1. Introduction
Cell suspensions are a convenient model to investigate the interactions of plants and microbes, including those existing for extended periods. Cell suspensions are widely used model systems to examine secondary metabolic pathways, enzyme induction, gene expression, degradation of alien compounds, and problems in cytology. Across the globe, more than a hundred plant species transferred to suspension cultures are used in the biosynthetic industry to produce economically important substances [
1]. Special requirements are placed on culture conditions, culture purity during transfers, and culture protection against microbial contamination.
It is important to ensure that the growing suspension culture is sterile. However, in the isolation of an explant and in its introduction into cultures, bacteria may remain, which initially coexist with the plant as endo- and exosymbionts. The bacteria may be transferred to the suspension culture and may latently coexist with it. The major sources of such contamination are the incorrect preparation of materials for culturing, errors during transfers, and the disruption of culture conditions [
2]. Some plant cell suspensions coexist with fungi and bacteria for a long time [
3]. In this context, of major importance is the question as to whether the bacteria found in plant suspension cultures under specific conditions are endo- or exosymbionts.
The making of artificial associations, in which cell systems are prepared by introducing microorganisms into populations of plant cells being cultured, is a comparatively young direction in cell engineering [
4]. It is presupposed that cells and their populations should acquire new, useful properties owing to microbial presence. Such plant–microbe associations can be intercellular (exosymbiotic) as well as intracellular (endosymbiotic).
Arabidopsis thaliana (
A. thaliana) is a model plant used commonly in genetic and molecular biological research. Many studies, especially those reporting the generation of transgenic plants, have used cultured isolated tissues of
A. thaliana [
5]. In the standard maintenance of an
A. thaliana suspension culture, transfers are made every 10 days, with no visual signs of bacterial or other contamination being observed [
6]. However, when growth is extended to 20 and more days or when the culture conditions (temperature, aeration, etc.) are disrupted, the medium thickens and gets turbid and suspended matter appears in the supernatant liquid. This makes it possible to assume that the culture contains a bacterial microflora at this stage.
Here, we tested a suspension culture of A. thaliana cells for the presence of bacteria. The tests were followed by the identification of the bacteria.
2. Experiments
Research object. The major research object were microorganisms isolated from a suspension culture of
A. thaliana (L.) Heynh. (wild type; Columbia ecotype, Col-0). The culture was supplied by the All-Russia Collection of Cell Cultures of Higher Plants (Institute of Plant Physiology, Russian Academy of Sciences). The culture was grown in the Shenk and Hildebrandt’s (1972) liquid medium, with an addition of 1 mg l-1 of 2,4-D, 0.1 mg l-1 of kinetin, and 100 mg l-1 of meso-inositol [
7]. Growth was conducted at 25 ± 1 °C in constantly stirred glass flasks in the dark. The passage duration was 10 days. All work with the suspension culture was carried out in a laminar box. Before culturing, the medium was autoclaved at 1.5 atm for 30 min. As the culture grew, the growth was controlled visually, and the presence of bacteria was checked by microscopy.
Confocal microscopy. Cell suspensions were observed with a TCS SP5 confocal laser scanning microscope (Leica, Microsystems, Wetzlar, Germany) by using the fluorescent dye FM1-43 (Invitrogen, Waltham, MA, USA) [
8]. Test tubes containing a 10-day-old culture of
A. thaliana received a 0.1% FM1-43 solution in 10 mM phosphate-buffered saline (PBS; pH 7.2). After staining for 2 min, the culture was washed with PBS. DNA-containing structures were detected with the fluorescent dye SYTO 17 (Invitrogen, Waltham, MA, USA).
Transmission electron microscopy (TEM). Samples for TEM were taken from a 10-day-old Arabidopsis culture. For negative staining, a 2% uranyl acetate solution was used. TEM was done with a Libra 120 transmission electron microscope (Carl Zeiss, Oberkochen, Germany).
Isolation of bacteria. A 10-day-old plant cell culture was filtered, and bacteria were isolated by centrifugation. After sedimentation with a 5810 R centrifuge (Eppendorf, Framingham, MA, USA) at 3000× g for 30 min, the obtained supernatant was centrifuged again (15,000× g; 30 min). The bacteria pellet was resuspended in PBS and kept in a 5% formaldehyde solution for further immunization.
