysteine: Impact on pregnancy outcome. Matern. Child Nutr. 2013, 9, 155–166. [Google Scholar] [CrossRef]
- Fareed, M.M.; Ullah, S.; Qasmi, M.; Shityakov, S. The Role of Vitamins in DNA Methylation as Dietary Supplements or Neutraceuticals: A Systematic Review. Curr. Mol. Med. 2023, 23, 1012–1027. [Google Scholar] [CrossRef]
- Niculescu, M.D.; Craciunescu, C.N.; Zeisel, S.H. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J. 2006, 20, 43–49. [Google Scholar] [CrossRef]
- Zeisel, S. Choline, Other Methyl-Donors and Epigenetics. Nutrients 2017, 9, 445. [Google Scholar] [CrossRef]
- Locker, J.; Reddy, T.V.; Lombardi, B. DNA methylation and hepatocarcinogenesis in rats fed a choline-devoid diet. Carcinogenesis 1986, 7, 1309–1312. [Google Scholar] [CrossRef]
- Veenendaal, M.V.E.; Painter, R.C.; de Rooij, S.R.; Bossuyt, P.M.M.; van der Post, J.A.M.; Gluckman, P.D.; Hanson, M.A.; Roseboom, T.J. Transgenerational effects of prenatal exposure to the 1944–45 Dutch famine. BJOG 2013, 120, 548–553. [Google Scholar] [CrossRef]
- Yatsenko, S.A.; Rajkovic, A. Genetics of human female infertility. Biol. Reprod. 2019, 101, 549–566. [Google Scholar] [CrossRef]
- Zorrilla, M.; Yatsenko, A.N. The Genetics of Infertility: Current Status of the Field. Curr. Genet. Med. Rep. 2013, 1, 247–260. [Google Scholar] [CrossRef]
- Layman, L.C. Human gene mutations causing infertility. J. Med. Genet. 2002, 39, 153–161. [Google Scholar] [CrossRef]
- Wu, J.; Li, D.; Liu, X.; Li, Q.; He, X.; Wei, J.; Li, X.; Li, M.; Rehman, A.U.; Xia, Y.; et al. IDDB: A comprehensive resource featuring genes, variants and characteristics associated with infertility. Nucleic Acids Res. 2021, 49, D1218–D1224. [Google Scholar] [CrossRef]
- Chon, S.J.; Umair, Z.; Yoon, M.-S. Premature Ovarian Insufficiency: Past, Present, and Future. Front. Cell Dev. Biol. 2021, 9, 672890. [Google Scholar] [CrossRef]
- Tran, S.; Wang, Y.; Lamba, P.; Zhou, X.; Boehm, U.; Bernard, D.J. The CpG island in the murine Foxl2 proximal promoter is differentially methylated in primary and immortalized cells. PLoS ONE 2013, 8, e76642. [Google Scholar] [CrossRef] [PubMed]
- Kober, P.; Rymuza, J.; Baluszek, S.; Maksymowicz, M.; Nyc, A.; Mossakowska, B.J.; Zieliński, G.; Kunicki, J.; Bujko, M. DNA Methylation Pattern in Somatotroph Pituitary Neuroendocrine Tumors. Neuroendocrinology 2024, 114, 51–63. [Google Scholar] [CrossRef]
- Li, X.; Wang, W.; Ding, X. Pan-cancer investigation of psoriasis-related BUB1B gene: Genetical alteration and oncogenic immunology. Sci. Rep. 2023, 13, 6058. [Google Scholar] [CrossRef] [PubMed]
- Almutairi, M.H.; Alrubie, T.M.; Alshareeda, A.T.; Albarakati, N.; Almotiri, A.; Alamri, A.M.; Almutairi, B.O.; Alanazi, M. Differential expression and regulation of ADAD1, DMRTC2, PRSS54, SYCE1, SYCP1, TEX101, TEX48, and TMPRSS12 gene profiles in colon cancer tissues and their in vitro response to epigenetic drugs. PLoS ONE 2024, 19, e0307724. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Pan, Z.; Zhang, Q.; Xie, Z.; Liu, H.; Li, Q. Differential mRNA expression and promoter methylation status of SYCP3 gene in testes of yaks and cattle-yaks. Reprod. Domest. Anim. 2012, 47, 455–462. [Google Scholar] [CrossRef]
- Zhou, Y.; Connor, E.E.; Bickhart, D.M.; Li, C.; Baldwin, R.L.; Schroeder, S.G.; Rosen, B.D.; Yang, L.; Van Tassell, C.P.; Liu, G.E. Comparative whole genome DNA methylation profiling of cattle sperm and somatic tissues reveals striking hypomethylated patterns in sperm. GigaScience 2018, 7, giy039. [Google Scholar] [CrossRef]
- Yu, M.; Wang, H.; Xu, H.; Lv, Y.; Li, Q. High MCM8 expression correlates with unfavorable prognosis and induces immune cell infiltration in hepatocellular carcinoma. Aging 2022, 14, 10027–10049. [Google Scholar] [CrossRef]
- Lu, Z.; Hou, G. Characterization of the function and clinical value of ERCC family genes in lung adenocarcinoma. Front. Oncol. 2024, 14, 1476100. [Google Scholar] [CrossRef]
- Terribas, E.; Bonache, S.; García-Arévalo, M.; Sánchez, J.; Franco, E.; Bassas, L.; Larriba, S. Changes in the expression profile of the meiosis-involved mismatch repair genes in impaired human spermatogenesis. J. Androl. 2010, 31, 346–357. [Google Scholar] [CrossRef]
- Yao, C.; Lu, L.; Ji, Y.; Zhang, Y.; Li, W.; Shi, Y.; Liu, J.; Sun, M.; Xia, F. Hypo-Hydroxymethylation of Nobox is Associated with Ovarian Dysfunction in Rat Offspring Exposed to Prenatal Hypoxia. Reprod. Sci. 2022, 29, 1424–1436. [Google Scholar] [CrossRef]
- Gamal, L.; Noshy, M.M.; Aboul-Naga, A.M.; Sabit, H.; El-Shorbagy, H.M. DNA methylation of GDF-9 and GHR genes as epigenetic regulator of milk production in Egyptian Zaraibi goat. Genes Genom. 2024, 46, 135–148. [Google Scholar] [CrossRef]
- Pan, Z.; Wang, X.; Di, R.; Liu, Q.; Hu, W.; Cao, X.; Guo, X.; He, X.; Lv, S.; Li, F.; et al. A 5-Methylcytosine Site of Growth Differentiation Factor 9 (GDF9) Gene Affects Its Tissue-Specific Expression in Sheep. Animals 2018, 8, 200. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Chao, H.; Chen, B.; Zhang, L.; Li, L.; Sun, X.; Shen, W. DNA methylation of germ-cell-specific basic helix-loop-helix (HLH) transcription factors, Sohlh2 and Figlα during gametogenesis. Mol. Hum. Reprod. 2011, 17, 550–561. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fan, H.; Cui, Z.; Zhang, H.; Mani, S.K.; Diab, A.; Lefrancois, L.; Fares, N.; Merle, P.; Andrisani, O. DNA demethylation induces SALL4 gene re-expression in subgroups of hepatocellular carcinoma associated with Hepatitis B or C virus infection. Oncogene 2017, 36, 2435–2445. [Google Scholar] [CrossRef] [PubMed]
