Abstract
Cutaneous melanoma (CM) is a public health issue and a significant challenge for scientists. At the dawn of the “omics era”, we witnessed groundbreaking advances in CM molecular stratification and therapeutic management assisted by genomic profiling and sequencing technologies. However, melanomagenesis is a complex and multifactorial process that cannot be restricted to genomic aspects, requiring investigation from a multi-omics perspective. Recently, droplet digital polymerase chain reaction (ddPCR) emerged as a powerful omics technology that can be used for absolute allele quantification, copy number variation (CNV) analysis, rare mutation and DNA methylation detection, genetic rearrangements, and transcriptomic evaluations in different types of biological samples, revolutionizing CM biomedical research and clinical care. This paper presents how ddPCR can complement existing approaches in the field to detect multiple types of alterations in both body fluids as well as formalin-fixed paraffin-embedded (FFPE) tissues harvested from CM patients and highlights how these findings may broaden our vision on CM diagnosis, prognosis and therapy in the context of precision medicine.
Keywords:
cutaneous melanoma; ddPCR; omics; biomarkers; ctDNA; liquid biopsy; targeted therapy; immunotherapy 1. Multi-Omics-Based Biomarkers: The Roadmap towards Personalized Care in CM
Skin melanoma is one of the most heterogeneous and metastatic malignancies, with an increasing incidence in fair-skinned populations [1]. Genomic interrogation, assisted by high-throughput sequencing technologies and microarrays, has guided the rudimentary stratification of these tumors and the therapeutic decisions [2]. However, it has been shown that the pathogenesis of CM is more complex than previously thought, involving dramatic transformations at almost all tumor levels: genomic, epigenomic, transcriptomic, proteomic, metabolomic, and so forth [3,4]. Therefore, to better capture and characterize biological events associated with prognosis or response to therapy in cancers, scientific research has undergone a remarkably swift transition from single-level tumor interrogation to multidimensional omics research. The recent advancements in technology and bioinformatics equipped us with data on multiple types of omics measurements, such as mRNA-gene expression, DNA methylation, microRNAs (miRNAs), copy-number variations (CNVs), and so on. These types of omics activities may be independent or overlap, reflecting distinctive patterns of the disease [5].
Recent studies have revealed the importance of multi-integration for precision medicine in CM. Several research groups have focused on the prognosis potential of multi-omics data. For instance, it has been proved that integrating gene expression, DNA methylation, and CNVs data may reveal new dysregulated signaling pathways in CM, with important implications for prognosis [3]. Similarly, an integrative study analyzing stage III patients confirmed the prognostic capabilities of gene expression, proteins, and microRNAs in correlation with clinical, pathological, and mutational data [6]. Other studies have focused on the predictive value of multi-omics analysis in CM. For example, in the HOPE project, whole-exome sequencing (WES) and gene expression profiling data revealed that complete remission under the clinically approved anti-PD1 antibody (e.g., Nivolumab, Opdivo) is associated with elevated levels of PDL-1 protein and an increased number of single nucleotide variants (SNVs) [7]. Therefore, in the future, evaluation of this disease from a multi-omics perspective is expected to provide a more specific molecular classification of CM and give clues about the mechanisms that drive tumorigenesis, metastasis, and resistance to therapy in these tumors [8].
Droplet digital polymerase chain reaction (ddPCR), an accurate and relatively inexpensive omics technology, is arousing considerable scientific interest in the field of biomarker research [9]. This technology enables the investigation and validation of several types of omics alterations detected by whole-genome screening (WGS) techniques, such as Next-Generation Sequencing (NGS). NGS provides a comprehensive picture of the mutational inventory of a tumor; however, the technology is not suitable for tracking mutations over time, as it is costly and meticulous [9]. Nevertheless, combining NGS with ddPCR can broaden the applications and increase the benefits of both techniques. For instance, once NGS identifies a set of mutations in ctDNA, researchers can exploit ddPCR, which is less expensive and laborious, to interrogate that set of biomarkers and obtain relevant information about tumor progression and response to therapy [10].
2. Droplet Digital PCR: A Versatile Omics Technology in Oncology
ddPCR, a highly sensitive and specific technology, relies on a water-oil emulsion droplet system that involves partitioning nucleic acid samples into 20,000 nanoliter-sized droplets that serve as independent test tubes or reactions [11]. By employing oil, water, and a chemical stabilizer emulsion, each sample is diluted into thousands of individual partitions, some of which do not contain template DNA and others containing one or more target sequences [12]. A PCR reaction takes place in each tube. Each partition is then examined for amplified target DNA by fluorescence so that the number of positive and negative droplets can be counted, facilitating the quantification of target molecules under the assumption of Poisson distribution [13]. The limit of detection (LoD) is about 0.005%, lower than that of RT-PCR (1%), pyrosequencing (5%), melting curve analysis (10%), and Sanger sequencing (20%) [14]. Through the partitioning process, ddPCR brings multiple advantages over traditional PCR techniques, that include: Absolute quantification of DNA copies in the input samples without the need for external calibration curves used in qPCR; low susceptibility to PCR inhibitors; increased accuracy, especially when working with low concentrations of samples or degraded samples, as well as reproducibility and increased sensitivity of the experiments [12]. Applications of this technology may include absolute allele quantification, CNVs analysis, rare mutations, DNA methylation detection, transcriptomic evaluations (mRNA, miRNA), and genetic rearrangements in various types of biological samples [12,15,16,17,18]. However, the use of ddPCR requires the identification of specific genetic alterations, which is why it almost always accompanies whole-genome profiling technologies such as NGS [19,20].