Cell size determination. Cell sizes were determined by TEM and dynamic light scattering (Zetasizer Nano-ZS, Malvern, UK). Samples were prepared by the stepwise centrifugation of a 10-day-old Arabidopsis culture. After the first centrifugation (3000× g; 30 min), the supernatant liquid was passed through a small-pore-size paper filter to clean it free of cellular debris. The culture was then centrifuged again (15,000× g; 30 min), and the sediment was resuspended in PBS.
Identification of bacteria. Bacteria were identified on the basis of their 16S rRNA sequence at the “Syntol science and production complex” (Russia) by polymerase chain reaction (PCR) with a set of universal primers.
Culturing of bacteria. For culturing the isolated cells and examining their properties, we used three media: medium no. 523, Luria–Bertani medium, and blood agar. The cultured cells were stained with Gram and Ziehl–Neelsen stain. The presence of bacterial spores in the suspension culture was checked by the method of Peshkov. Bacteria were viewed with a DMI 3000B inverted microscope (Leica, Germany) fitted with an optical system for integrated modulation contrast.
Immunochemical identification of bacteria. For the specific detection of bacteria, we used rabbit polyclonal antibodies raised against the strain under study after fixation with 3% formaldehyde. Rabbits were immunized intramuscularly three times at 2-week intervals with Freund’s complete adjuvant (
v:
v, 1:1). A week after the last immunization, the animals were bled. The immunoglobulin fraction of the antiserum was obtained by ammonium sulfate precipitation (30%–70% of the final saturation); this was followed by ion-exchange chromatography on a Mono-Q anion-exchange column (Pharmacia, Uppsala, Sweden). The performance of the resultant antibodies was tested by dot assay. Two microliters of successive twofold dilutions of the bacterial sample were applied to a nitrocellulose membrane. The attachment of the bacteria to the membrane was tested by nonspecific staining with Ponso S. When the result was positive, the nitrocellulose was placed in a blocking buffer (2% skimmed milk powder in PBS) for 15 min. After blocking, the nitrocellulose strips were washed three times with PBS. Next, the samples were placed in an antiserum (2 µg mL
−1) solution and were incubated at 37 ± 2 °C for 1 h. Antigen–antibody binding was detected with colloidal-gold-conjugated goat antirabbit antibodies [
9].
3. Results and Discussion
To test the assumption that the suspension culture of Arabidopsis contained contaminant bacteria, we used confocal microscopy, because it permits the preparation of thin high-resolution optical sections and the three-dimensional (3D) reconstruction of the resultant images. It was found that the plant cells had a prolate shape and a size of about 10–40 µm. In the field of view, there also were smaller objects (about 1 µm in size). In the aging culture at the stage of degeneration and decrease in cytoplasmic volume, these particles moved actively between the cell wall and plasmalemma during plasmolysis. Such motility is untypical of plant cells or their organelles. Subsequent staining with SYTO 17 (staining of DNA/RNA) helped to detect, alongside Arabidopsis cell nuclei, structures that contained nucleic acids. The structures were untypical of plant cells, as they were round and about 1 µm in diameter.
To test for the presence of bacteria, we used several microbiological staining procedures. Peshkov staining enabled the detection of bacterial spores in the culture liquid of Arabidopsis cells. Gram staining enabled the bacteria to be characterized as gram-positive. Ziehl–Neelsen staining showed that the bacteria were not acid-tolerant.