- Li, L.C.; Okino, S.T.; Dahiya, R. DNA methylation in prostate cancer. Biochim. Biophys. Acta BBA Rev. Cancer 2004, 1704, 87–102. [Google Scholar] [CrossRef]
- Griswold, M.D.; Kim, J.S. Site-specific methylation of the promoter alters deoxyribonucleic acid-protein interactions and prevents follicle-stimulating hormone receptor gene transcription. Biol. Reprod. 2001, 64, 602–610. [Google Scholar] [CrossRef][Green Version]
- Li, X.; Liao, J.; Guo, Z. Detection value of FOXO1 gene methylation, blood glucose and lipids in patients with type 2 diabetic kidney disease. Medicine 2022, 101, e31663. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, M.; Hu, D. Identification of Prognostic Immune-Related Genes by Integrating mRNA Expression and Methylation in Lung Adenocarcinoma. Int. J. Genom. 2020, 20, 9548632. [Google Scholar] [CrossRef]
- Spitschak, M.; Vanselow, J. Bovine large luteal cells show increasing de novo DNA methylation of the main ovarian CYP19A1 promoter P2. Gen. Comp. Endocrinol. 2012, 178, 37–45. [Google Scholar] [CrossRef]
- Grelet, S.; Andries, V.; Polette, M.; Gilles, C.; Staes, K.; Martin, A.P.; Kileztky, C.; Terryn, C.; Dalstein, V.; Cheng, C.W.; et al. The human NANOS3 gene contributes to lung tumour invasion by inducing epithelial-mesenchymal transition. J. Pathol. 2015, 237, 25–37. [Google Scholar] [CrossRef]
- Maekawa, R.; Mihara, Y.; Sato, S.; Okada, M.; Tamura, I.; Shinagawa, M.; Shirafuta, Y.; Takagi, H.; Taketani, T.; Tamura, H.; et al. Aberrant DNA methylation suppresses expression of estrogen receptor 1 (ESR1) in ovarian endometrioma. J. Ovarian Res. 2019, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Richards, J.S.; Shimada, M. The Cell Type—Specific Expression of Lhcgr in Mouse Ovarian Cells: Evidence for a DNA-Demethylation—Dependent Mechanism. Endocrinology 2018, 159, 2062–2074. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Lin, Z.; Ye, Z.; Liang, J.; Yu, R.; Wan, Z.; Hou, J. Development of a prognostic model for early-stage gastric cancer-related DNA methylation-driven genes and analysis of immune landscape. Front. Mol. Biosci. 2024, 11, 1455890. [Google Scholar] [CrossRef]
- Yu, J.; Liang, Q.; Wang, J.; Wang, K.; Gao, J.; Zhang, J.; Zeng, Y.; Chiu, P.W.; Ng, E.K.; Sung, J.J. REC8 functions as a tumor suppressor and is epigenetically downregulated in gastric cancer, especially in EBV-positive subtype. Oncogene 2017, 36, 182–193. [Google Scholar] [CrossRef]
- Topham, L.; Gregoire, S.; Kang, H.; Salmon-Divon, M.; Lax, E.; Millecamps, M.; Szyf, M.; Stone, L. The methyl donor S-adenosyl methionine reverses the DNA methylation signature of chronic neuropathic pain in mouse frontal cortex. Pain Rep. 2021, 6, e944. [Google Scholar] [CrossRef]
- Shao, K.; Pu, W.; Zhang, J.; Guo, S.; Qian, F.; Glurich, I.; Jin, Q.; Ma, Y.; Ju, S.; Zhang, Z.; et al. DNA hypermethylation contributes to colorectal cancer metastasis by regulating the binding of CEBPB and TFCP2 to the CPEB1 promoter. Clin. Epigenet. 2021, 13, 89. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.R.; In, Y.H.; Park, J.; Park, T.; Jung, K.H.; Chai, J.C.; Chung, M.K.; Lee, Y.S.; Chai, Y.G. Genome-scale DNA methylation pattern profiling of human bone marrow mesenchymal stem cells in long-term culture. Exp. Mol. Med. 2012, 44, 503–512. [Google Scholar] [CrossRef]
- Zhu, X.; Jiang, P.; Ying, X.; Tang, X.; Deng, Y.; Gao, X.; Yang, X. Pregnancy induced hypertension and umbilical cord blood DNA methylation in newborns: An epigenome-wide DNA methylation study. BMC Pregnancy Childbirth 2024, 24, 433. [Google Scholar] [CrossRef]
- Spindola, L.M.; Santoro, M.L.; Pan, P.M.; Ota, V.K.; Xavier, G.; Carvalho, C.M.; Talarico, F.; Sleiman, P.; March, M.; Pellegrino, R.; et al. Detecting multiple differentially methylated CpG sites and regions related to dimensional psychopathology in youths. Clin. Epigenet. 2019, 11, 146. [Google Scholar] [CrossRef]
- Zhang, J.; Cai, X.; Cui, W.; Wei, Z. Bioinformatics and Experimental Analyses Reveal MAP4K4 as a Potential Marker for Gastric Cancer. Genes 2022, 13, 1786. [Google Scholar] [CrossRef]
- Mijnes, J.; Veeck, J.; Gaisa, N.T.; Burghardt, E.; de Ruijter, T.C.; Gostek, S.; Dahl, E.; Pfister, D.; Schmid, S.C.; Knüchel, R.; et al. Promoter methylation of DNA damage repair (DDR) genes in human tumor entities: RBBP8/CtIP is almost exclusively methylated in bladder cancer. Clin. Epigenet. 2018, 10, 15. [Google Scholar] [CrossRef]
- Vasilyev, S.A.; Skryabin, N.A.; Kashevarova, A.A.; Tolmacheva, E.N.; Savchenko, R.R.; Vasilyeva, O.Y.; Lopatkina, M.E.; Zarubin, A.A.; Fishman, V.S.; Belyaeva, E.O.; et al. Differential DNA Methylation of the IMMP2L Gene in Families with Maternally Inherited 7q31.1 Microdeletions is Associated with Intellectual Disability and Developmental Delay. Cytogenet. Genome Res. 2021, 161, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Liu, G.; Jin, H.; Chen, X.; He, J.; Xiao, J.; Qin, Y.; Mao, Y.; Zhao, L. The Dysregulation of SOX Family Correlates with DNA Methylation and Immune Microenvironment Characteristics to Predict Prognosis in Hepatocellular Carcinoma. Dis. Markers 2022, 2022, 2676114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, X.Q.; Ling, X.Z.; Zhao, X.H.; Zhou, K.Z.; Wang, J.Y.; Zhang, G.X. Prediction of the Effect of Methylation in the Promoter Region of ZP2 Gene on Egg Production in Jinghai Yellow Chickens. Vet. Sci. 2022, 9, 570. [Google Scholar] [CrossRef]