ddPCR is useful both for the analysis of archived tumor tissues, which are degraded and have a limited concentration of DNA, and for the analysis of biological fluids [21]. In recent years, liquid biopsy (LB), based on the analysis of circulating components derived from tumors—circulating tumor DNA (ctDNA), RNA, extracellular vesicles (EVs), or tumor cells (CTCs) has gained tremendous attention because of its potential to provide in real-time an accurate description of the genetic landscape of a tumor [22]. LB has proven to be more informative than tissue biopsy, which is spatially limited and ineloquent for tumor evolution [22]. Thus, only by analyzing the biological material from patients’ body fluids, researchers and clinicians can obtain valuable information on the dynamics of the genetic profile of the tumor, which can be integrated and used to guide the therapeutic management for each patient [20]. Due to its versatility and ability to operate with small amounts of biological material, ddPCR is an ideal methodology for analyzing LBs (Figure 1) [21]. Several blood-based biomarkers interrogated by ddPCR have also found diagnostic, predictive, and monitoring applications in certain malignancies [21]. Novel approaches use some other body fluids such as cerebrospinal fluid (CSF) or urine to screen for and validate biomarkers in cancer patients [21]. However, in this article, we aim to briefly present the applications of ddPCR for biomarker research in CM.
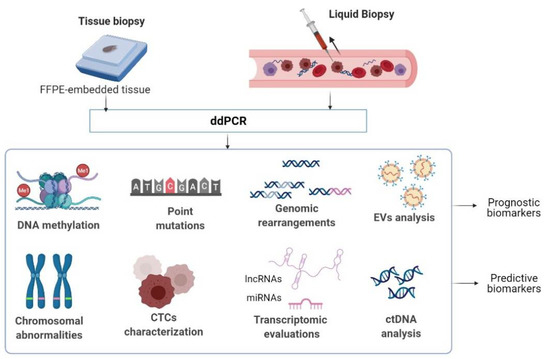
Figure 1.
Applications of ddPCR for biomarker research in oncology. FFPE tissue- formalin-fixed paraffin-embedded tissue; CTCs- circulating tumor cells; EVs-extracellular vesicles; ctDNA- circulating tumor DNA; miRNAs- microRNAs; lncRNAs- long non-coding RNAs.
3. Interrogating CM Mutational landscape via ddPCR
3.1. Screening for Biomarkers in Tissue Biopsies
Although highly invasive and ineloquent for tumor heterogeneity, tissue biopsy remains the gold standard for clinical molecular analyzes in cancer. Particularly for CM, tissue biopsy studies focus on hotspot mutations such as BRAF and KRAS, which are critical for guiding therapeutic decisions in clinical management. Remarkably, ddPCR showed enhanced sensitivity compared to the widely used Cobas® 4800 system based on real-time PCR amplification, Sanger sequencing, and allele-specific PCR (AS-PCR) (35.6% vs. 9.2%, 26.4%, and 26.4%) in the detection of BRAF V600E mutations in FFPE tissues from 87 CM patients diagnosed in different Breslow stages [23]. In a group of eight patients in the clinical cohort, the BRAF V600E mutation was detectable only by ddPCR; notably, these patients could have benefited from vemurafenib [23]. Five out of these eight patients who tested BRAF V600E positive only through ddPCR later developed sentinel lymph node metastases, highlighting that ddPCR should be the primary method for detecting and monitoring BRAF V600E mutant melanomas [23]. In parallel, another study demonstrated the superiority of ddPCR compared to Sanger sequencing and pyrosequencing in detecting common BRAF, NRAS, and TERT promoter mutations in 40 archived melanoma tissues [14]. Overall, ddPCR was much more sensitive, detecting mutations in 12.5% and 23% of tumors classified as wild-type by pyrosequencing and Sanger sequencing. The sensitivity of ddPCR was also much higher in tumors with <50% tumor cellularity, providing a rationale for using ddPCR in the detection and monitoring of human melanomas [14].
Recently, Salgado et al. used ddPCR to investigate the molecular mechanisms associated with TERT reactivation in human melanomas. CpG methylation in the TERT promoter (TERTp) was related to TERT mRNA expression [24]. Hence, two hotspot mutations in TERTp termed C228T and C250T, have been documented to facilitate the binding of transcription factor E26 transformation-specific/ternary complex factor (ETS/TCF) and subsequent TERT induction. To elucidate the genetic and epigenetic mechanisms regulating TERT gene expression in CM, Salgado et al. designated a ddPCR protocol to assess TERTp methylation fraction (MF), alongside C228T and C250T TERTp mutations in 44 healthy, benign and malignant tumor samples [24]. They observed that TERTp methylation is correlated with chromatin accessibility and TERT expression levels in melanoma cell lines; thus, due to increased TERTp methylation, TERT expression requires an open chromatin state in TERTp-wild type samples or a combination of moderate MF and chromatin accessibility in the presence of C228T/C250T hotspot mutations [24]. Given that TERTp hypermethylation has been proposed as an indicator of a poor patient prognosis in CM and certain other tumors, quantifying TERT methylation by ddPCR may open new perspectives for the prognosis and monitoring of CM patients [24].
3.2. Searching for Biomarkers in Liquid Biopsies
In the first instance, ddPCR has proven to be an ideal methodology for the analysis of cell-free ctDNA in CM patients’ plasma. ctDNAs are short DNA fragments (134–144 base pairs) derived from tumor cells that have undergone necrosis or apoptosis [25]. These fragments are released into the bloodstream and are actively investigated as surrogate biomarkers for the analysis of genomic and epigenomic alterations in primary tumors and metastatic lesions [26]. By far, ctDNA mutations (mt) are of considerable interest for CM research. It has been reported that BRAF mt may be reliable indicators for response to targeted therapy in CM, being detectable in 70% of patients in the non-responsive group and only in 10% of patients in the responsive group [27]. In line with these observations, Tsao et al. confirmed that the screening of BRAF and NRAS mt in ctDNA by ddPCR is an effective method to monitor the response to targeted therapy in stage IV melanoma patients, being even more informative than LDH, a blood-based biomarker correlated with disease relapse [28]. In parallel, other studies have shown that lower baseline ctDNA mt detected through ddPCR, including BRAF V600E and BRAF V600K mt, may associate with higher response rates and improved survival in CM patients treated with BRAFi [29,30]. Furthermore, it has been suggested that the basal levels of ctDNA, which can also be assessed through ddPCR, may be important predictive biomarkers in CM patients who underwent immunotherapy; briefly, patients with undetectable ctDNA at baseline had superior progression-free survival (PFS) and overall survival (OS) rates than those with detectable ctDNA [31]. Nonetheless, other research groups revealed that besides the baseline ctDNA status, the dynamics of ctDNA may provide valuable information on the patient’s clinical outcome following immunotherapy [32].
