Detection and Identification of Lactic Acid Bacteria in Semi-Finished Beer Products Using Molecular Techniques †
Abstract
1. Introduction
2. Materials and Methods
2.1. Beer Samples, Transportation and Fermentation Procedures
2.2. Microbiological Analyses
2.3. Molecular Analyses
3. Results and Discussion
3.1. Microbiological Results
3.2. Molecular Identification of LAB
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| LAB | Lactic Acid Bacteria |
| TVC | Total Viable Count |
| PCA | Plate Count Agar |
| RBC | Rose Bengal Chloramphenicol |
| MRS | Man Rogosa Sharpe |
| PCR | Polymerase Chain Reaction |
| CFU | Colony Forming Unit |
| VBNC | Viable But Non-Culturable |
References
- Farber, M.; Barth, R. Mastering Brewing Science; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2019; pp. 1–4. [Google Scholar]
- Hill, E.A. Brewing Microbiology—Managing Microbes, Ensuring Quality and Valorising Waste; Woodhead Publishing Limited: Shaxton, UK; CRC Press LCC: Boca Raton, FL, USA, 2015; pp. 6–7. [Google Scholar]
- Briggs, D.E.; Boulton, C.A.; Brooks, P.A.; Stevens, R. Brewing: Science and Practice; Woodhead Publishing Limited: Shaxton, UK; CRC Press LCC: Boca Raton, FL, USA, 2004; pp. 5–6. [Google Scholar]
- Suzuki, K.; Iijima, K.; Sakamoto, K.; Sami, M.; Yamashita, H. A review of hop resistance in beer spoilage lactic acid bacteria. J. Inst. Brew. 2006, 112, 173–191. [Google Scholar] [CrossRef]
- Suzuki, K. Gram-positive spoilage bacteria in brewing. Brew. Microbiol. 2015, 89, 141–173. [Google Scholar]
- Barroso, E.; van de Wiele, T.; Jiménez-Girón, A.; Muñoz-González, I.; Martín-Alvarez, P.J.; Moreno-Arribas, M.V.; Bartolomé, B.; Peláez, C.; Martínez-Cuesta, M.C.; Requena, T. Lactobacillus plantarum IFPL935 impacts colonic metabolism in a simulator of the human gut microbiota during feeding with red wine polyphenols. Appl. Microbiol. Biotechnol. 2014, 98, 6805–6815. [Google Scholar] [CrossRef] [PubMed]