We next chose nutrient media to culture the bacteria. Bacteria grew only when the
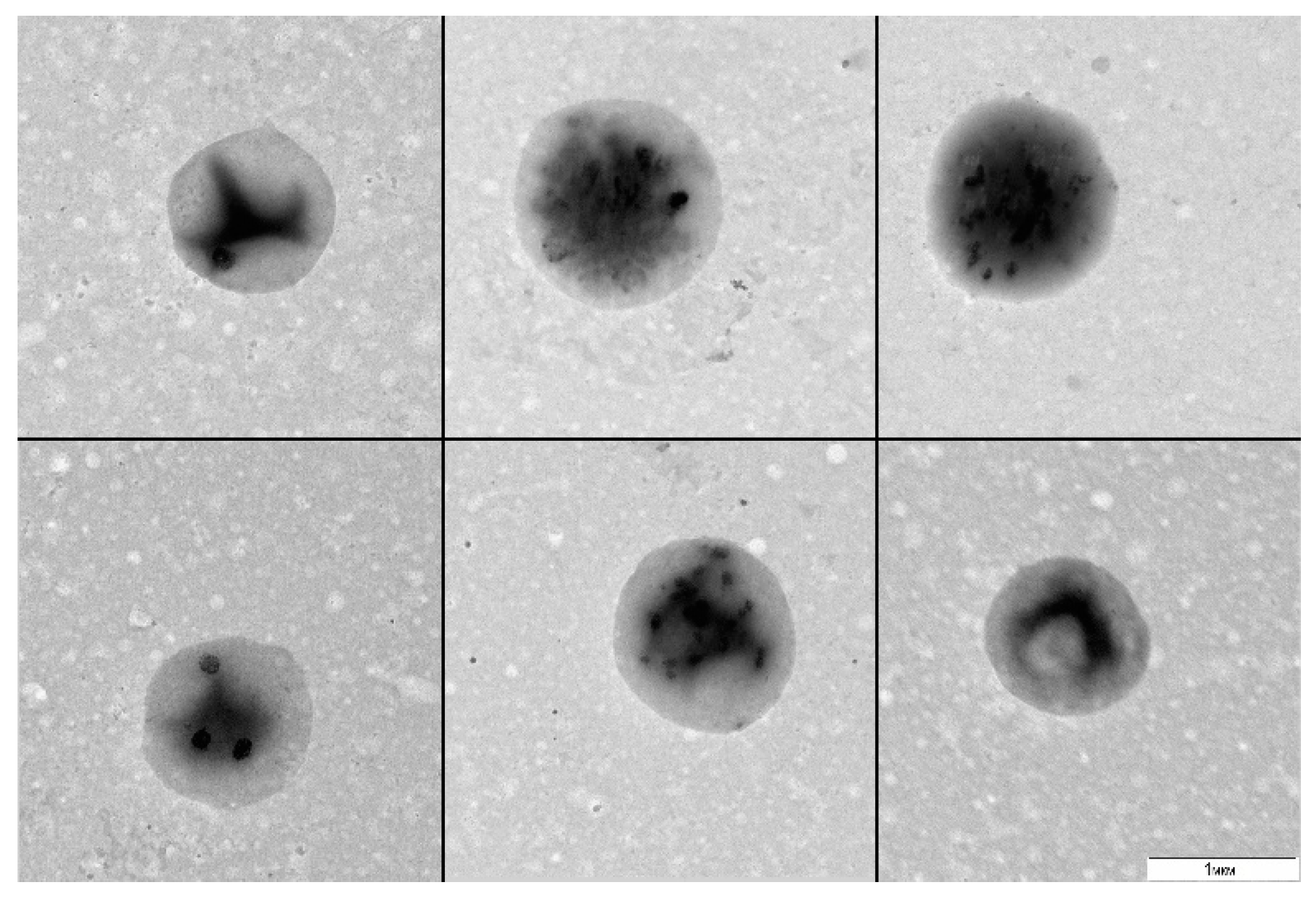
A. thaliana cell suspension was cultured without stirring or transfers for 20 and more days (stress conditions) and was subsequently plated in a liquid 523 medium. After 30 days of culturing, we observed visual signs of bacterial growth: the medium got turbid. The examination of the resultant material by TEM revealed 1 µm round bacteria with a narrow size distribution (
Figure 1).
