Study on Yield Variability in Oil Palm Progenies and Their Genetic Origins †
Abstract
:1. Introduction
Hypothesis of the Study
- H0. There is no significant variation in YFFB yield performance among the progenies.H1.The performance of the progenies in YFFB yield varies significantly.
- H0. There is no significant variation in their performance for YFFB yield.H1.There is significant variation in genetic origins’ performance for YFFB yield.
- H0. The genetic variance cannot influence low YFFB yield in oil palm progenies.H1.The genetic variance could influence low YFFB yield in oil palm progenies.
- H0.There is no significant variation in yearly performance for YFFB.H1.There is significant variation in yearly performance for YFFB.
2. Materials and Method
2.1. Data Collection
2.2. Statistical Analysis
3. Results and Discussion
3.1. Experimental Findings on Yield Traits in DP Progenies
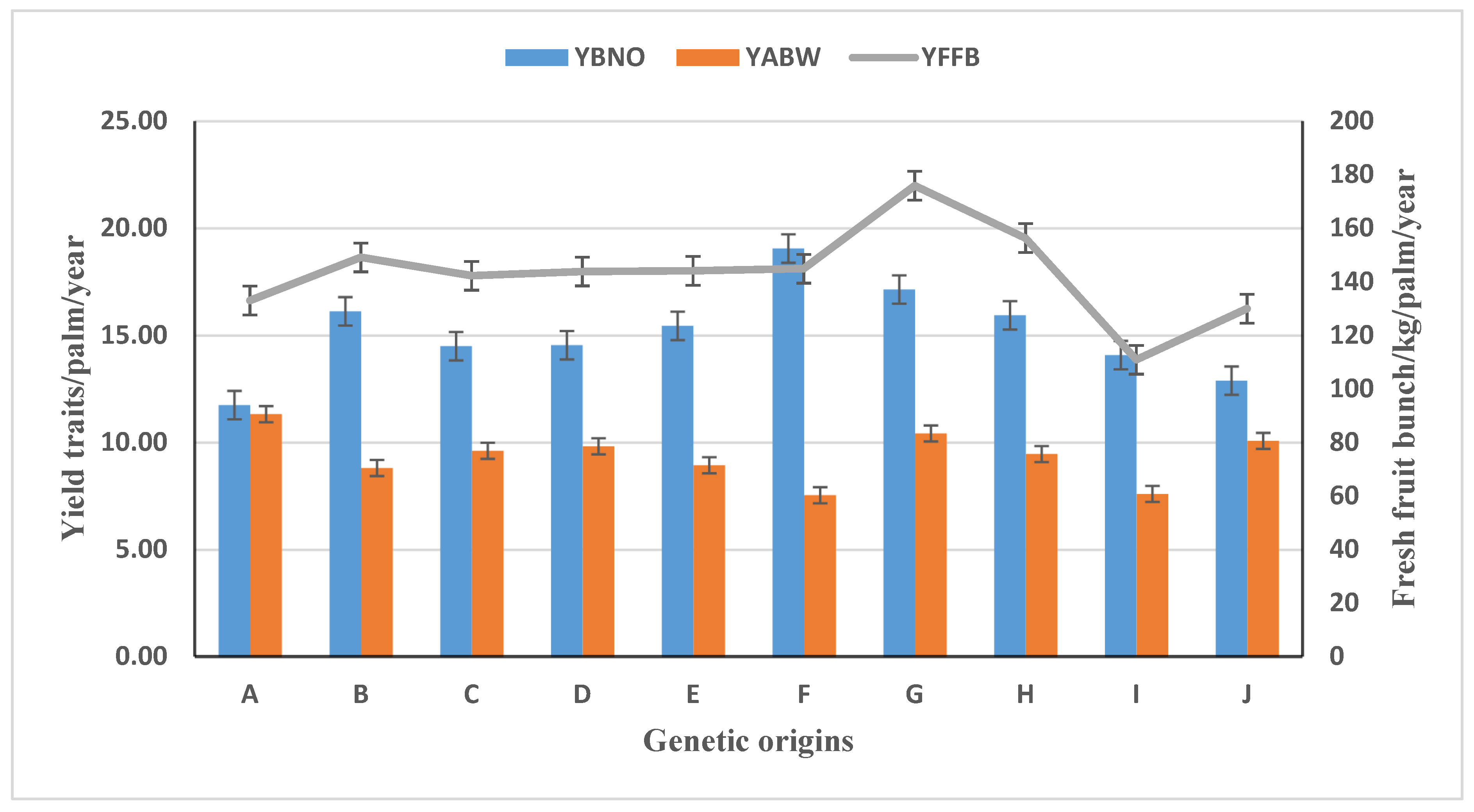
3.2. Parental Origins’ Performance in Oil Palm Yield and Yield Traits
4. Conclusions
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maluin, F.N.; Hussein, M.Z.; Idris, A.S. An overview of the oil palm industry: Challenges and some emerging opportunities for nanotechnology development. Agronomy 2020, 10, 356. [Google Scholar] [CrossRef] [Green Version]
- Bakoumé, C.; Wickneswari, R.; Siju, S.; Rajanaidu, N.; Kushairi, A.; Billotte, N. Genetic diversity of the world’s largest oil palm (Elaeis guineensis Jacq.) field gene bank accessions using microsatellite markers. Genet. Resour. Crop Evol. 2015, 62, 349–360. [Google Scholar] [CrossRef]
- Feintrenie, L.; Levang, P. Sumatra’s rubber agroforests: Advent, rise and fall of a sustainable cropping system. Small-Scale For. 2009, 8, 323–335. [Google Scholar] [CrossRef]
- Swaray, S.; Rafii, M.Y.; Din Amiruddin, M.; Firdaus Ismail, M.; Jamian, S.; Jalloh, M.; Oladosu, Y.; Mustakim Mohamad, M.; Marjuni, M.; Kolapo, O.K.; et al. Assessment of oil palm pollinating weevil (Elaeidobius kamerunicus) population density in biparental dura × pisifera hybrids on deep peat-soil in Perak state, Malaysia. Insects 2021, 12, 221. [Google Scholar] [CrossRef]