- Filannino, P.; Bai, Y.; Di Cagno, R.; Gobbetti, M.; Gänzle, M.G. Metabolism of phenolic compounds by Lactobacillus spp. during fermentation of cherry juice and broccoli puree. Food Microbiol. 2015, 46, 272–279. [Google Scholar] [CrossRef] [PubMed]
- HBA, S.A. (Hellenic Breweries of Atalanti S.A.). Brewing Steps. Available online: https://www.eza.gr/en/brewing/ (accessed on 7 September 2021).
- Gevers, D.; Huys, G.; Swings, J. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 2001, 205, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Devolli, A.; Kodra, M.; Shahinasi, E.; Stafasani, M.; Dara, F. Determination of Optiman Kieselguhr Doses to Improve Beer Filtration. In Proceedings of the UBT International Conference, Durres, Durrës, Albania, 28–30 October 2016; pp. 11–16. [Google Scholar]
- Geissler, A.J.; Behr, J.; Kamp, K.v.; Vogel, R.F. Metabolic strategies of beer spoilage lactic acid bacteria in beer. Int. J. Food Microbiol. 2016, 216, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Maifreni, M.; Frigo, F.; Bartolomeoli, I.; Buiatti, S.; Picon, S.; Marino, M. Bacterial biofilm as a possible source of contamination in the microbrewery environment. Food Control 2015, 50, 809–814. [Google Scholar] [CrossRef]
- Liu, J.; Deng, Y.; Li, L.; Li, B.; Li, Y.; Zhou, S.; Shirtliff, M.E.; Xu, Z.; Peters, B.M. Discovery and control of culturable and viable but non-culturable cells of a distinctive Lactobacillus harbinensis strain from spoiled beer. Sci. Rep. 2018, 8, 11446. [Google Scholar] [CrossRef] [PubMed]
- Back, W. Brewery. In Colour Atlas and Handbook of Beverage Biology; Back, W., Ed.; Verlag Hans Carl: Nürnberg, Germany, 2005. [Google Scholar]