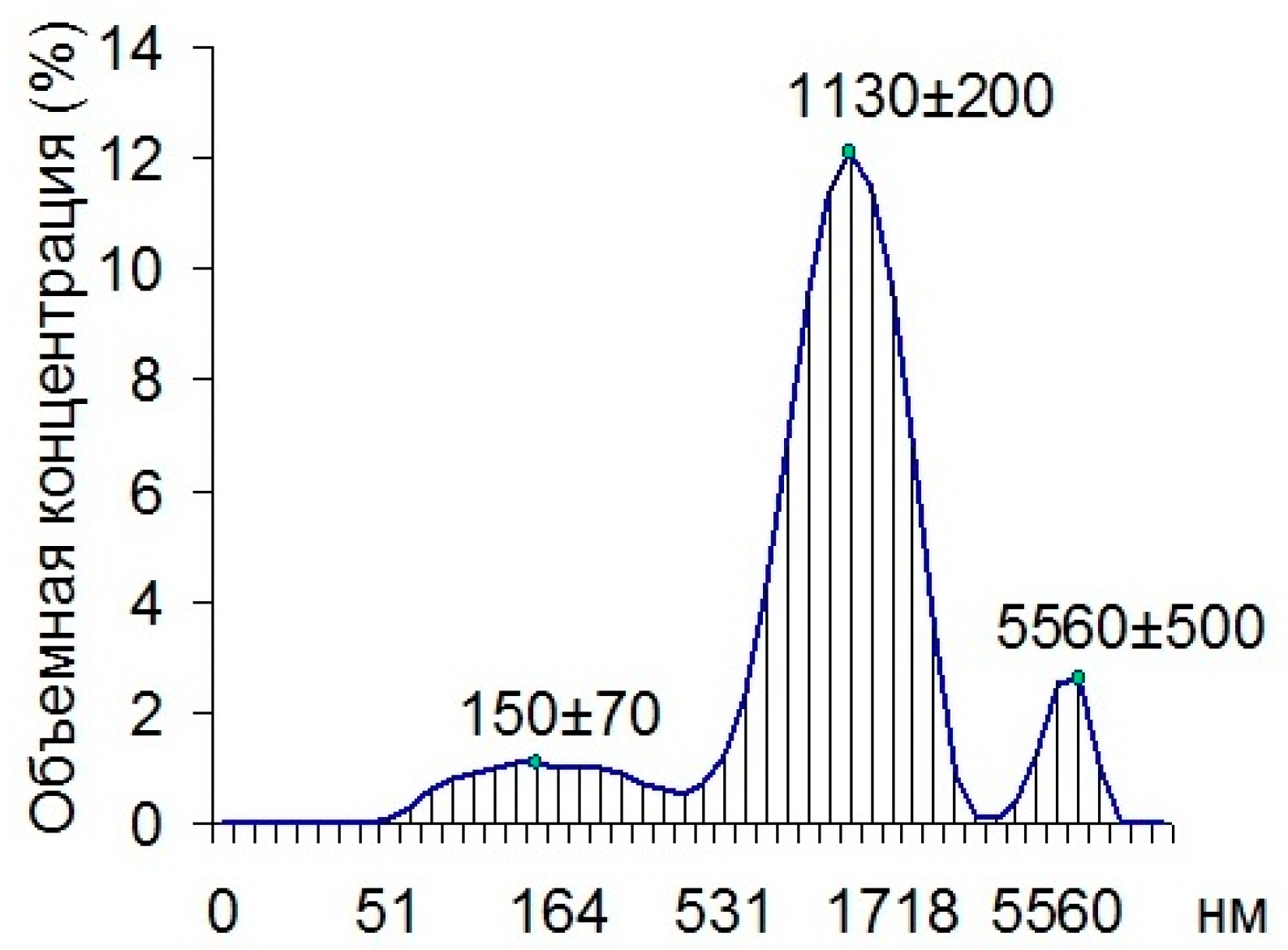
Using dynamic light scattering, we found that 80.7% of all the particles in the cuvette with the material being examined were composed of 1130 nm particles, 11.4% of the particles were composed of 150 nm particles, and 7.9% of the particles were made up of 5560 nm particles (
Figure 2).
The identification was conducted by PCR on the basis of 16S rRNA by using a set of universal primers. The resultant sequence was as follows:
TGGGTCTAATACCGGATATGACAAGGAACCGCATGGTTTTTTGTGGAAAGGGTTTGTACTGGTTTTAGATGGGCTCACGGCCTATCAGCTTGTTGGTGGGGTAATGGCTCACCAAGGCGACGACGGGTAGCCGGCCTGAGAGGGTGACCGGCCACACTGGGACTGAGACACGGCCCARACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCGACGCCGCGTGAGGGATGACGGCCTTCGGGTTGTAAACCTCTTTCAGCAGGGAAGAAGCGAAAGTGACGGTACCTGCAGAAGAAGCGCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGCGCGAGCGTTGTCCGGAATTATTGGGCGTAAAGAGCTTGTAGGCGGTTTGTTGCGTCTGCTGTGAAAGACCGGGGCTTAACCCCGGTATTGCAGTGGGTACGGGCAGACTAGAGTGCAGTAGGGGAGACTGGAATTCCTGGTGTAGCGGTGAAATGCGCAGATATCAGGAGGAACACCGATGGCGAAGGCAGGTCTCTGGGCTGTAACTGACGCTGAGAAGCGAAAGCATGGGGAGCGAACAGGATTAGATACCCTGGTAGTCCATGCCGTAAACGTTGGGCACTAGGTGTgGGGGGACATTCCACGTTTTCCGCGCCGTAGCTAACGCATTAAGTGCCCCGCCTGGGGAGTACGGCCGCAAGGCTWAAACTCAAAGGAATTGACGGGGGCCCGCACAAGCGGCGGAGCATGCGGATTAATTCGATGCAACGCGAAGAACCTTACCAAGGCTTGACATATACTGGATCGCCTCAGAGATGGGGTTTCCCTTCGGGGCTGGTATACAGGTGGTGCATGGTTGTCGTCAGCTCGTGTCGTGAGATGTTGGGTTAAGTCCCGCAACGAGCGCAACCCTCGTTYTATGTTGCCAGCACGTTATGGTGGGGACTCATAGGAGACTGCCGGGGTCAACTCGGAGGAAGGTGGGGATGACGTCAAATCATCATGCCCCTTATGTCTTGGGCTTCACGCATGCTACAATGGCCGGTACAAAGGGTTGCGATACTGTGAGGTTGAGCTAATCCC.
