Abstract
Grain sorghum (Sorghum bicolor (L.) Moench) is one of the major crops used for various purposes, including animal and human nutrition. The most relevant strategy for creating new sorghum hybrids is the use of lines with cytoplasmic male sterility (CMS). However, the process of creating sterile lines and lines that restore fertility is very laborious and time-consuming. The breeders need to know the genotype of the original parental forms of sorghum by the presence of the main genes that control fertility to speed up breeding work. One of these is the Rf1 gene. Our study aimed to identify alleles of the Rf1 gene in collection samples of grain sorghum (Sorghum bicolor (L.) Moench) adapted to the arid conditions of southern Russia. The studies were carried out in southern Russia (FSBSI “ARC “Donskoy”, Zernograd, Russia) between 2018–2019. The presence of alleles of the Rf1 fertility gene using the Xtxp18 SSR marker by PCR analysis in collection samples of grain sorghum (313 pcs.) was studied. Some samples were crossed with two sterile lines: “Demetra S” and “Dzhetta S” (developed in FSBSI “ARC “Donskoy”) in 2019. The assessment of the fertility of self-pollinated lines was carried out in the field using a 3-point scale. The polymorphism of the Xtxp18 marker allowed for the identification of a wide allelic diversity of the Rf1 gene in collection samples of sorghum, and the association of the identified Rf1 alleles with the fertility and sterility of self-pollinated hybrids of grain sorghum as a result of the study was performed. This result will make it possible to deepen understanding of the influence of the Rf1 gene alleles on the level of fertility of sorghum plants in the future, as well as to accelerate the breeding process to create sterile lines and their fertile analogues for further obtaining commercial hybrids.
1. Introduction
Grain sorghum (Sorghum bicolor (L.) Moench) is one of the major crops used for various purposes, including animal and human nutrition [1]. The most relevant strategy for creating new sorghum hybrids is the use of lines with cytoplasmic male sterility (CMS) [2]. However, the process of creating sterile lines and lines that restore fertility is very laborious and time-consuming [3]. Breeders need to know the genotype of the original parental forms of sorghum by the presence of the main genes that control fertility to speed up breeding work. One of these is the Rf1 gene [4].
The genetic background of our collection of samples of grain sorghum adapted to the arid climate of southern Russia has not been studied before.
Our study aimed to identify alleles of the Rf1 gene in collection samples of grain sorghum (Sorghum bicolor (L.) Moench) adapted to the arid conditions of southern Russia.
2. Experiments
The studies were carried out on southern Russia (FSBSI “ARC “Donskoy”, Zernograd, Russia) between 2018–2019 (46°50′42″ N, 40°18′30″ E).
We studied 313 collection samples of grain sorghum (Sorghum bicolor (L.) Moench) in laboratory conditions, and 106 hybrid combinations (53 combinations each with sterile lines of Demetra S and Dzhetta S) in the field in two repetitions.
The genomic DNA was isolated from the leaves and grains Sorghum bicolor using the modified CTAB protocol [5].
The homogenization of grains and leaves of sorghum was done using a Bertin Precellys24 homogenizer (France) in 2 mL test tubes with the addition of 6 pcs of 2.8 mm zirconium oxide beads, in the presence of 200–250 μL of DNA isolation buffer with the following program: 6500 rpm—30 s, pause 5 s, 6500 rpm—30 s.
The presence of alleles of the Rf1 fertility gene was assessed using the Xtxp18 SSR marker by PCR analysis [6]. The estimation of the allele size of the Rf1 gene was carried out using electrophoresis on 2% agarose gel, a Bio-Rad GelDoc XR+ device and the Bio-Rad ImageLab 6.0.1 software.
This study crossed 53 samples with two sterile lines, “Demetra S” and “Dzhetta S” (developed in FSBSI “ARC “Donskoy”), which was done in 2018 and 2019. The assessment of the fertility of self-pollinated lines was carried out in the field using a 3-point scale: 0—sterile line, 1—semi-sterile line, 2—fertile line. Data analyses was carried out in Microsoft Excel.
3. Results
As a result of the assessment of the collection samples of grain sorghum for the alleles of the Rf1 gene, a wide genetic diversity was revealed and many different alleles were identified. The percentage distribution of grain sorghum samples by the presence of Rf1 gene alleles is shown in Figure 1.
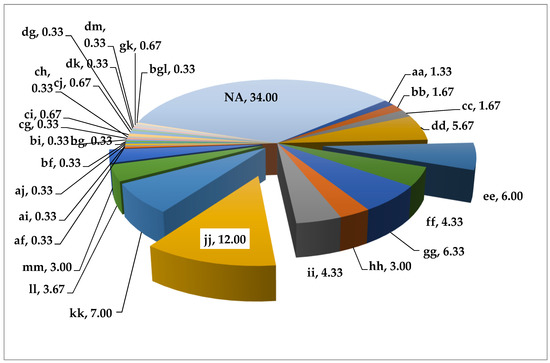
Figure 1.