ddPCR can also be used to assess the methylation status of ctDNA. DNA methylation patterns are highly dynamic during tumor progression, so that DNA methylation analysis can provide important information about disease evolution and response to treatment [26]. Although several methylation-based ctDNA LB assays have been validated for human cancers (liver, lung, and colorectal cancer), currently for CM there is no specific methylation panel that includes biomarkers associated with tumor progression or resistance to therapy [33]. However, detection of certain hypermethylated tumor suppressor genes, such as PTEN, CDKN2A, RASSF1A, and MGMT in plasma has been reported to have diagnostic value in CM patients [34]. Additionally, it was found that the methylation level of transposable element LINE-1 may be associated with poor OS in stage III CM patients [34]. Given this information, it would be interesting to verify these putative blood biomarkers through ddPCR, in order to obtain more accurate clinical information on the prognosis and disease evolution in CM patients.
Additionally, ddPCR may also be a promising methodology for evaluating miRNA expression in CM patients’ body fluids [26]. miRNAs are single-stranded RNA molecules (18–22 nucleotides in length) that can regulate the expression of their target genes by binding the complementary mRNA sequences at the 3 ’untranslated region (3′ UTR) [26]. Circulating miRNAs, released from tumor cells into the bloodstream are stable and highly accessible molecules and may be exploited as promising biomarkers in CM. Certain molecules identified through conventional methods (RT-qPCR) in the sera of CM patients, such as miR-221, miR-199a-5p, miR-33a, miR-424, and miR-206 have been reported to play valuable prognosis and diagnosis roles in CM clinical management [35]. At the moment, there are no published studies evaluating CM-related miRNAs in patients’ plasma by ddPCR; there were just a few studies with melanoma cell lines [36]. However, ddPCR has been recently applied to assess miRNA-34b/c methylation status in cfDNA in malignant pleural mesothelioma [37]. Therefore, although a nascent domain, we believe that ddPCR evaluation of CM-associated miRNAs in patients’ blood may open new perspectives in the clinical management of CM tumors.
Furthermore, ddPCR can be extremely useful in analyzing and characterizing CTCs. Dissociated either from the primary tumor or metastatic compartments into the blood, these cancer cells may offer valuable information on the tissue of origin, but also on the prognosis and clinical outcome of patients [26]. The first prospective application of ddPCR in the analysis of melanoma CTCs consists in quantifying specific melanoma-associated antigens (MAAs), such as MAGE-A3, PAX3, and MART-1, which are strong predictive biomarkers [26]. The second prospective application of this technique focuses on the digital quantification of CTC-derived transcripts, which will help predict the response to targeted therapies and immunotherapies in CM [38]. A recent study, led by Hong et al., highlighted that ddPCR evaluation of 19 melanoma CTC-derived transcripts facilitates non-invasive monitoring of tumor burden in CM patients, supporting the rational application of immunotherapy in these subjects [38]. Furthermore, they also reported that a decline in CTC score at 7 weeks is positively associated with improved OS, whereas a rise in CTC score led to poor survival in 53% of patients. Given that there is virtually no blood-based biomarker for tumor burden and the neural crest origin of melanoma cells provides unique RNA transcripts that help distinguish CTCs from normal blood cells, this ddPCR protocol offers new hopes in the fight against drug resistance in CM patients [38].
Another methodology for validating biomarkers in CM refers to the identification of CM-associated mutations in EVs through ddPCR. Released from both cancer and stromal cells, EVs are cup-shaped nanovesicles containing (mi)RNAs, DNAs, and proteins, that have key roles in metastatic niche preparation [39]. Zocco et al. used ddPCR technology to assess the BRAF V600E prognostic power in EV-derived DNA [40]. Similar to other reports, the authors found that BRAF V600E copy levels above 50 copies/mL of plasma are suggestive of a poor prognosis and that the dynamics of BRAF V600E copy numbers may be relevant for monitoring the response to BRAF inhibitors (BRAFi) in CM patients [40]. Briefly, BRAF V600E copy levels have been found to become almost undetectable after exposure to BRAFi, but grow rapidly once the tumors become refractory to therapy and the disease progresses [40]. Additionally, Clark et al. developed a ddPCR protocol for the detection of BRAF splicing variants p61, p55, p48, and p41, in cell-free RNA (cfRNA) derived from CM patients’ plasma [41]. Notably, 24 of 38 patients enrolled in the study and treated with BRAF/MEK inhibitor showed an increase in ctDNA levels as a sign of disease progression after treatment initiation. BRAF splicing variants were detected in three of these 38 patients: two patients harbored the BRAF p61 variant, while one presented the p55 variant. Remarkably, RNA isolation and analysis of EVs from CM resistant cell lines and patient plasma showed that BRAF splicing variants are specific to EVs, suggesting that besides ctDNA, RNA encapsulated in EVs may provide specific information about the tumor [41]. In parallel, another study reported that certain patients might present mutations in EVs that are undetectable in tissue, suggesting that screening of EVs-derived nucleic acids by ddPCR may provide clues on the occurrence of BRAF/MEK inhibitor therapy resistance before radiological evaluation of the tumor [39].