On the basis of its 16S rRNA sequence (identity: 99.73%), the isolate corresponded to the species
Rothia amarae (
R. amarae). The strain used for comparison was strain J18, which, according to GenBank, is a type strain of
R. amarae [
10].
After the isolate was identified, it was cultured on blood agar—the optimal medium for
R. amarae growth [
10]. The obtained isolate was loop-seeded on blood agar and was placed in a thermostat set at 25 ± 1 °C (a temperature necessary for plant cell growth) and also at 30 ± 2 °C [
10]. In either case, the result was positive—the bacteria grew.
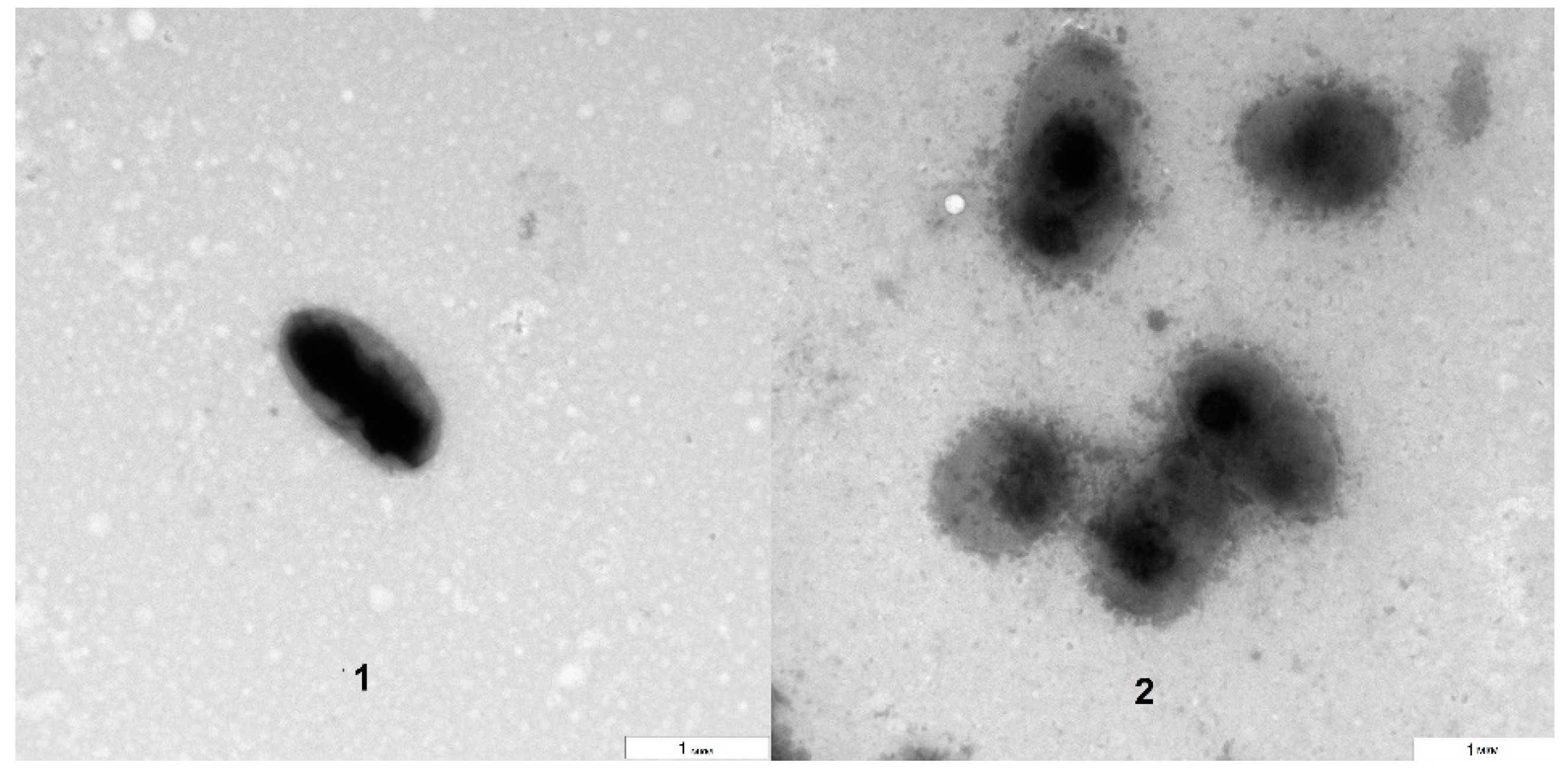
The TEM analysis showed that the bacteria grown on blood agar were morphologically similar to those isolated directly from the
Arabidopsis suspension culture (
Figure 3). However, the electron densities of the bacterial surfaces differed to some extent. The approximate size of the bacteria (about 1 µm) was preserved, as was their aggregation as dyads and tetrads. The obtained results agreed with the literature data that describe