- Breton, C.V.; Salam, M.T.; Gilliland, F.D. Heritability and role for the environment in DNA methylation in AXL receptor tyrosine kinase. Epigenetics 2011, 6, 895–898. [Google Scholar] [CrossRef]
- Mudduluru, G.; Allgayer, H. The human receptor tyrosine kinase Axl gene-promoter characterization and regulation of constitutive expression by Sp1, Sp3 and CpG methylation. Biosci. Rep. 2008, 28, 161–176. [Google Scholar] [CrossRef] [PubMed]
- Motta, A.B. Epigenetic Marks in Polycystic Ovary Syndrome. Curr. Med. Chem. 2020, 27, 6727–6743. [Google Scholar] [CrossRef]
- Bar-Sadeh, B.; Pnueli, L.; Keestra, S.; Bentley, G.R.; Melamed, P. Srd5a1 is Differentially Regulated and Methylated During Prepubertal Development in the Ovary and Hypothalamus. J. Endocr. Soc. 2023, 7, bvad108. [Google Scholar] [CrossRef]
- Shi, D.; Zhou, X.; Cai, L.; Wei, X.; Zhang, L.; Sun, Q.; Zhou, F.; Sun, L. Placental DNA methylation analysis of selective fetal growth restriction in monochorionic twins reveals aberrant methylated CYP11A1 gene for fetal growth restriction. FASEB J. 2023, 37, e23207. [Google Scholar] [CrossRef]
- Horning, A.M.; Awe, J.A.; Wang, C.M.; Liu, J.; Lai, Z.; Wang, V.Y.; Jadhav, R.R.; Louie, A.D.; Lin, C.L.; Kroczak, T.; et al. DNA methylation screening of primary prostate tumors identifies SRD5A2 and CYP11A1 as candidate markers for assessing risk of biochemical recurrence. Prostate 2015, 75, 1790–1801. [Google Scholar] [CrossRef]
- Kandi, V.; Vadakedath, S. Effect of DNA Methylation in Various Diseases and the Probable Protective Role of Nutrition: A Mini-Review. Cureus 2015, 7, e309. [Google Scholar] [CrossRef]
- Kuroda, A.; Rauch, T.A.; Todorov, I.; Ku, H.T.; Al-Abdullah, I.H.; Kandeel, F.; Mullen, Y.; Pfeifer, G.P.; Ferreri, K. Insulin gene expression is regulated by DNA methylation. PLoS ONE 2009, 4, e6953. [Google Scholar] [CrossRef]
- Zhong, X.; Jin, F.; Huang, C.; Du, M.; Gao, M.; Wei, X. DNA methylation of AMHRII and INSR gene is associated with the pathogenesis of Polycystic Ovary Syndrome (PCOS). Technol. Health Care 2021, 29 (Suppl. S1), 11–25. [Google Scholar] [CrossRef] [PubMed]
- Canivell, S.; Ruano, E.G.; Sisó-Almirall, A.; Kostov, B.; González-de Paz, L.; Fernandez-Rebollo, E.; Hanzu, F.A.; Párrizas, M.; Novials, A.; Gomis, R. Differential methylation of TCF7L2 promoter in peripheral blood DNA in newly diagnosed, drug-naïve patients with type 2 diabetes. PLoS ONE 2014, 9, e99310. [Google Scholar] [CrossRef] [PubMed]
- Smail, H.O.; Mohamad, D.A. Identification of DNA methylation of CAPN10 gene changes in the patients with type 2 diabetes mellitus as a predictive biomarker instead of HbA1c, random blood sugar, lipid profile, kidney function test, and some risk factors. Endocr. Regul. 2023, 57, 221–234. [Google Scholar] [CrossRef]
- Yu, J.T.; Hu, X.W.; Chen, H.Y.; Yang, Q.; Li, H.D.; Dong, Y.H.; Zhang, Y.; Wang, J.N.; Jin, J.; Wu, Y.G.; et al. DNA methylation of FTO promotes renal inflammation by enhancing m6A of PPAR-α in alcohol-induced kidney injury. Pharmacol. Res. 2021, 163, 105286. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.N.; Qin, Y.; Wu, B.; Peng, H.; Li, M.; Luo, H.; Liu, L.L. DNA methylation in polycystic ovary syndrome: Emerging evidence and challenges. Reprod. Toxicol. 2022, 111, 11–19. [Google Scholar] [CrossRef]
- Diboun, I.; Wani, S.; Ralston, S.H.; Albagha, O.M.E. Epigenetic DNA Methylation Signatures Associated with the Severity of Paget’s Disease of Bone. Front. Cell Dev. Biol. 2022, 10, 903612. [Google Scholar] [CrossRef]
- Ragusa, M.A.; Naselli, F.; Cruciata, I.; Volpes, S.; Schimmenti, C.; Serio, G.; Mauro, M.; Librizzi, M.; Luparello, C.; Chiarelli, R.; et al. Indicaxanthin Induces Autophagy in Intestinal Epithelial Cancer Cells by Epigenetic Mechanisms Involving DNA Methylation. Nutrients 2023, 15, 3495. [Google Scholar] [CrossRef]
- Zhao, G.; Zhou, J.; Gao, J.; Liu, Y.; Gu, S.; Zhang, X.; Su, P. Genome-wide DNA methylation analysis in permanent atrial fibrillation. Mol. Med. Rep. 2017, 16, 5505–5514. [Google Scholar] [CrossRef]
- Xu, S.; Furukawa, T.; Kanai, N.; Sunamura, M.; Horii, A. Abrogation of DUSP6 by hypermethylation in human pancreatic cancer. J. Hum. Genet. 2005, 50, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.S.; Li, L.; Ji, M.; Cheng, Y.; Ying, J.; Fan, Y.; Zhong, L.; Liu, X.; Tsao, S.W.; Chan, A.T.; et al. FEZF2, a novel 3p14 tumor suppressor gene, represses oncogene EZH2 and MDM2 expression and is frequently methylated in nasopharyngeal carcinoma. Carcinogenesis 2013, 34, 1984–1993. [Google Scholar] [CrossRef]
- Salpea, P.; Russanova, V.R.; Hirai, T.H.; Sourlingas, T.G.; Sekeri-Pataryas, K.E.; Romero, R.; Epstein, J.; Howard, B.H. Postnatal development- and age-related changes in DNA-methylation patterns in the human genome. Nucleic Acids Res. 2012, 40, 6477–6494. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.; Ma, Y.; Xu, J.; Chen, G.; Mahato, R.K. DNA methylation-mediated FGFR1 silencing enhances NF-κB signaling: Implications for asthma pathogenesis. Front. Mol. Biosci. 2024, 11, 1433557. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, L.; Monti, P.; Favero, C.; Carugno, M.; Tarantini, L.; Maggioni, C.; Bonzini, M.; Pesatori, A.C.; Bollati, V. Association between night shift work and methylation of a subset of immune-related genes. Front. Public Health 2023, 10, 1083826. [Google Scholar] [CrossRef]