4. Discussion
Skin melanoma remains a devastating neoplasm. In the last decade, considerable efforts have been devoted to identify novel prognostic and predictive biomarkers and optimize CM’s therapeutic protocols [42]. The development of ddPCR, considered the latest generation of PCR, has allowed the accurate detection and quantification of low-abundance nucleic acids, opening new perspectives in the clinical management of skin tumors [43]. Research presented at American Society of Clinical Oncology (ASCO) meetings describes novel applications of this technology, mainly when used in conjunction with LB [44,45,46].
In recent years, LB, based on the analysis of circulating components derived from tumors: CTCs, exosomes, or ctDNA/ctRNA, has gained tremendous attention because of its potential to provide, in real-time, an accurate description of the genetic landscape of a tumor. This procedure can be effective in screening and diagnosis, and monitoring tumor response to systemic therapies in CM patients [47]. Liquid biopsy is also expected to replace the tissue sampling procedure since it is non-invasive, effective in the scenery of an inaccessible tumor, and eloquent for the intratumoral heterogeneity of solid tumors [48]. Increasing evidence suggests that ctDNA recapitulates the genomic complexity of the tumor, and, therefore, it might represent a non-invasive tool for assessing its omic profile [49]. Thus, ddPCR-based analysis of LB may be a promising strategy in the clinical context to improve the accuracy of diagnosis, monitoring, and the therapeutic benefit of cancer patients.
A significant area of application of ddPCR is to monitor patients for resistance mutations and changes in ctDNA levels after exposure to a particular treatment. Currently, there are no validated circulating biomarkers in CM to assess the therapeutic response in patients with advanced-stage disease [50]. However, Syeda et al. observed that pretreatment and on-treatment BRAF V600-mutant ctDNA levels may be used as predictive biomarkers of clinical outcome following targeted therapy [51]. Increased levels of BRAF V600-mutant ctDNA before and during treatment (week 4) with dabrafenib or dabrafenib/trametinib were associated with poor clinical outcomes in CM patients. A ctDNA cutoff point of ≥64 copies/mL stratified patients as high risk for shortened survival PFS (HR = 1.74, p < 0.0001) and OS (HR = 2.23, p < 0.0001), making ctDNA a valuable biomarker in CM clinical care [51]. In parallel, other studies have shown that lower baseline ctDNA mt detected through ddPCR, including BRAF V600E and BRAF V600K mt, may associate with higher response rates and improved survival in CM patients treated with BRAFi [29,30].
Furthermore, ddPCR may be used to assess the basal levels of ctDNA, which is also an important predictive biomarker in CM patients who underwent immunotherapy; briefly, patients with undetectable ctDNA at baseline had superior PFS and OS rates than those with detectable ctDNA [31]. Nonetheless, other research groups used the ddPCR assay to distinguish between tumor growth and pseudo-progression, an odyssey in cancer immunotherapy [45]. Tumors seem to grow before they are grounded by treatment, so failure to detect pseudoprogression can often lead to discontinuation of immunotherapy, even though it is effective on the tumor [45]. Lee et al. used ddPCR to quantify BRAF and NRAS mt in the ctDNA of 29 CM melanoma patients at baseline and during the first 12 wks of immunotherapy treatment. They found that all nine subjects with pseudoprogression harbored a significant decrease or undetectable ctDNA levels upon treatment exposure, whereas 18 of 20 patients with progressing neoplasms showed an increase or no change in their ctDNA levels [45]. Thus, this study suggests that the dynamics of the ctDNA levels assessed through ddPCR may provide valuable information on the patient’s clinical outcome following immunotherapy.
ddPCR is currently applied to study the biology of metastatic brain tumors. Lee and colleagues employed this assay to analyze ctDNA levels in the blood of 48 patients with advanced melanoma and brain metastases receiving immunotherapy [52]. Eight patients presented with melanoma brain metastases. Interestingly, researchers noticed that the absence of baseline ctDNA before treatment initiation was a good prognostic factor, but this did not apply to subjects harboring brain metastases. They further observed that brain metastases were smaller than those localized in other body sites and postulated that the blood-brain barrier could have filtered out the ctDNA preventing it from reaching into circulation [52]. Although the study highlighted that ddPCR-based blood ctDNA analysis might help monitor the therapeutic responses in metastatic melanoma, it may not be potent in detecting melanomas metastasizing to the brain or monitoring whether the brain tumors respond to immunotherapies [52]. However, Parietti et al. has recently highlighted an alternative route that may be used to detect the leptomeningeal dissemination of skin tumors [53]. They showed for the first time that the identification of a BRAF mt in the CSF of a melanoma patient could be suggestive for the diagnosis of leptomeningeal metastasis as the first and only site of dissemination and in the context of a normal brain magnetic resonance imaging (MRI) [53]. Thus, all this information suggests that the CSF mt analysis via ddPCR may be a valuable strategy for detecting and monitoring highly aggressive skin melanomas, even in the presence of a negative brain MRI.
The applications of ddPCR are currently expanding. Often, the ddPCR can be used to corroborate data obtained with different technologies, especially NGS and microarrays [54,55,56]. For instance, Diefenbach et al. showed that ddPCR experiments might be highly efficient to correlate ctDNA methylation data from bisulfite amplicon sequencing with ctDNA copy number variations to demonstrate that paraoxonase 3 (PON3) methylation may be a valuable biomarker of prognosis in CM in the absence of tumor mutation data for BRAF, RAS or EGFR genes [18]. ddPCR was also used to validate data obtained with microarrays in different experiments [57].