Rothia cells as being gram-positive, round, and dyad- and tetrad-forming and their colonies as being milky white, slimy and having a smooth concave surface [
11]. Thus, we have shown that it is possible to grow the isolated bacteria outside a plant suspension culture, specifically on blood agar in Petri dishes.
The correspondence between the bacteria grown on blood agar and those isolated from the suspension culture was confirmed by raising polyclonal antibodies in rabbits. For immunization, we used a suspension of fixed cells that were washed free of formaldehyde.
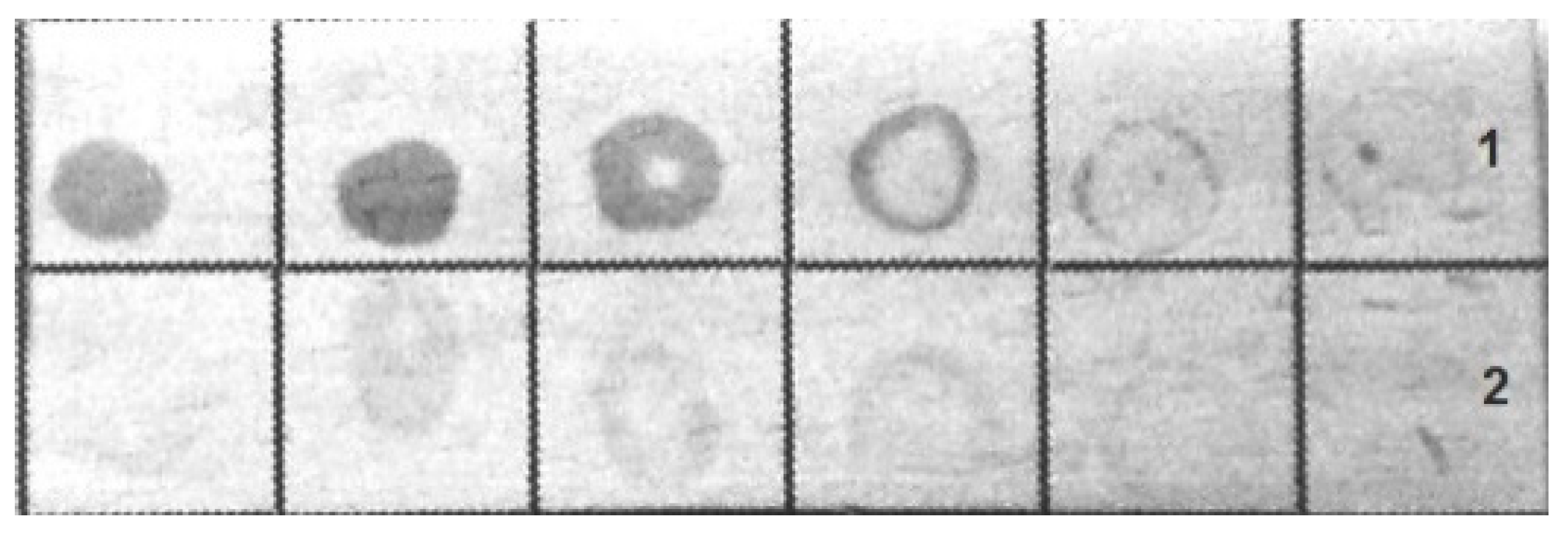
The bacteria were immunochemically identified by an indirect dot assay with colloidal-gold-labeled antirabbit immunoglobulins as the secondary antibodies. Both fixed bacteria used for immunization and bacteria grown on blood agar were included in the assay.