- Alvarado, S.G.; Lenkov, K.; Williams, B.; Fernald, R.D. Social Crowding during Development Causes Changes in GnRH1 DNA Methylation. PLoS ONE 2015, 10, e0142043. [Google Scholar] [CrossRef]
- Bui, C.; Ouzzine, M.; Talhaoui, I.; Sharp, S.; Prydz, K.; Coughtrie, M.W.; Fournel-Gigleux, S. Epigenetics: Methylation-associated repression of heparan sulfate 3-O-sulfotransferase gene expression contributes to the invasive phenotype of H-EMC-SS chondrosarcoma cells. FASEB J. 2010, 24, 436–450. [Google Scholar] [CrossRef]
- Brockman, Q.R.; Rytlewski, J.D.; Milhem, M.; Monga, V.; Dodd, R.D. Integrated Epigenetic and Transcriptomic Analysis Identifies Interleukin 17 DNA Methylation Signature of Malignant Peripheral Nerve Sheath Tumor Progression and Metastasis. JCO Precis. Oncol. 2024, 8, e2300325. [Google Scholar] [CrossRef]
- Wyatt, A.K.; Zavodna, M.; Viljoen, J.L.; Stanton, J.A.; Gemmell, N.J.; Jasoni, C.L. Changes in methylation patterns of Kiss1 and Kiss1r gene promoters across puberty. Genet. Epigenet. 2013, 5, 51–62. [Google Scholar] [CrossRef]
- Demanelis, K.; Argos, M.; Tong, L.; Shinkle, J.; Sabarinathan, M.; Rakibuz-Zaman, M.; Sarwar, G.; Shahriar, H.; Islam, T.; Rahman, M.; et al. Association of Arsenic Exposure with Whole Blood DNA Methylation: An Epigenome-Wide Study of Bangladeshi Adults. Environ. Health Perspect. 2019, 127, 57011. [Google Scholar] [CrossRef]
- Shi, J.; Xu, J.; Chen, Y.E.; Li, J.S.; Cui, Y.; Shen, L.; Li, J.J.; Li, W. The concurrence of DNA methylation and demethylation is associated with transcription regulation. Nat. Commun. 2021, 12, 5285. [Google Scholar] [CrossRef] [PubMed]
- Beetch, M.; Lubecka, K.; Shen, K.; Flower, K.; Harandi-Zadeh, S.; Suderman, M.; Flanagan, J.M.; Stefanska, B. Stilbenoid-Mediated Epigenetic Activation of Semaphorin 3A in Breast Cancer Cells Involves Changes in Dynamic Interactions of DNA with DNMT3A and NF1C Transcription Factor. Mol. Nutr. Food Res. 2019, 63, e1801386. [Google Scholar] [CrossRef]
- Stuckel, A.J.; Zeng, S.; Lyu, Z.; Zhang, W.; Zhang, X.; Dougherty, U.; Mustafi, R.; Khare, T.; Zhang, Q.; Joshi, T.; et al. Sprouty4 is epigenetically upregulated in human colorectal cancer. Epigenetics 2023, 18, 2145068. [Google Scholar] [CrossRef]
- Singh, V.; Singh, L.C.; Vasudevan, M.; Chattopadhyay, I.; Borthakar, B.B.; Rai, A.K.; Phukan, R.K.; Sharma, J.; Mahanta, J.; Kataki, A.C.; et al. Esophageal Cancer Epigenomics and Integrome Analysis of Genome-Wide Methylation and Expression in High Risk Northeast Indian Population. OMICS 2015, 19, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Huang, Z.; Zeng, Z.; Li, J.; Xie, H.; Xie, C. An integrative analysis of DNA methylation and gene expression to predict lung adenocarcinoma prognosis. Front. Genet. 2022, 13, 970507. [Google Scholar] [CrossRef] [PubMed]
- Nobile, V.; Pucci, C.; Chiurazzi, P.; Neri, G.; Tabolacci, E. DNA Methylation, Mechanisms of FMR1 Inactivation and Therapeutic Perspectives for Fragile X Syndrome. Biomolecules 2021, 11, 296. [Google Scholar] [CrossRef]
- Xu, C.J.; Bonder, M.J.; Söderhäll, C.; Bustamante, M.; Baïz, N.; Gehring, U.; Jankipersadsing, S.A.; van der Vlies, P.; van Diemen, C.C.; van Rijkom, B.; et al. The emerging landscape of dynamic DNA methylation in early childhood. BMC Genom. 2017, 18, 25. [Google Scholar] [CrossRef]
- Zhang, H.; Dong, P.; Fan, H.; Liang, H.; Zhang, K.; Zhao, Y.; Guo, S.; Schrodi, S.J.; Fan, Y.; Zhang, D. Gene body hypomethylation of pyroptosis-related genes NLRP7, NLRP2, and NLRP3 facilitate non-invasive surveillance of hepatocellular carcinoma. Funct. Integr. Genom. 2023, 23, 198. [Google Scholar] [CrossRef]
- Dietrich, D.; Hasinger, O.; Liebenberg, V.; Field, J.K.; Kristiansen, G.; Soltermann, A. DNA methylation of the homeobox genes PITX2 and SHOX2 predicts outcome in non-small-cell lung cancer patients. Diagn. Mol. Pathol. 2012, 21, 93–104. [Google Scholar] [CrossRef]
- Luzón-Toro, B.; Villalba-Benito, L.; Fernández, R.M.; Torroglosa, A.; Antiñolo, G.; Borrego, S. RMRP, RMST, FTX and IPW: Novel potential long non-coding RNAs in medullary thyroid cancer. Orphanet J. Rare Dis. 2021, 16, 4. [Google Scholar] [CrossRef]
- Anvar, Z.; Chakchouk, I.; Demond, H.; Sharif, M.; Kelsey, G.; Van den Veyver, I.B. DNA Methylation Dynamics in the Female Germline and Maternal-Effect Mutations That Disrupt Genomic Imprinting. Genes 2021, 12, 1214. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wu, W.; Luo, H.; Liu, Z.; Liu, H.; Li, Q.; Pan, Z. Molecular characterization and epigenetic regulation of Mei1 in cattle and cattle-yak. Gene 2015, 573, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fukui, N.; Yahata, M.; Katsuragawa, Y.; Tashiro, T.; Ikegawa, S.; Lee, M.T. Genome-wide DNA methylation profile implicates potential cartilage regeneration at the late stage of knee osteoarthritis. Osteoarthr. Cartil. 2016, 24, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Fridley, B.L.; Song, H.; Lawrenson, K.; Cunningham, J.M.; Ramus, S.J.; Cicek, M.S.; Tyrer, J.; Stram, D.; Larson, M.C.; et al. Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat. Commun. 2013, 4, 1628. [Google Scholar] [CrossRef]
- Hu, X.; Wang, Y.; Zhang, X.; Li, C.; Zhang, X.; Yang, D.; Liu, Y.; Li, L. DNA methylation of HOX genes and its clinical implications in cancer. Exp. Mol. Pathol. 2023, 134, 104871. [Google Scholar] [CrossRef]