Another potential application of ddPCR is DNA methylation analysis. A relatively recent study highlighted the prognostic and predictive value of hypermethylated RASSF1A assessed through ddPCR as a circulating tumor biomarker in patients with pediatric solid tumors [58]. Some other authors employed ddPCR assay to explore circulating miRNA signatures that could be potential biomarkers in cancers [59,60]. In addition, a series of experiments used ddPCR to assess the molecular features of CTCs, highlighting that it may be a valuable strategy for monitoring disease progression and even response to immune checkpoint therapy in CM metastatic patients [38,61]. Finally, ddPCR is helpful for discriminating splicing/transcript variants in cancers, often linked to disease progression or drug resistance. Clark et al. developed a ddPCR protocol for detecting BRAF splicing variants p61, p55, p48, and p41, in cfRNA derived from CM patients’ plasma [41]. Notably, 24 of 38 patients enrolled in the study and treated with BRAF/MEK inhibitor showed an increase in ctDNA levels as a sign of disease progression following treatment initiation. BRAF splicing variants were detected in three of these 38 patients: two patients harbored the BRAF p61 variant, while one presented the p55 variant [41]. Remarkably, RNA isolation and analysis of EVs from CM resistant cell lines and patient plasma showed that BRAF splicing variants are specific to EVs, suggesting that besides ctDNA, RNA encapsulated in EVs may provide detailed information about tumor evolution [41]. In parallel, another study reported that certain patients might present mutations in EVs that are undetectable in tissue, suggesting that screening of EVs-derived nucleic acids by ddPCR may provide clues on the occurrence of BRAF/MEK inhibitor therapy resistance before radiological evaluation of the tumor [39].
5. Conclusions
The clinical management of CM has evolved in recent years towards a more personalized approach that requires an accurate assessment of the molecular alterations associated with tumor growth and evolution, as well as resistance to therapy [41]. Suitable for both archived and LB samples, ddPCR can be used for numerous omics evaluations, including absolute allele quantification, CNVs analysis, rare mutations, DNA methylation detection, transcriptomic evaluations (mRNA, miRNA), and genomic rearrangements, being of great interest to CM research [21]. Given that multi-omics integration is essential for improving the performance of prognostic and predictive models in CM, the validation of novel multi-omics biomarkers through ddPCR can play an important role in this direction [6]. As ddPCR offers outstanding opportunities for CM biomarker research, it is expected that this technology to open new avenues for precision medicine in this difficult to treat skin tumor.
Author Contributions
Conceptualization, E.-G.D. and M.N.; writing—original draft preparation, E.-G.D.; writing—review and editing, M.N.; supervision, M.N.; funding acquisition, M.N. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Core Program, implemented with the support of NASR, project PN 19.29.01.01.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Grzywa, T.M.; Paskal, W.; Włodarski, P.K. Intratumor and Intertumor Heterogeneity in Melanoma. Transl. Oncol. 2017, 10, 956–975. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.A.; Nathanson, K.L. Molecular testing in melanoma. Cancer J. 2012, 18, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Shi, X.; Zhao, Q.; Krauthammer, M.; Rothberg, B.E.G.; Ma, S. Integrated analysis of multidimensional omics data on cutaneous melanoma prognosis. Genomics 2016, 107, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Asmis, R.; Hawkins, G.A.; Howard, T.D.; Cox, L.A. The Need for Multi-Omics Biomarker Signatures in Precision Medicine. Int. J. Mol. Sci. 2019, 20, 4781. [Google Scholar] [CrossRef]
- Zhao, Q.; Shi, X.; Xie, Y.; Huang, J.; Shia, B.; Ma, S. Combining multidimensional genomic measurements for predicting cancer prognosis: Observations from TCGA. Brief. Bioinform. 2015, 16, 291–303. [Google Scholar] [CrossRef]
- Jayawardana, K.; Schramm, S.-J.; Haydu, L.; Thompson, J.F.; Scolyer, R.A.; Mann, G.J.; Müller, S.; Yang, J.Y.H. Determination of prognosis in metastatic melanoma through integration of clinico-pathologic, mutation, mRNA, microRNA, and protein information. Int. J. Cancer 2015, 136, 863–874. [Google Scholar] [CrossRef]
- Yoshikawa, S.; Kiyohara, Y.; Otsuka, M.; Kondou, R.; Nonomura, C.; Miyata, H.; Iizuka, A.; Ohshima, K.; Urakami, K.; Nagashima, T.; et al. Multi-omics Profiling of Patients with Melanoma Treated with Nivolumab in Project HOPE. Anticancer Res. 2017, 37, 1321–1328. [Google Scholar] [CrossRef][Green Version]