Figure 4 shows that the antibodies bound to the bacteria were grown on blood agar, whereas the formaldehyde-fixed bacteria had a less pronounced coloration in the dot assay.
Thus, the bacteria grown on blood agar and those obtained directly from a living suspension culture of Arabidopsis were found to be morphologically and immunochemically identical.
4. Conclusions
This is the first time that a nonpathogenic bacterial microflora has been found in a suspension culture of A. thaliana cells. The 16S rRNA gene sequencing showed that the isolate belonged to R. amarae. The growth of the isolate on blood agar preserves the morphological and immunochemical properties of the isolate from the plant cell suspension culture.
Whether the isolated strain is a contaminant or a true symbiont remains an open question. It is known that
Rothia bacteria live mostly in oceanic and waste water and in benthos. Members of
Rothia are part of the normal microflora of the oral cavity, respiratory tract, and stomach of humans [
12].
Rothia bacteria have also been detected as endophytes in association with associated with Magellan sphagnum (
Sphagnum magellanicum Brid.), stellate pohostemon (
Dysophylla stellata (Lour.) Benth.), pointed banana (
Musa acuminate Colla), alder (
Alnus), and
Seidlitzia rosmarinus Ehrenb. ex Boiss. [
13,
14,
15,
16,
17]. Endophytic
Rothia are inhibitory against several pathogenic fungi and bacteria. Notably,
Rothia was detected in both roots and shoots, indicating the possible migration of some species from roots to shoots. The root-associated bacteria showed higher levels of indole-3-acetic acid synthesis compared with those isolated from the shoots, as well as the higher production of 1-aminocyclopropane-1-carboxylate deaminase [
17]. In addition, some actinobacteria, including members of
Rothia, are nitrogen fixers [
18]. It cannot be ruled out that the
R. amarae strain isolated in this work can be endosymbiotic with a suspension culture of
A. thaliana. The bacterial “inclusions” found by us in a suspension culture of
A. thaliana merit further investigation to identify them more deeply and clarify their symbiotic properties.
Author Contributions
A.S. and O.S. conceived and designed the experiments; A.S. and L.D. performed the experiments; A.G., L.D. and O.S. analyzed the data and wrote the paper. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki. Animal care complied with the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes, the Guide for the Care and Use of Laboratory Animals, and Russian legislation.
Acknowledgments
We are grateful to M.K.S. and A.M.B. for help with microscopy. This work was supported in part by the Russian Foundation for Basic Research (grant no. 18-04-00469).