- Gao, P.; Sun, N.; Zhao, T.; Sun, Y.; Gu, J.; Ma, D.; Tian, H.; Peng, Z.; Zhang, Y.; Han, F.; et al. Identification of prognostic indicators, diagnostic markers, and possible therapeutic targets among LIM homeobox transcription factors in breast cancer. Cancer Innov. 2022, 1, 252–269. [Google Scholar] [CrossRef]
- Wang, C.; Xing, Q.; Song, B.; Li, G.; Xu, Z.; Wang, T.; Chen, Y.; Xu, Y.; Cao, Y. Aberrant DNA methylation in the PAX2 promoter is associated with Müllerian duct anomalies. Arch. Gynecol. Obstet. 2020, 301, 1455–1461. [Google Scholar] [CrossRef]
- Yu, H.; Pask, A.J.; Shaw, G.; Renfree, M.B. Comparative analysis of the mammalian WNT4 promoter. BMC Genom. 2009, 10, 416. [Google Scholar] [CrossRef]
- Yamashita, S.; Tsujino, Y.; Moriguchi, K.; Tatematsu, M.; Ushijima, T. Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2′-deoxycytidine treatment and oligonucleotide microarray. Cancer Sci. 2006, 97, 64–71. [Google Scholar] [CrossRef]
- Inagaki, H.; Ota, S.; Nishizawa, H.; Miyamura, H.; Nakahira, K.; Suzuki, M.; Nishiyama, S.; Kato, T.; Yanagihara, I.; Kurahashi, H. Obstetric complication-associated ANXA5 promoter polymorphisms may affect gene expression via DNA secondary structures. J. Hum. Genet. 2019, 64, 459–466. [Google Scholar] [CrossRef]
- Di Dalmazi, G.; Morandi, L.; Rubin, B.; Pilon, C.; Asioli, S.; Vicennati, V.; De Leo, A.; Ambrosi, F.; Santini, D.; Pagotto, U.; et al. DNA Methylation of Steroidogenic Enzymes in Benign Adrenocortical Tumors: New Insights in Aldosterone-Producing Adenomas. J. Clin. Endocrinol. Metab. 2020, 105, dgaa585. [Google Scholar] [CrossRef] [PubMed]
- Hou, B.; Shu, M.; Liu, C.; Du, Y.; Xu, C.; Jiang, H.; Hou, J.; Chen, X.; Wang, L.; Wu, X. Unveiling the role of UPF3B in hepatocellular carcinoma: Potential therapeutic target. Cancer Sci. 2024, 115, 2646–2658. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wu, M.; Hu, X.; Yang, J.; Han, R.; Ma, Y.; Zhang, X.; Yuan, Y.; Liu, R.; Jiang, G.; et al. Ankylosing spondylitis is associated with aberrant DNA methylation of IFN regulatory factor 8 gene promoter region. Clin. Rheumatol. 2019, 38, 2161–2169. [Google Scholar] [CrossRef]
- Do, W.L.; Conneely, K.; Gabram-Mendola, S.; Krishnamurti, U.; D’Angelo, O.; Miller-Kleinhenz, J.; Gogineni, K.; Torres, M.; McCullough, L.E. Obesity-associated methylation in breast tumors: A possible link to disparate outcomes? Breast Cancer Res. Treat. 2020, 181, 135–144. [Google Scholar] [CrossRef]
- Adkins, R.M.; Thomas, F.; Tylavsky, F.A.; Krushkal, J. Parental ages and levels of DNA methylation in the newborn are correlated. BMC Med. Genet. 2011, 12, 47. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Wang, W.; Tang, Y.; Zhou, D.; Gao, Y.; Zhang, Q.; Zhou, X.; Zhu, H.; Xing, L.; Yu, J. mRNA and methylation profiling of radioresistant esophageal cancer cells: The involvement of Sall2 in acquired aggressive phenotypes. J. Cancer 2017, 8, 646–656. [Google Scholar] [CrossRef]
- Qin, R.; Cao, L.; Wang, J.; Liu, J. Promoter Methylation of Ezrin and its Impact on the Incidence and Prognosis of Cervical Cancer. Cell. Physiol. Biochem. 2018, 50, 277–287. [Google Scholar] [CrossRef]
- Gong, G.; Lin, T.; Yuan, Y. Integrated analysis of gene expression and DNA methylation profiles in ovarian cancer. J. Ovarian Res. 2020, 13, 30. [Google Scholar] [CrossRef]
- Dubovsky, J.A.; McNeel, D.G.; Power, J.J.; Gordon, J.; Sotomayor, E.M.; Pinilla-Ibarz, J.A. Treatment of chronic lymphocytic leukemia with a hypomethylating agent induces expression of NXF2, an immunogenic cancer testis antigen. Clin. Cancer Res. 2009, 15, 3406–3415. [Google Scholar] [CrossRef]
- Sung, H.Y.; Choi, E.N.; Jo, S.A.; Oh, S.; Ahn, J.-H. Amyloid protein-mediated differential DNA methylation status regulates gene expression in Alzheimer’s disease model cell line. Biochem. Biophys. Res. Commun. 2011, 414, 700–705. [Google Scholar] [CrossRef]
- Lindqvist, B.M.; Farkas, S.A.; Wingren, S.; Nilsson, T.K. DNA methylation pattern of the SLC25A43 gene in breast cancer. Epigenetics 2012, 7, 300–306. [Google Scholar] [CrossRef][Green Version]
- Liang, Y.; Zeng, J.; Luo, B.; Li, W.; He, Y.; Zhao, W.; Hu, N.; Jiang, N.; Luo, Y.; Xian, Y.; et al. TET2 promotes IL-1β expression in J774.1 cell through TLR4/MAPK signaling pathway with demethylation of TAB2 promoter. Mol. Immunol. 2020, 126, 136–142. [Google Scholar] [CrossRef]
- Ishido, M.; Higashi, K.; Mori, H.; Ueno, M.; Kurokawa, K. DNA methylation profiles of transgenerational rat hyperactivity primed by silver nanoparticles: Comparison with valproate model rats of autism. Behav. Brain Res. 2025, 477, 115293. [Google Scholar] [CrossRef]
- Nishino, K.; Hattori, N.; Tanaka, S.; Shiota, K. DNA methylation-mediated control of Sry gene expression in mouse gonadal development. J. Biol. Chem. 2004, 279, 22306–22313. [Google Scholar] [CrossRef]
- Topalovic, V.; Krstic, A.; Schwirtlich, M.; Dolfini, D.; Mantovani, R.; Stevanovic, M.; Mojsin, M. Epigenetic regulation of human SOX3 gene expression during early phases of neural differentiation of NT2/D1 cells. PLoS ONE 2017, 12, e0184099. [Google Scholar] [CrossRef]
- Zhou, X.; He, Y.; Li, N.; Bai, G.; Pan, X.; Zhang, Z.; Zhang, H.; Li, J.; Yuan, X. DNA methylation mediated RSPO2 to promote follicular development in mammals. Cell Death Dis. 2021, 12, 653. [Google Scholar] [CrossRef]