- Hoffmann, M.P.; Vera, A.C.; Van Wijk, M.T.; Giller, K.E.; Oberthür, T.; Donough, C.; Whitbread, A.M. Simulating potential growth and yield of oil palm (Elaeis guineensis) with PALMSIM: Model description, evaluation and application. Agric. Syst. 2014, 131, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Nanda, D.; Kansci, G.; Rafflegeau, S.; Bourlieu, C.; Ebongue, G.N.; Genot, C. Impact of post-harvest storage and freezing of palm fruits on the extraction yield and quality of African crude palm oil extracted in the laboratory. OCL Oilseeds Fats Crops Lipids 2020, 27, 52. [Google Scholar] [CrossRef]
- Rébéna, A.; Rafflegeau, S.; Kansci, G.; Nanda, D.; Genot, C. Surveys on the consumption, perception and uses of red palm oil among housewives and restaurateurs of Yaoundé, Cameroon. Cah. Agric. 2019, 28, 27. [Google Scholar] [CrossRef]
- Dong, S.; Xia, H.; Wang, F.; Sun, G. The effect of red palm oil on vitamin A deficiency: A meta-analysis of randomized controlled trials. Nutrients 2017, 9, 1281. [Google Scholar] [CrossRef] [Green Version]
- Ishak, Z.; Hashim, A.T.; Rosli, S.K.; Bakar, D.A.; Ooi, S.E.; Mohd, N.; Ong-Abdullah, M. Oil Palm Tissue Culture: Fast Tracking Elite Commercial Lines. In The Oil Palm Genome; Springer: Cham, Switzerland, 2020; pp. 47–68. [Google Scholar]
- MPOB. Insect Pollination Efficiency in Oil Palm; Workshop; MPOB: Selangor, Malaysia, 2018. [Google Scholar]
- Kushairi, A.; Ong-Abdullah, M.; Nambiappan, B.; Hishamuddin, E.; Bidin, M.N.I.Z.; Ghazali, R.; Subramaniam, V.; Sundram, S.; Parveez, G.K.A. Oil palm economic performance in Malaysia and R&D progress in 2018. J. Oil Palm Res. 2019, 31, 165–194. [Google Scholar] [CrossRef]
- Oil World. Independent Global Market Analyses & Forecasts Since 1958; ISTA Mielke Gmbh: Hamburg, Germany, 2018; Available online: https://www.oilworld.biz/t/publications/data-base (accessed on 15 June 2018).