- Dumitru, C.; Constantin, C.; Popp, C.; Cioplea, M.; Zurac, S.; Vassu, T.; Neagu, M. Innovative array-based assay for omics pattern in melanoma. J. Immunoass. Immunochem. 2017, 38, 343–354. [Google Scholar] [CrossRef]
- Huerta, M.; Roselló, S.; Sabater, L.; Ferrer, A.; Tarazona, N.; Roda, D.; Gambardella, V.; Alfaro-Cervelló, C.; Garcés-Albir, M.; Cervantes, A.; et al. Circulating Tumor DNA Detection by Digital-Droplet PCR in Pancreatic Ductal Adenocarcinoma: A Systematic Review. Cancers 2021, 13, 994. [Google Scholar] [CrossRef]
- Ding, P.N.; Becker, T.; Bray, V.; Chua, W.; Ma, Y.; Xu, B.; Lynch, D.; de Souza, P.; Roberts, T. Plasma next generation sequencing and droplet digital PCR-based detection of epidermal growth factor receptor (EGFR) mutations in patients with advanced lung cancer treated with subsequent-line osimertinib. Thorac. Cancer 2019, 10, 1879–1884. [Google Scholar] [CrossRef]
- Mao, X.; Liu, C.; Tong, H.; Chen, Y.; Liu, K. Principles of digital PCR and its applications in current obstetrical and gynecological diseases. Am. J. Transl. Res. 2019, 11, 7209–7222. [Google Scholar] [PubMed]
- Manoj, P. Droplet digital PCR technology promises new applications and research areas. Mitochondrial DNA 2016, 27, 742–746. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Shen, S.; Jiang, H.; Chen, Z. Application of Digital PCR in Detecting Human Diseases Associated Gene Mutation. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 43, 1718–1730. [Google Scholar] [CrossRef]
- McEvoy, A.C.; Wood, B.A.; Ardakani, N.M.; Pereira, M.R.; Pearce, R.; Cowell, L.; Robinson, C.; Grieu-Iacopetta, F.; Spicer, A.J.; Amanuel, B.; et al. Droplet Digital PCR for Mutation Detection in Formalin-Fixed, Paraffin-Embedded Melanoma Tissues: A Comparison with Sanger Sequencing and Pyrosequencing. J. Mol. Diagn. 2018, 20, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, P.D.R.; Margiotti, K.; Mesoraca, A.; Giorlandino, C. Quantification of circulating microRNAs by droplet digital PCR for cancer detection. BMC Res. Notes 2020, 13, 351. [Google Scholar] [CrossRef]
- Demaree, B.; Weisgerber, D.; Dolatmoradi, A.; Hatori, M.; Abate, A.R. Direct quantification of EGFR variant allele frequency in cell-free DNA using a microfluidic-free digital droplet PCR assay. Methods Cell Biol. 2018, 148, 119–131. [Google Scholar] [CrossRef]
- Preobrazhenskaya, E.V.; Bizin, I.V.; Kuligina, E.S.; Shleykina, A.Y.; Suspitsin, E.N.; Zaytseva, O.A.; Anisimova, E.I.; Laptiev, S.A.; Gorodnova, T.V.; Belyaev, A.M.; et al. Detection of BRCA1 gross rearrangements by droplet digital PCR. Breast Cancer Res. Treat. 2017, 165, 765–770. [Google Scholar] [CrossRef]
- Diefenbach, R.; Lee, J.; Chandler, D.; Wang, Y.; Pflueger, C.; Long, G.; Scolyer, R.; Carlino, M.; Menzies, A.; Kefford, R.; et al. Hypermethylation of Circulating Free DNA in Cutaneous Melanoma. Appl. Sci. 2019, 9, 5074. [Google Scholar] [CrossRef]
- Reinert, T.; Schøler, L.V.; Thomsen, R.; Tobiasen, H.; Vang, S.; Nordentoft, I.; Lamy, P.; Kannerup, A.-S.; Mortensen, F.V.; Stribolt, K.; et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016, 65, 625–634. [Google Scholar] [CrossRef]
- Busser, B.; Lupo, J.; Sancey, L.; Mouret, S.; Faure, P.; Plumas, J.; Chaperot, L.; Leccia, M.T.; Coll, J.L.; Hurbin, A.; et al. Plasma Circulating Tumor DNA Levels for the Monitoring of Melanoma Patients: Landscape of Available Technologies and Clinical Applications. Biomed. Res. Int. 2017, 2017, 5986129. [Google Scholar] [CrossRef]
- Olmedillas-López, S.; García-Arranz, M.; García-Olmo, D. Current and Emerging Applications of Droplet Digital PCR in Oncology. Mol. Diagn. Ther. 2017, 21, 493–510. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.K.; Enderle, D.; Noerholm, M.; Breakefield, X.O.; Skog, J.K. Exosome-based liquid biopsies in cancer: Opportunities and challenges. Ann. Oncol. 2021, 32, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Malicherova, B.; Burjanivova, T.; Grendar, M.; Minarikova, E.; Bobrovska, M.; Vanova, B.; Jasek, K.; Jezkova, E.; Kapinova, A.; Antosova, M.; et al. Droplet digital PCR for detection of BRAF V600E mutation in formalin-fixed, paraffin-embedded melanoma tissues: A comparison with Cobas(®) 4800, Sanger sequencing, and allele-specific PCR. Am. J. Transl. Res. 2018, 10, 3773–3781. [Google Scholar] [PubMed]
- Salgado, C.; Roelse, C.; Nell, R.; Gruis, N.; van Doorn, R.; van der Velden, P. Interplay between TERT promoter mutations and methylation culminates in chromatin accessibility and TERT expression. PLoS ONE 2020, 15, e0231418. [Google Scholar] [CrossRef] [PubMed]
- Burjanivova, T.; Malicherova, B.; Grendar, M.; Minarikova, E.; Dusenka, R.; Vanova, B.; Bobrovska, M.; Pecova, T.; Homola, I.; Lasabova, Z.; et al. Detection of BRAFV600E Mutation in Melanoma Patients by Digital PCR of Circulating DNA. Genet. Test. Mol. Biomarkers 2019, 23, 241–245. [Google Scholar] [CrossRef]