Percentage distribution of grain sorghum samples by the presence of Rf1 gene alleles: aa—266 bp, bb—262 bp, cc—258 bp, dd—248 bp, ee—238 bp (associated with Rf1), ff—236 bp, gg—232 bp, hh—230 bp, ii—228 bp, jj—220 bp (associated with Rf1), kk—210 bp, ll—200 bp, mm—190 bp, af—266 bp + 236 bp, ai—266 bp + 228 bp, aj—266 bp + 220 bp, bf—262 bp + 236 bp, bg—262 bp + 232 bp, bi—262 bp + 228 bp, cg—258 bp + 232 bp, ch—258 bp + 230 bp, ci—258 bp + 228 bp, cj—258 bp + 220 bp, dg—248 bp + 232 bp, dk—248 bp + 210 bp, dm—248 bp + 190 bp, gk—232 bp + 210 bp, bgl—262 bp + 232 bp + 200 bp, NA—null-allele.
We identified 36 samples of grain sorghum with the j allele associated with fertility, 18 samples with the e allele associated with sterility, as well as samples with previously unknown alleles, which we temporarily named k—22 samples, l—11 samples, and m—9 samples. All identified genotypes are presented in Table 1.

Table 1.
Identified alleles in collection samples of grain sorghum.
In the field, we assessed self-pollinated lines of grain sorghum on a 3-point scale—fertile, semi-sterile and sterile. The number of identified fertile and sterile hybrid combinations is presented in Table 2.

Table 2.
The identified fertile and sterile hybrid combinations of grain sorghum.
Most of the combinations are fertile. For paternal forms with a null allele (NA) and allele “e”, this may be due to the influence of other genes that control fertility. One semi-sterile line of Demetra S/ZSK 163/17 has been identified. Sterility in 2 repetitions was revealed in crossing combinations Demetra S/ZSK 1530/15, Dzhetta S/Zernogradskoe 204w4, Demetra S/LBK 28, Demetra S//Svetloje/Belozernoje 100, and Dzhetta S/Svetloje/Belozernoje 100; hence, their paternal forms are sterility fixers and can be used in further breeding work.
4. Discussion
As a result of our study, the genetic background of 313 collection samples of grain sorghum was identified by the presence of Rf1 gene alleles. The use of the Xtxp18 marker made it possible to identify a wide variety of samples, in particular to identify those that carry alleles associated with sterility and fertility. This result is consistent with data obtained by Klein et al. [6]. In addition, we identified three new allelic variants, temporarily designated by us as k, l, and m, which were not reported by other researchers. These allelic variants may serve as a basis for future studies of mutations in grain sorghum.
Analysis of self-pollinated grain sorghum lines obtained from crossing with sterile lines Demetra S and Dzhetta S revealed 4 paternal samples that can be used in the breeding process as sterility fixers, and therefore further obtaining new commercial hybrids in the future.
This result will make it possible to deepen understanding of the influence of the Rf1 gene alleles on the level of fertility of sorghum plants in the future, as well as to accelerate the breeding process to create sterile lines and their fertile analogues for further obtaining commercial hybrids.
Supplementary Materials
The poster presentation is available online at https://www.mdpi.com/article/10.3390/IECPS2020-08710/s1.
Author Contributions
Conceptualization, N.V. and E.I.; writing—original draft, N.V., funding acquisition, A.P.; analyzed the data, N.V.; performed the experiments, N.V. and V.K. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
This work was funded by Russian Academy of Sciences (0706-2015-0001; 0706-2019-0003).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| FSBSI “ARC “Donskoy” | Federal State Budgetary Scientific Institution “Agricultural Research Center “Donskoy” |
| bp | base pairs |
References
- Dahlberg, J.; Berenji, J.; Sikora, V.; Latković, D. Assessing sorghum [Sorghum bicolor (L) Moench] germplasm for new traits: Food, fuels & unique uses. Maydica 2011, 56, 85–92. [Google Scholar]
- Ashok Kumar, A.; Reddy, V.S.; Reddy, S.; Ramaiah, B. Development of Male-Sterile Lines in Sorghum; International Crops Research Institute for the Semi-Arid Tropics: Patancheru, Andhra Pradesh, India, 2008; pp. 72–78. ISBN 978-92-9066-512-0. [Google Scholar]
- Rakshit, S.; Bellundagi, A. Conventional Breeding Techniques in Sorghum. In Breeding Sorghum for Diverse End Uses; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Kiyosawa, A.; Yonemaru, J.I.; Kawahigashi, H.; Goto, K. Analysis of quantitative trait loci for fertility restoration in seven F2 populations derived from sorghum F1 hybrids bred in Japan. Breed. Sci. 2020, 70, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, R.R.; Klein, P.E.; Chhabra, A.K.; Dong, J.; Pammi, S.; Childs, K.L.; Mullet, J.E.; Rooney, W.L.; Schertz, K.F. Molecular mapping of the Rf1 gene for pollen fertility restoration in sorghum (Sorghum bicolor L.). Theor. Appl. Genet. 2001, 102, 1206–1212. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).