- Ordway, E.M.; Naylor, R.L.; Nkongho, R.N.; Lambin, E.F. Oil palm expansion in Cameroon: Insights into sustainability opportunities and challenges in Africa. Glob. Environ. Chang. 2017, 47, 190–200. [Google Scholar] [CrossRef]
- Ordway, E.M.; Naylor, R.L.; Nkongho, R.N.; Lambin, E.F. Oil palm expansion and deforestation in Southwest Cameroon associated with proliferation of informal mills. Nat. Commun. 2019, 10, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cochard, B.; Adon, B.; Kouame, R.K.; Durand-Gasselin, T.; Amblard, P. Advantages of improved commercial palm oil (Elaeis guineensis Jacq.) seeds. OCL Ol. Corps Gras Lipides 2001, 8, 654–658. [Google Scholar] [CrossRef] [Green Version]
- Rafflegeau, S.; Michel-Dounias, I.; Tailliez, B.; Ndigui, B.; Papy, F. Unexpected N and K nutrition diagnosis in oil palm smallholdings using references of high-yielding industrial plantations. Agron. Sustain. Dev. 2010, 30, 777–787. [Google Scholar] [CrossRef] [Green Version]
- Rafflegeau, S.; Nanda, D.; Genot, C. Artisanal mills and local production of palm oil by smallholders. In Achieving Sustainable Cultivation of Oil Palm; Volum 2: Diseases, Pests quality and sustainability; Burleigh Dodds Science Publishing: Cambridge, UK, 2018. [Google Scholar] [CrossRef] [Green Version]
- Ghulam, K.A.P.; Elina, H.; Soh, K.L.; Meilina, O.A.; Kamalrudin, M.S.; Mohd, N.I.Z.B.; Shamala, S.; Zafarizal, A.A.H.; Zainab, I. Oil palm economic performance in Malaysia and R&D progress in 2019. J. Oil Palm Res. 2020, 32, 159–190. [Google Scholar] [CrossRef]
- Sarkar, M.S.K.; Begum, R.A.; Pereira, J.J. Impacts of climate change on oil palm production in Malaysia. Environ. Sci. Pollut. Res. 2020, 27, 9760–9770. [Google Scholar] [CrossRef] [PubMed]
- Rafii, M.Y.; Rajanaidu, N.; Jalani, B.S.; Kushairi, A. Performance and heritability estimations on oil palm progenies tested in different environments. J. Oil Palm Res. 2002, 14, 15–24. [Google Scholar]
- Rafii, M.Y.; Isa, Z.A.; Kushairi, A.; Saleh, G.B.; Latif, M.A. Variation in yield components and vegetative traits in Malaysian oil palm (Elaeis guineensis jacq.) dura × pisifera hybrids under various planting densities. Ind. Crops Prod. 2013, 46, 147–157. [Google Scholar] [CrossRef]
- Shabanimofrad, M.; Rafii, M.Y.; Wahab, P.M.; Biabani, A.R.; Latif, M.A. Phenotypic, genotypic and genetic divergence found in 48 newly collected Malaysian accessions of Jatropha curcas L. Ind. Crops Prod. 2013, 42, 543–551. [Google Scholar] [CrossRef] [Green Version]
- Burton, G.W. Quantitative inheritance in grasses. In Proceedings of the 6th International Grassland Congress, State College, PA, USA, 17–23 August 1952; Volume 1, pp. 277–283. [Google Scholar]
- Burton, G.W.; De Vane, E.H. Estimating heritability in tall fesscusce from replicated clone natural materials. Agron. J. 1952, 45, 171–181. [Google Scholar]
- Singh, R.K.; Chaudhary, B.D. Biometrical Methods in Quantitative Genetic Analysis; Line x tester analysis; Kalyani publishers: New Delhi, India, 1985; Volume 3, pp. 215–223. [Google Scholar]
- Johnson, H.W.; Robinson, H.F.; Comstock, R.E. Estimates of genetic and environmental variability in soybeans. Agron. J. 1955, 47, 314–318. [Google Scholar] [CrossRef]
- Robinson, H.F.; Comstock, R.E.; Harvey, P.H. Genotypic and phenotypic correlation in corn and their implications in selection. Agron. J. 1949, 43, 282–287. [Google Scholar] [CrossRef] [Green Version]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Abdul Malek, M.; Rahim, H.A.; Hussin, G.; Latif, M.A.; Kareem, I. Genetic variability and selection criteria in rice mutant lines as revealed by quantitative traits. Sci. World J. 2014, 2014, 190531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arolu, I.W.; Rafii, M.Y.; Marjuni, M.; Hanafi, M.M.; Sulaiman, Z.; Rahim, H.A.; Abidin, M.I.Z.; Amiruddin, M.D.; Din, A.K.; Nookiah, R. Breeding of high yielding and dwarf oil palm planting materials using Deli dura × Nigerian pisifera population. Euphytica 2017, 213, 154. [Google Scholar] [CrossRef]
- Oettli, P.; Behera, S.K.; Yamagata, T. Climate based predictability of oil palm tree yield in Malaysia. Sci. Rep. 2018, 8, 2271. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, F.; Biemans, H.; Jacobs, C.; Supit, I.; Van Diepen, C.A.; Fawell, J.; Capri, E.; Steduto, P. Water Use of Oil Crops: Current Water Use and Future Outlooks; ILSI Europe Aisbl: Brussels, Belgium, 2011; Available online: https://library.wur.nl/WebQuery/wurpubs/fulltext/193177 (accessed on 19 August 2020).