- Huang, S.K.; Hoon, D.S.B. Liquid biopsy utility for the surveillance of cutaneous malignant melanoma patients. Mol. Oncol. 2016, 10, 450–463. [Google Scholar] [CrossRef]
- Shinozaki, M.; O’Day, S.J.; Kitago, M.; Amersi, F.; Kuo, C.; Kim, J.; Wang, H.-J.; Hoon, D.S.B. Utility of circulating B-RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clin. Cancer Res. 2007, 13, 2068–2074. [Google Scholar] [CrossRef]
- Tsao, S.C.-H.; Weiss, J.; Hudson, C.; Christophi, C.; Cebon, J.; Behren, A.; Dobrovic, A. Monitoring response to therapy in melanoma by quantifying circulating tumour DNA with droplet digital PCR for BRAF and NRAS mutations. Sci. Rep. 2015, 5, 11198. [Google Scholar] [CrossRef]
- Santiago-Walker, A.; Gagnon, R.; Mazumdar, J.; Casey, M.; Long, G.V.; Schadendorf, D.; Flaherty, K.; Kefford, R.; Hauschild, A.; Hwu, P.; et al. Correlation of BRAF Mutation Status in Circulating-Free DNA and Tumor and Association with Clinical Outcome across Four BRAFi and MEKi Clinical Trials. Clin. Cancer Res. 2016, 22, 567–574. [Google Scholar] [CrossRef]
- Sanmamed, M.F.; Fernández-Landázuri, S.; Rodríguez, C.; Zárate, R.; Lozano, M.D.; Zubiri, L.; Perez-Gracia, J.L.; Martín-Algarra, S.; González, A. Quantitative cell-free circulating BRAFV600E mutation analysis by use of droplet digital PCR in the follow-up of patients with melanoma being treated with BRAF inhibitors. Clin. Chem. 2015, 61, 297–304. [Google Scholar] [CrossRef]
- Seremet, T.; Jansen, Y.; Planken, S.; Njimi, H.; Delaunoy, M.; El Housni, H.; Awada, G.; Schwarze, J.K.; Keyaerts, M.; Everaert, H.; et al. Undetectable circulating tumor DNA (ctDNA) levels correlate with favorable outcome in metastatic melanoma patients treated with anti-PD1 therapy. J. Transl. Med. 2019, 17, 303. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Long, G.V.; Boyd, S.; Lo, S.; Menzies, A.M.; Tembe, V.; Guminski, A.; Jakrot, V.; Scolyer, R.A.; Mann, G.J.; et al. Circulating tumour DNA predicts response to anti-PD1 antibodies in metastatic melanoma. Ann. Oncol. 2017, 28, 1130–1136. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, R.J.; Lee, J.H.; Rizos, H. Methylated circulating tumor DNA as a biomarker in cutaneous melanoma. Melanoma Manag. 2020, 7, MMT46. [Google Scholar] [CrossRef] [PubMed]
- Micevic, G.; Theodosakis, N.; Bosenberg, M. Aberrant DNA methylation in melanoma: Biomarker and therapeutic opportunities. Clin. Epigenetics 2017, 9, 34. [Google Scholar] [CrossRef]
- Neagu, M.; Constantin, C.; Cretoiu, S.M.; Zurac, S. miRNAs in the Diagnosis and Prognosis of Skin Cancer. Front. cell Dev. Biol. 2020, 8, 71. [Google Scholar] [CrossRef]
- Poenitzsch Strong, A.M.; Setaluri, V.; Spiegelman, V.S. MicroRNA-340 as a modulator of RAS-RAF-MAPK signaling in melanoma. Arch. Biochem. Biophys. 2014, 563, 118–124. [Google Scholar] [CrossRef]
- Sato, H.; Soh, J.; Aoe, K.; Fujimoto, N.; Tanaka, S.; Namba, K.; Torigoe, H.; Shien, K.; Yamamoto, H.; Tomida, S.; et al. Droplet digital PCR as a novel system for the detection of microRNA-34b/c methylation in circulating DNA in malignant pleural mesothelioma. Int. J. Oncol. 2019, 54, 2139–2148. [Google Scholar] [CrossRef]
- Hong, X.; Sullivan, R.J.; Kalinich, M.; Kwan, T.T.; Giobbie-Hurder, A.; Pan, S.; LiCausi, J.A.; Milner, J.D.; Nieman, L.T.; Wittner, B.S.; et al. Molecular signatures of circulating melanoma cells for monitoring early response to immune checkpoint therapy. Proc. Natl. Acad. Sci. USA 2018, 115, 2467–2472. [Google Scholar] [CrossRef]
- Yap, S.A.; Münster-Wandowski, A.; Nonnenmacher, A.; Keilholz, U.; Liebs, S. Analysis of cancer-related mutations in extracellular vesicles RNA by Droplet DigitalTM PCR. Biotechniques 2020, 69, 99–107. [Google Scholar] [CrossRef]
- Zocco, D.; Bernardi, S.; Novelli, M.; Astrua, C.; Fava, P.; Zarovni, N.; Carpi, F.M.; Bianciardi, L.; Malavenda, O.; Quaglino, P.; et al. Isolation of extracellular vesicles improves the detection of mutant DNA from plasma of metastatic melanoma patients. Sci. Rep. 2020, 10, 15745. [Google Scholar] [CrossRef]
- Clark, M.E.; Rizos, H.; Pereira, M.R.; McEvoy, A.C.; Marsavela, G.; Calapre, L.; Meehan, K.; Ruhen, O.; Khattak, M.A.; Meniawy, T.M.; et al. Detection of BRAF splicing variants in plasma-derived cell-free nucleic acids and extracellular vesicles of melanoma patients failing targeted therapy therapies. Oncotarget 2020, 11, 4016–4027. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, P.F.; Cocorocchio, E. Novel Biomarkers and Druggable Targets in Advanced Melanoma. Cancers 2022, 14, 81. [Google Scholar] [CrossRef] [PubMed]
- Syeda, M.M.; Wiggins, J.M.; Corless, B.; Spittle, C.; Karlin-Neumann, G.; Polsky, D. Validation of Circulating Tumor DNA Assays for Detection of Metastatic Melanoma. Methods Mol. Biol. 2020, 2055, 155–180. [Google Scholar] [CrossRef]