- Giordano, A.; Pignolet, B.; Mascia, E.; Clarelli, F.; Sorosina, M.; Misra, K.; Bucciarelli, F.; Ferrè, L.; Moiola, L.; Liblau, R.; et al. DNA Methylation in the Anti-Mullerian Hormone Gene and the Risk of Disease Activity in Multiple Sclerosis. Ann. Neurol. 2024, 96, 289–301. [Google Scholar] [CrossRef]
- Yu, Y.Y.; Sun, C.X.; Liu, Y.K.; Li, Y.; Wang, L.; Zhang, W. Genome-wide screen of ovary-specific DNA methylation in polycystic ovary syndrome. Fertil. Steril. 2015, 104, 145–153.e6. [Google Scholar] [CrossRef]
- Dhawan, S.; Georgia, S.; Tschen, S.I.; Fan, G.; Bhushan, A. Pancreatic β cell identity is maintained by DNA methylation-mediated repression of Arx. Dev. Cell 2011, 20, 419–429. [Google Scholar] [CrossRef]
- Li, S.; Zhu, D.; Duan, H.; Ren, A.; Glintborg, D.; Andersen, M.; Skov, V.; Thomassen, M.; Kruse, T.; Tan, Q. Differential DNA methylation patterns of polycystic ovarian syndrome in whole blood of Chinese women. Oncotarget 2017, 8, 20656–20666. [Google Scholar] [CrossRef]
- Tanaka, Y.; Kanda, M.; Sugimoto, H.; Shimizu, D.; Sueoka, S.; Takami, H.; Ezaka, K.; Hashimoto, R.; Okamura, Y.; Iwata, N.; et al. Translational implication of Kallmann syndrome-1 gene expression in hepatocellular carcinoma. Int. J. Oncol. 2015, 46, 2546–2554. [Google Scholar] [CrossRef]
- Xiang, Y.; Cheng, Y.; Li, X.; Li, Q.; Xu, J.; Zhang, J.; Liu, Y.; Xing, Q.; Wang, L.; He, L.; et al. Up-regulated expression and aberrant DNA methylation of LEP and SH3PXD2A in pre-eclampsia. PLoS ONE 2013, 8, e59753. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, Y.; Wu, J.; Qiao, M.; Zhou, J.; Xu, Z.; Li, Z.; Sun, H.; Peng, X.; Mei, S. Comprehensive Analysis of Methylome and Transcriptome to Identify Potential Genes Regulating Porcine Testis Development. Int. J. Mol. Sci. 2024, 25, 9105. [Google Scholar] [CrossRef]
- Fujii, S.; Srivastava, V.; Hegde, A.; Kondo, Y.; Shen, L.; Hoshino, K.; Gonzalez, Y.; Wang, J.; Sasai, K.; Ma, X.; et al. Regulation of AURKC expression by CpG island methylation in human cancer cells. Tumour Biol. 2015, 36, 8147–8158. [Google Scholar] [CrossRef]
- Namous, H.; Braz, C.U.; Wang, Y.; Khatib, H. The Activation of Protamine 1 Using Epigenome Editing Decreases the Proliferation of Tumorigenic Cells. Front. Genome Ed. 2022, 4, 844904. [Google Scholar] [CrossRef]
- Sujit, K.M.; Pallavi, S.; Singh, V.; Andrabi, S.W.; Trivedi, S.; Sankhwar, S.N.; Gupta, G.; Rajender, S. SPATA16 promoter hypermethylation and downregulation in male infertility. Andrologia 2022, 54, e14548. [Google Scholar] [CrossRef]
- Nanavaty, V.; Abrash, E.W.; Hong, C.; Park, S.; Fink, E.E.; Li, Z.; Sweet, T.J.; Bhasin, J.M.; Singuri, S.; Lee, B.H.; et al. DNA Methylation Regulates Alternative Polyadenylation via CTCF and the Cohesin Complex. Mol. Cell 2020, 78, 752–764.e6. [Google Scholar] [CrossRef]
- Yang, Q.; Xie, Y.; Pan, B.; Cheng, Y.; Zhu, Y.; Fei, X.; Li, X.; Yu, J.; Chen, Z.; Li, J.; et al. The Expression and Epigenetic Characteristics of the HSF2 Gene in Cattle-Yak and the Correlation with Its Male Sterility. Animals 2024, 14, 1410. [Google Scholar] [CrossRef]
- Ivascu, C.; Wasserkort, R.; Lesche, R.; Dong, J.; Stein, H.; Thiel, A.; Eckhardt, F. DNA methylation profiling of transcription factor genes in normal lymphocyte development and lymphomas. Int. J. Biochem. Cell Biol. 2007, 39, 1523–1538. [Google Scholar] [CrossRef]
- Coppens, G.; Vanhorebeek, I.; Güiza, F.; Derese, I.; Wouters, P.J.; Téblick, A.; Dulfer, K.; Joosten, K.F.; Verbruggen, S.C.; Berghe, G.V.D. Abnormal DNA methylation within HPA-axis genes years after paediatric critical illness. Clin. Epigenet. 2024, 16, 31. [Google Scholar] [CrossRef]
- Amor, H.; Zeyad, A.; Hammadeh, M.E. Tobacco smoking and its impact on the expression level of sperm nuclear protein genes: H2BFWT, TNP1, TNP2, PRM1 and PRM2. Andrologia 2021, 53, e13964. [Google Scholar] [CrossRef]
- Gross, N.; Peñagaricano, F.; Khatib, H. Integration of whole-genome DNA methylation data with RNA sequencing data to identify markers for bull fertility. Anim. Genet. 2020, 51, 502–510. [Google Scholar] [CrossRef]
- Han, W.; Xue, Q.; Li, G.; Yin, J.; Zhang, H.; Zhu, Y.; Xing, W.; Cao, Y.; Su, Y.; Wang, K.; et al. Genome-wide analysis of the role of DNA methylation in inbreeding depression of reproduction in Langshan chicken. Genomics 2020, 112, 2677–2687. [Google Scholar] [CrossRef]
- Zhu, L.; Li, X.; Yuan, Y.; Dong, C.; Yang, M. APC Promoter Methylation in Gastrointestinal Cancer. Front. Oncol. 2021, 11, 653222. [Google Scholar] [CrossRef]
- Dobrovic, A.; Simpfendorfer, D. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 1997, 57, 3347–3350. [Google Scholar] [PubMed]
- Rauch, T.A.; Wang, Z.; Wu, X.; Kernstine, K.H.; Riggs, A.D.; Pfeifer, G.P. DNA methylation biomarkers for lung cancer. Tumour Biol. 2012, 33, 287–296. [Google Scholar] [CrossRef]
- Pan, X.; Chen, Z.; Huang, R.; Yao, Y.; Ma, G. Transforming growth factor β1 induces the expression of collagen type I by DNA methylation in cardiac fibroblasts. PLoS ONE 2013, 8, e60335. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Li, G.; Wang, G.; Huo, Z.; Feng, X.; Du, L.; Li, Y.; Yang, Q.; Ma, X.; Yu, B.; et al. Development of a Risk Score Model for Osteosarcoma Based on DNA Methylation-Driven Differentially Expressed Genes. J. Oncol. 2022, 2022, 7596122. [Google Scholar] [CrossRef]
- Tompa, M.; Kajtar, B.; Galik, B.; Gyenesei, A.; Kalman, B. DNA methylation and protein expression of Wnt pathway markers in progressive glioblastoma. Pathol. Res. Pract. 2021, 222, 153429. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Zhu, Y.; Yu, H.; Zhou, C.; Kijlstra, A.; Yang, P. Dynamic DNA Methylation Changes of Tbx21 and Rorc during Experimental Autoimmune Uveitis in Mice. Mediat. Inflamm. 2018, 2018, 9129163. [Google Scholar] [CrossRef]