- Kern, C.C.; Vogel, R.F.; Behr, J. Differentiation of Lactobacillus brevis strains using Matrix-Assisted-Laser-Desorption-Ionization-Time-of-Flight Mass Spectrometry with respect to their beer spoilage potential. Food Microbiol. 2014, 40, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Hutzler, M.; Müller-Auffermann, K.; Koob, J.; Riedl, R.; Jacob, F. Beer spoiling microorganisms—A current overview. Brauwelt Int. 2013, 1, 23–25. [Google Scholar]
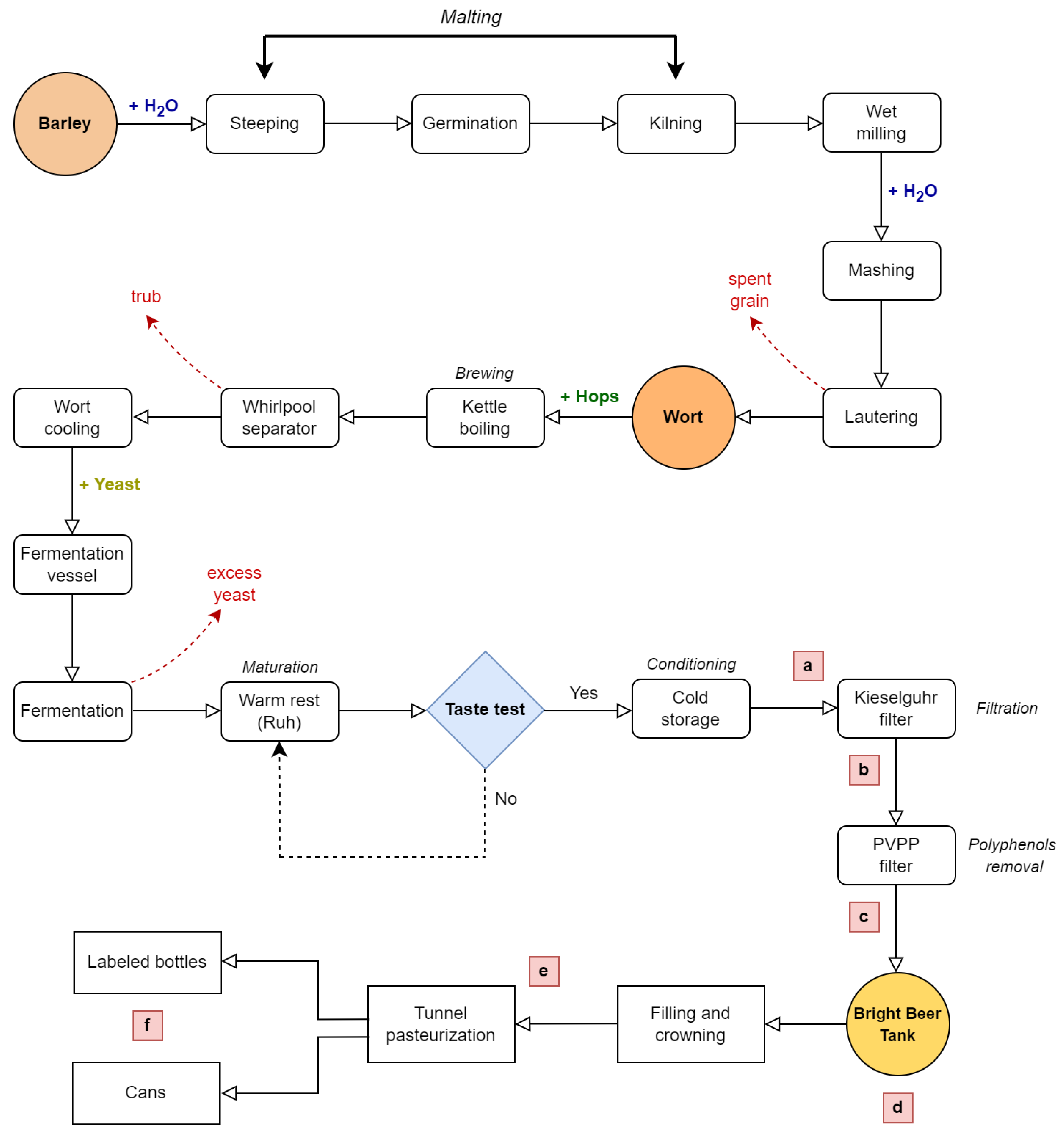
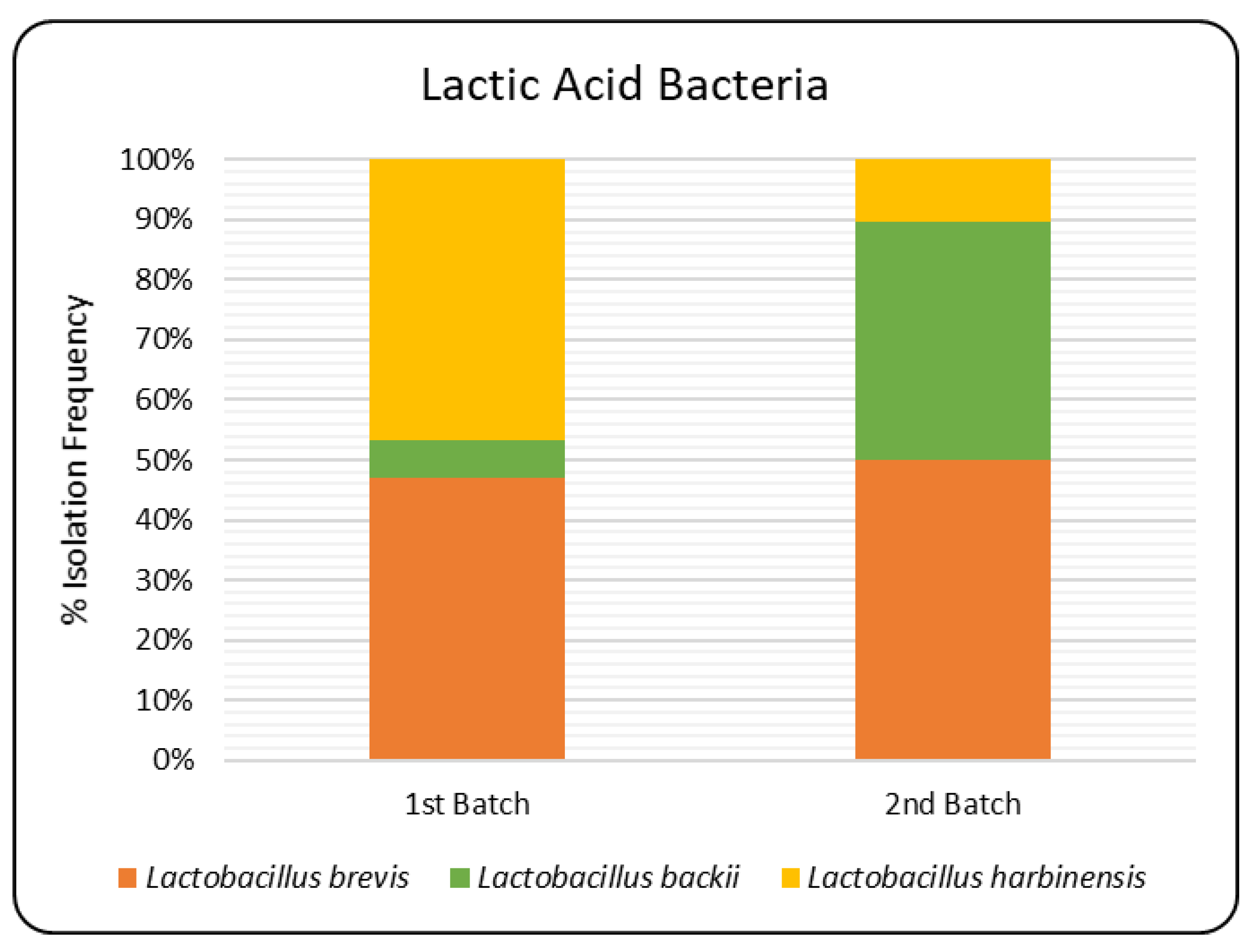
| Microorganism | Batch | Sampling Points (log CFU/mL) | |||||
|---|---|---|---|---|---|---|---|
| a * | b | c | d | e | f | ||
| Yeasts | 1st | 5.40 | <2.0 | <2.0 | <2.0 | <2.0 | <2.0 |
| 2nd | 4.98 | 2.36 | <1.0 | <1.0 | <1.0 | <1.0 | |
| LAB | 1st | 1.52 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 |
| 2nd | 3.44 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsekouras, G.; Tryfinopoulou, P.; Panagou, E.Z. Detection and Identification of Lactic Acid Bacteria in Semi-Finished Beer Products Using Molecular Techniques. Biol. Life Sci. Forum 2021, 6, 122. https://doi.org/10.3390/Foods2021-11046
Tsekouras G, Tryfinopoulou P, Panagou EZ. Detection and Identification of Lactic Acid Bacteria in Semi-Finished Beer Products Using Molecular Techniques. Biology and Life Sciences Forum. 2021; 6(1):122. https://doi.org/10.3390/Foods2021-11046
Chicago/Turabian StyleTsekouras, Georgios, Paschalitsa Tryfinopoulou, and Efstathios Z. Panagou. 2021. "Detection and Identification of Lactic Acid Bacteria in Semi-Finished Beer Products Using Molecular Techniques" Biology and Life Sciences Forum 6, no. 1: 122. https://doi.org/10.3390/Foods2021-11046
APA StyleTsekouras, G., Tryfinopoulou, P., & Panagou, E. Z. (2021). Detection and Identification of Lactic Acid Bacteria in Semi-Finished Beer Products Using Molecular Techniques. Biology and Life Sciences Forum, 6(1), 122. https://doi.org/10.3390/Foods2021-11046