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| PBS | phosphate-buffered saline |
| TEM | transmission electron microscopy |
References
- Eibl, R.; Meier, P.; Stutz, I.; Schildberger, D.; Hühn, T.; Eibl, D. Plant cell culture technology in the cosmetics and food industries: Current state and future trends. Appl. Microbiol. Biotechnol. 2018, 102, 8661–8675. [Google Scholar] [CrossRef]
- Buckley, P.M.; DeWilde, T.N.; Reed, B.M. Characterization and identification of bacteria isolated from micropropagated mint plants. In Vitro Cell Dev. Biol. Plant. 1995, 31, 58–64. [Google Scholar] [CrossRef]
- Habiba, U.; Reza, S.; Saha, M.L.; Khan, M.R.; Hadiuzzaman, S. Endogenous bacterial contamination during in vitro culture of table banana: Identification and prevention. Plant. Tissue Cult. Biotechnol. 2002, 12, 117–124. [Google Scholar]
- Baulina, O.I.; Lobakova, E.S.; Korzhenevskaya, T.G.; Butenko, R.G.; Gusev, M.V. Ultrastructure of ginseng cells and the cyanobacteria Chlorogloeopsis fritschii in the association cultivated in the dark. Moscow. Univ. Biol. Sci. Bull. 1995, 50, 1–11. [Google Scholar]
- Barkla, B.J.; Vera-Estrella, R.; Pantoja, O. Growing Arabidopsis in vitro: Cell suspensions, in vitro culture, and regeneration. Methods Mol. Biol. 2014, 1062, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Stepanchenko, N.S.; Fomenkov, A.A.; Moshkov, I.E.; Rakitin, V.Y.; Novikova, G.V.; Nosov, A.V. Phytohormone interplay controls proliferation of in vitro cultivated cells of Arabidopsis thaliana ethylene-insensitive mutants. Dokl. Biol. Sci. 2012, 442, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Shenk, R.U.; Hildebrandt, A.C. Medium and techniques for induction and growth of monocotyledonous and dicotyledonous plant cell cultures. Can. J. Bot. 1972, 50. [Google Scholar] [CrossRef]
- Nishikawa, S.; Sasaki, F. Internalization of styryl dye FM1-43 in the hair cells of lateral line organs in Xenopus larvae. J. Histochem. Cytochem. 1996, 44, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Dykman, L.A.; Khlebtsov, N.G. Gold nanoparticles in biomedical applications: Recent advances and perspectives. Chem. Soc. Rev. 2012, 41, 2256–2282. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Jin, Z.; Tong, J.; Li, W.; Pasciak, M.; Gamian, A.; Liu, Z.; Huang, Y. Rothia amarae sp. nov., from sludge of a foul water sewer. Int. J. Syst. Evol. Microbiol. 2002, 52, 2257–2260. [Google Scholar] [CrossRef] [PubMed]
- Ko, K.S.; Lee, M.Y.; Park, Y.K.; Peck, K.R.; Song, J.-H. Molecular identification of clinical Rothia isolates from human patients: Proposal of a novel Rothia species, Rothia arfidiae sp. nov. J. Bacteriol. Virol. 2009, 39, 159–164. [Google Scholar] [CrossRef]
- Nardone, G.; Compare, D. The human gastric microbiota: Is it time to rethink the pathogenesis of stomach diseases? United Eur. Gastroenterol. J. 2015, 3, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Opelt, K.; Berg, C.; Berg, G. The bryophyte genus Sphagnum is a reservoir for powerful and extraordinary antagonists and potentially facultative human pathogens. FEMS Microbiol. Ecol. 2007, 61, 38–53. [Google Scholar] [CrossRef] [PubMed]
- Sekhar, A.C.; Thomas, P. Isolation and identification of shoot-tip associated endophytic bacteria from banana cv. Grand Naine and testing for antagonistic activity against Fusarium oxysporum f. sp. cubense. Am. J. Plant. Sci. 2015, 6, 943–954. [Google Scholar] [CrossRef][Green Version]
- Xiong, Z.-J.; Zhang, J.-L.; Zhang, D.-F.; Zhou, Z.-L.; Liu, M.-J.; Zhu, W.-Y.; Zhao, L.-X.; Xu, L.-H.; Li, W.-J. Rothia endophytica sp. nov., an actinobacterium isolated from Dysophylla stellata (Lour.) Benth. Int. J. Syst. Evol. Microbiol. 2013, 63, 3964–3969. [Google Scholar] [CrossRef] [PubMed]
- Davis, I.; Sevigny, J.; Kleiner, V.; Mercurio, K.; Pesce, C.; Swanson, E.; Thomas, W.K.; Tisa, L.S. Draft genome sequences of 10 bacterial strains isolated from root nodules of Alnus trees in New Hampshire. Microbiol. Resour. Announc. 2020, 9, e01440-19. [Google Scholar] [CrossRef] [PubMed]
- Shurigin, V.; Egamberdieva, D.; Li, L.; Davranov, K.; Panosyan, H.; Birkeland, N.-K.; Wirth, S.; Bellingrath-Kimura, S.D. Endophytic bacteria associated with halophyte Seidlitzia rosmarinus Ehrenb. ex Boiss. from saline soil of Uzbekistan and their plant beneficial traits. J. Arid Land 2020, 12, 730–740. [Google Scholar] [CrossRef]
- Gtari, M.; Ghodhbane-Gtari, F.; Nouioui, I.; Beauchemin, N.; Tisa, L.S. Phylogenetic perspectives of nitrogen-fixing actinobacteria. Arch. Microbiol. 2012, 194, 3–11. [Google Scholar] [CrossRef] [PubMed]
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).