- Cui, B.; Xiao, X.; Wang, J.; Wang, H.; Wu, C.; Yan, Y.; Zheng, J.; Wang, J.; Zong, Y.; Zhang, Y.; et al. Low THRB (thyroid hormone receptor beta) Promoter Methylation Levels in Peripheral Blood Leukocytes Induced by Systematic Inflammation Are Involved in Low Thyroid Hormone Function in Metabolic Syndrome. Hypertension 2021, 78, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sang, M.; Li, Y.; Li, Y.; Yuan, E.; Yang, L.; Shi, W.; Yuan, Y.; Yang, B.; Yang, P.; et al. WNT3 hypomethylation counteracts low activity of the Wnt signaling pathway in the placenta of preeclampsia. Cell. Mol. Life Sci. 2021, 78, 6995–7008. [Google Scholar] [CrossRef]
- Gonseth, S.; Shaw, G.M.; Roy, R.; Segal, M.R.; Asrani, K.; Rine, J.; Wiemels, J.; Marini, N.J. Epigenomic profiling of newborns with isolated orofacial clefts reveals widespread DNA methylation changes and implicates metastable epiallele regions in disease risk. Epigenetics 2019, 14, 198–213. [Google Scholar] [CrossRef]
- Mathios, D.; Hwang, T.; Xia, Y.; Phallen, J.; Rui, Y.; See, A.P.; Maxwell, R.; Belcaid, Z.; Casaos, J.; Burger, P.C.; et al. Genome-wide investigation of intragenic DNA methylation identifies ZMIZ1 gene as a prognostic marker in glioblastoma and multiple cancer types. Int. J. Cancer 2019, 145, 3425–3435. [Google Scholar] [CrossRef] [PubMed]
- Chim, C.-S.; Wong, S.-Y.; Pang, A.; Chu, P.; Lau, J.S.; Wong, K.-F.; Kwong, Y.-L. Aberrant promoter methylation of the retinoic acid receptor alpha gene in acute promyelocytic leukemia. Leukemia 2005, 19, 2241–2246. [Google Scholar] [CrossRef] [PubMed]
- Simonova, O.A.; Kuznetsova, E.B.; Tanas, A.S.; Rudenko, V.V.; Poddubskaya, E.V.; Kekeeva, T.V.; Trotsenko, I.D.; Larin, S.S.; Kutsev, S.I.; Zaletaev, D.V.; et al. Abnormal Hypermethylation of CpG Dinucleotides in Promoter Regions of Matrix Metalloproteinases Genes in Breast Cancer and Its Relation to Epigenomic Subtypes and HER2 Overexpression. Biomedicines 2020, 8, 116. [Google Scholar] [CrossRef]
- Da Silva Melo, A.R.; Barroso, H.; De Araújo, D.U.; Pereira, F.R.; De Oliveira, N.F.P. The influence of sun exposure on the DNA methylation status of MMP9, miR-137, KRT14 and KRT19 genes in human skin. Eur. J. Dermatol. 2015, 25, 436–443. [Google Scholar] [CrossRef]
- Strelnikov, V.V.; Kuznetsova, E.B.; Tanas, A.S.; Rudenko, V.V.; Kalinkin, A.I.; Poddubskaya, E.V.; Kekeeva, T.V.; Chesnokova, G.G.; Trotsenko, I.D.; Larin, S.S.; et al. Abnormal promoter DNA hypermethylation of the integrin, nidogen, and dystroglycan genes in breast cancer. Sci. Rep. 2021, 11, 2264. [Google Scholar] [CrossRef]
- Wang, R.; Zhu, H.; Yang, M.; Zhu, C. DNA methylation profiling analysis identifies a DNA methylation signature for predicting prognosis and recurrence of lung adenocarcinoma. Oncol. Lett. 2019, 18, 5831–5842. [Google Scholar] [CrossRef]
- Zhang, W.; Li, X.; Yu, U.; Huang, X.; Wang, H.; Lu, Y.; Liu, S.; Zhang, J. Genome-wide methylation and gene-expression analyses in thalassemia. Aging 2024, 16, 11591–11605. [Google Scholar] [CrossRef]
- Légaré, C.; Overend, G.; Guay, S.P.; Monckton, D.G.; Mathieu, J.; Gagnon, C.; Bouchard, L. DMPK gene DNA methylation levels are associated with muscular and respiratory profiles in DM1. Neurol Genet. 2019, 5, e338. [Google Scholar] [CrossRef] [PubMed]
- Pagliara, D.; Ciolfi, A.; Pedace, L.; Haghshenas, S.; Ferilli, M.; Levy, M.A.; Miele, E.; Nardini, C.; Cappelletti, C.; Relator, R.; et al. Identification of a robust DNA methylation signature for Fanconi anemia. Am. J. Hum. Genet. 2023, 110, 1938–1949. [Google Scholar] [CrossRef] [PubMed]
- Adeoye, O.; Olawumi, J.; Opeyemi, A.; Christiania, O. Review on the role of glutathione on oxidative stress and infertility. JBRA Assist. Reprod. 2018, 22, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Franzago, M.; Santurbano, D.; Vitacolonna, E.; Stuppia, L. Genes and Diet in the Prevention of Chronic Diseases in Future Generations. Int. J. Mol. Sci. 2020, 21, 2633. [Google Scholar] [CrossRef]
- Bekdash, R.A. Epigenetics, Nutrition, and the Brain: Improving Mental Health through Diet. Int. J. Mol. Sci. 2024, 25, 4036. [Google Scholar] [CrossRef]
- Bekdash, R.A. Early Life Nutrition and Mental Health: The Role of DNA Methylation. Nutrients 2021, 13, 3111. [Google Scholar] [CrossRef]
- Puche-Juarez, M.; Toledano, J.M.; Moreno-Fernandez, J.; Gálvez-Ontiveros, Y.; Rivas, A.; Diaz-Castro, J.; Ochoa, J.J. The Role of Endocrine Disrupting Chemicals in Gestation and Pregnancy Outcomes. Nutrients 2023, 15, 4657. [Google Scholar] [CrossRef]
- Erdoğan, K.; Sanlier, N.T.; Sanlier, N. Are epigenetic mechanisms and nutrition effective in male and female infertility? J. Nutr. Sci. 2023, 12, e103. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies. Scientific opinion on Dietary Reference Values for choline. EFSA J. 2016, 14, e04484. [Google Scholar] [CrossRef]
- EFSA Panel on Nutrition, Novel Foods and Food Allergens; Turck, D.; Castenmiller, J.; De Henauw, S.; Hirsch-Ernst, K.I.; Kearney, J.; Maciuk, A.; Mangelsdorf, I.; McArdle, H.J.; Naska, A.; et al. Scientific Opinion on the safety of betaine as a novel food pursuant to Regulation (EU) 2015/2283. EFSA J. 2019, 17, e05658. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies. Scientific Opinion on Dietary Reference Values for folate. EFSA J. 2014, 12, 3893. [Google Scholar] [CrossRef]