- Murad, A.M.; Carneiro, J.G.; Casali-da-Rocha, J.C. A single institution experience with droplet digital polymerase chain reaction (dd-PCR) liquid biopsy (LB) for therapeutic decision in advanced solid tumors. J. Clin. Oncol. 2021, 39, 3038. [Google Scholar] [CrossRef]
- Lee, J.H.J.; Long, G.V.; Menzies, A.M.; Guminski, A.D.; Kefford, R.; Rizos, H.; Carlino, M.S. Analysis of circulating tumor DNA (ctDNA) in pseudoprogression in anti-PD1 treated metastatic melanoma (MM). J. Clin. Oncol. 2017, 35, 9546. [Google Scholar] [CrossRef]
- Johann, D.J.; Shin, I.J.; Peterson, E.; Steliga, M.V.; Muesse, J.; Marino, K.; Laun, S.; Greisman, V.; Emmert-Buck, M.; Tangrea, M. Synergizing microdissection with ddPCR to advance precision oncology. J. Clin. Oncol. 2021, 39, e15083. [Google Scholar] [CrossRef]
- Kamińska, P.; Buszka, K.; Zabel, M.; Nowicki, M.; Alix-Panabières, C.; Budna-Tukan, J. Liquid Biopsy in Melanoma: Significance in Diagnostics, Prediction and Treatment Monitoring. Int. J. Mol. Sci. 2021, 22, 9714. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef]
- Sacco, A.; Forgione, L.; Carotenuto, M.; De Luca, A.; Ascierto, P.A.; Botti, G.; Normanno, N. Circulating Tumor DNA Testing Opens New Perspectives in Melanoma Management. Cancers 2020, 12, 2914. [Google Scholar] [CrossRef]
- Dobre, E.-G.; Constantin, C.; Costache, M.; Neagu, M. Interrogating Epigenome toward Personalized Approach in Cutaneous Melanoma. J. Pers. Med. 2021, 11, 901. [Google Scholar] [CrossRef]
- Syeda, M.M.; Wiggins, J.M.; Corless, B.C.; Long, G.V.; Flaherty, K.T.; Schadendorf, D.; Nathan, P.D.; Robert, C.; Ribas, A.; Davies, M.A.; et al. Circulating tumour DNA in patients with advanced melanoma treated with dabrafenib or dabrafenib plus trametinib: A clinical validation study. Lancet Oncol. 2021, 22, 370–380. [Google Scholar] [CrossRef]
- Lee, J.H.J.; Menzies, A.M.; Carlino, M.S.; Kefford, R.; Scolyer, R.A.; Long, G.V.; Rizos, H. Circulating tumor DNA (ctDNA) in metastatic melanoma (MM) patients (pts) with brain metastases (mets). J. Clin. Oncol. 2019, 37, 9581. [Google Scholar] [CrossRef]
- Parietti, M.; Marra, E.; Ribero, S.; Abate, S.O.; Francia di Celle, P.; Rudà, R.; Quaglino, P.; Fierro, M.T. Leptomeningeal dissemination as a first sign of progression in metastatic melanoma: A diagnostic lesson. Melanoma Res. 2022, 32, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Coccaro, N.; Tota, G.; Anelli, L.; Zagaria, A.; Specchia, G.; Albano, F. Digital PCR: A Reliable Tool for Analyzing and Monitoring Hematologic Malignancies. Int. J. Mol. Sci. 2020, 21, 3141. [Google Scholar] [CrossRef]
- Boulos, H.; Tell, R.; Beaubier, N.; Blidner, R. Greater than two coexisting mutations in KRAS and NRAS identified in the circulating tumor DNA fraction of liquid biopsies by NGS and confirmed with ddPCR. J. Clin. Oncol. 2020, 38, e15563. [Google Scholar] [CrossRef]
- Galbiati, S.; Damin, F.; Ferraro, L.; Soriani, N.; Burgio, V.; Ronzoni, M.; Gianni, L.; Ferrari, M.; Chiari, M. Microarray Approach Combined with ddPCR: An Useful Pipeline for the Detection and Quantification of Circulating Tumour dna Mutations. Cells 2019, 8, 769. [Google Scholar] [CrossRef]
- Villegas-Ruíz, V.; Olmos-Valdez, K.; Castro-López, K.A.; Saucedo-Tepanecatl, V.E.; Ramírez-Chiquito, J.C.; Pérez-López, E.I.; Medina-Vera, I.; Juárez-Méndez, S. Identification and Validation of Novel Reference Genes in Acute Lymphoblastic Leukemia for Droplet Digital PCR. Genes 2019, 10, 376. [Google Scholar] [CrossRef]
- van Zogchel, L.M.J.; Lak, N.S.M.; Verhagen, O.J.H.M.; Tissoudali, A.; Gussmalla Nuru, M.; Gelineau, N.U.; Zappeij-Kannengieter, L.; Javadi, A.; Zijtregtop, E.A.M.; Merks, J.H.M.; et al. Novel Circulating Hypermethylated RASSF1A ddPCR for Liquid Biopsies in Patients With Pediatric Solid Tumors. JCO Precis. Oncol. 2021, 1738–1748. [Google Scholar] [CrossRef]
- Crimi, S.; Falzone, L.; Gattuso, G.; Grillo, C.M.; Candido, S.; Bianchi, A.; Libra, M. Droplet Digital PCR Analysis of Liquid Biopsy Samples Unveils the Diagnostic Role of hsa-miR-133a-3p and hsa-miR-375-3p in Oral Cancer. Biology 2020, 9, 379. [Google Scholar] [CrossRef]
- Laprovitera, N.; Riefolo, M.; Porcellini, E.; Durante, G.; Garajova, I.; Vasuri, F.; Aigelsreiter, A.; Dandachi, N.; Benvenuto, G.; Agostinis, F.; et al. MicroRNA expression profiling with a droplet digital PCR assay enables molecular diagnosis and prognosis of cancers of unknown primary. Mol. Oncol. 2021, 15, 2732–2751. [Google Scholar] [CrossRef]
- Aya-Bonilla, C.A.; Morici, M.; Hong, X.; McEvoy, A.C.; Sullivan, R.J.; Freeman, J.; Calapre, L.; Khattak, M.A.; Meniawy, T.; Millward, M.; et al. Detection and prognostic role of heterogeneous populations of melanoma circulating tumour cells. Br. J. Cancer 2020, 122, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).