- Crider, K.S.; Yang, T.P.; Berry, R.J.; Bailey, L.B. Folate and DNA methylation: A review of molecular mechanisms and the evidence for folate’s role. Adv. Nutr. 2012, 3, 21–38. [Google Scholar] [CrossRef]
- Nazni, P. Association of western diet & lifestyle with decreased fertility. Indian J. Med. Res. 2014, 140 (Suppl. S1), S78–S81. [Google Scholar]
- Ling, C.; Rönn, T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab. 2019, 29, 1028–1044. [Google Scholar] [CrossRef]
- Fernandez-Twinn, D.S.; Hjort, L.; Novakovic, B.; Ozanne, S.E.; Saffery, R. Intrauterine programming of obesity and type 2 diabetes. Diabetologia 2019, 62, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, K.M.; Reynolds, R.M.; Prescott, S.L.; Nyirenda, M.; Jaddoe, V.W.; Eriksson, J.G.; Broekman, B.F. Influence of maternal obesity on the long-term health of offspring. Lancet Diabetes Endocrinol. 2017, 5, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, S. Dietary trends and the decline in male reproductive health. Hormones 2023, 22, 165–197. [Google Scholar] [CrossRef]
- Salvaleda-Mateu, M.; Rodríguez-Varela, C.; Labarta, E. Do Popular Diets Impact Fertility? Nutrients 2024, 16, 1726. [Google Scholar] [CrossRef]
- Chiu, Y.H.; Chavarro, J.E.; Souter, I. Diet and female fertility: Doctor, what should I eat? Fertil. Steril. 2018, 110, 560–569. [Google Scholar] [CrossRef]
- Kirkwood, T.B.L. A systematic look at an old problem. Nature 2008, 451, 644–647. [Google Scholar] [CrossRef]
- Kenyon, C.J. The genetics of ageing. Nature 2010, 464, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Vellai, T.; Takács-Vellai, K. Regulation of protein turnover by longevity pathways. Adv. Exp. Med. Biol. 2010, 694, 69–80. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Vellai, T. How the amino acid leucine activates the key cell-growth regulator mTOR. Nature 2021, 596, 192–194. [Google Scholar] [CrossRef]
- Vellai, T.; Takács-Vellai, K.; Sass, M.; Klionsky, D.J. The regulation of aging: Does autophagy underlie longevity? Trends Cell Biol. 2009, 19, 487–494. [Google Scholar] [CrossRef]
- Hotzi, B.; Kosztelnik, M.; Hargitai, B.; Takács-Vellai, K.; Barna, J.; Bördén, K.; Málnási-Csizmadia, A.; Lippai, M.; Ortutay, C.; Bacquet, C.; et al. Sex-specific regulation of aging in Caenorhabditis elegans. Aging Cell 2018, 17, e12724. [Google Scholar] [CrossRef]
- Vellai, T.; Tóth, M.L.; Kovács, A.L. Janus-faced autophagy: A dual role of cellular self-eating in neurodegeneration? Autophagy 2007, 3, 461–463. [Google Scholar] [CrossRef]
- Billes, V.; Kovács, T.; Hotzi, B.; Manzéger, A.; Tagscherer, K.; Komlós, M.; Tarnóci, A.; Pádár, Z.; Erdős, A.; Bjelik, A.; et al. AUTEN-67 (Autophagy Enhancer-67) Hampers the Progression of Neurodegenerative Symptoms in a Drosophila model of Huntington’s Disease. J. Huntingt. Dis. 2016, 5, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Sigmond, T.; Barna, J.; Tóth, M.L.; Takács-Vellai, K.; Pásti, G.; Kovács, A.L.; Vellai, T. Autophagy in Caenorhabditis elegans. Methods Enzymol. 2008, 451, 521–540. [Google Scholar] [CrossRef]
- Sturm, Á.; Vellai, T. How does maternal age affect genomic stability in the offspring? Aging Cell 2022, 21, e13612. [Google Scholar] [CrossRef]
- Gorbunova, V.; Seluanov, A.; Mita, P.; McKerrow, W.; Fenyö, D.; Boeke, J.D.; Linker, S.B.; Gage, F.H.; Kreiling, J.A.; Petrash, A.P.; et al. The role of retrotransposable elements in ageing and age-associated diseases. Nature 2021, 596, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Moghadam, A.R.E.; Moghadam, M.T.; Hemadi, M.; Saki, G. Oocyte quality and aging. JBRA Assist. Reprod. 2022, 26, 105–122. [Google Scholar] [CrossRef] [PubMed]
- Pino, V.; Sanz, A.; Valdés, N.; Crosby, J.; Mackenna, A. The effects of aging on semen parameters and sperm DNA fragmentation. JBRA Assist. Reprod. 2020, 24, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Koubova, J.; Guarente, L. How does calorie restriction work? Genes Dev. 2003, 17, 313–321. [Google Scholar] [CrossRef]
- Mattson, M.P.; de Cabo, R. Effects of Intermittent Fasting on Health, Aging, and Disease. N. Engl. J. Med. 2020, 382, 1773–1774. [Google Scholar] [CrossRef]
- Guarente, L.; Picard, F. Calorie restriction—The SIR2 connection. Cell 2005, 120, 473–482. [Google Scholar] [CrossRef]
- Bordone, L.; Guarente, L. Calorie restriction, SIRT1 and metabolism: Understanding longevity. Nat. Rev. Mol. Cell Biol. 2005, 6, 298–305. [Google Scholar] [CrossRef]
- Niculescu, M.D.; Zeisel, S.H. Diet, methyl donors and DNA methylation: Interactions between dietary folate, methionine and choline. J. Nutr. 2002, 132 (Suppl. S8), 2333S–2335S. [Google Scholar] [CrossRef]
- Mikkelsen, K.; Trapali, M.; Apostolopoulos, V. Role of Vitamin B in Healthy Ageing and Disease. Subcell. Biochem. 2024, 107, 245–268. [Google Scholar] [CrossRef]
- Morgan, D.K.; Whitelaw, E. The case for transgenerational epigenetic inheritance in humans. Mamm. Genome 2008, 19, 394–397. [Google Scholar] [CrossRef]
- Horsthemke, B. A critical view on transgenerational epigenetic inheritance in humans. Nat. Commun. 2018, 9, 2973. [Google Scholar] [CrossRef] [PubMed]
- Whitelaw, N.C.; Whitelaw, E. Transgenerational epigenetic inheritance in health and disease. Curr. Opin. Genet. Dev. 2008, 18, 273–279. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).