- Myint, K.A.; Amiruddin, M.D.; Rafii, M.Y.; Samad, M.Y.A.; Ramlee, S.I.; Yaakub, Z.; Oladosu, Y. Genetic diversity and selection criteria of MPOB-Senegal oil palm (Elaeis guineensis Jacq.) germplasm by quantitative traits. Ind. Crops Prod. 2019, 139, 111558. [Google Scholar] [CrossRef]
- Swaray, S.; Din Amiruddin, M.; Rafii, M.Y.; Jamian, S.; Ismail, M.F.; Jalloh, M.; Marjuni, M.; Mustakim Mohamad, M.; Yusuff, O. Influence of parental dura and pisifera genetic origins on oil palm fruit set ratio and yield components in their D × P progenies. Agronomy 2020, 10, 1793. [Google Scholar] [CrossRef]
- Yusop, M.R.; Sukaimi, J.; Amiruddin, M.D.; Jalloh, M.; Swaray, S.; Yusuff, O.; Chukwu, S.C. Genetic improvement of oil palm through recurrent selection. In The Oil Palm Genome; Springer: Cham, Switzerland, 2020; pp. 35–46. [Google Scholar] [CrossRef]
- Bhagasara, V.K.; Ranwah, B.R.; Meena, B.L.; Khan, R. Estimation of GCV, PCV, heritability and genetic gain for yield and its related components in sorghum [Sorghum bicolor (L.) Moench]. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 1015–1024. [Google Scholar] [CrossRef] [Green Version]
- Amiruddin, M.D.; Bakar, N.A.A.; Malike, F.A.; Mustaffa, S.; Abdullah, N.; Marjuni, M.; Mohamad, M.M.; Hassan, M.Y.; Kushairi, A.; Nookiah, R. Wild and Advanced Resources of Elaeis guineensis and Elaeis oleifera. In The Oil Palm Genome; Springer: Cham, Switzerland, 2020; pp. 9–23. [Google Scholar] [CrossRef]
| Source of Variation | DF | YFFB (kg/Palm/Year) | YBON (Bunch/Palm/Year) | YABW (kg/Palm/Year) |
|---|---|---|---|---|
| Replications (R) | 3 | 318.74 ns | 1.28 ns | 1.06 ns |
| Years (Y) | 2 | 1973.68 ** | 775.92 ** | 142.38 ** |
| Progenies (G) | 23 | 7102.22 ** | 88.14 ** | 23.99 ** |
| Y∗G | 46 | 507.90 * | 5.23 * | 1.57 ns |
| Error | 186 | 557.42 | 3.60 | 1.23 |
| Variance component | ||||
| σ2g | 645.85 | 7.60 | 2.21 | |
| (61.83) | (65.37) | (62.60) | ||
| σ2yg | 41.43 | 0.45 | 0.10 | |
| (3.97) | (3.87) | (2.83) | ||
| σ2e | 357.22 | 3.58 | 1.22 | |
| (34.20) | (30.76) | (34.58) | ||
| Mean | 145.41 | 15.41 | 9.34 | |
| SE | 2.08 | 2.26 | 0.12 | |
| CV | 23.16 | 27.16 | 22.50 | |
| YEAR | YFFB (kg/Palm/Year) | YBON (Bunch/Palm/Year) | YABW (kg/Bunch) |
|---|---|---|---|
| YEAR1 (2017) | 153.69a | 18.89a | 8.00c |
| YEAR2 (2018) | 128.14b | 13.35c | 9.45b |
| YEAR3 (2019) | 154.40a | 14.00b | 10.58a |
| Progeny | |||
| HPDP415 | 129.55i | 11.78ij | 11.19ab |
| HPDP500 | 191.74a | 20.52a | 9.48efg |
| HPDP550 | 136.35hi | 11.73ij | 11.47a |
| HPDP618 | 142.22fghi | 14.50efg | 9.62defg |
| PKDP4118 | 136.75hi | 14.12fhg | 9.85def |
| PKDP4465 | 138.22hi | 15.60ef | 9.09fgh |
| PKDP4474 | 144.25fghi | 20.65a | 6.71jk |
| PKDP4482 | 89.78k | 12.37hij | 6.07k |
| PKDP4504 | 154.69defgh | 15.25ef | 9.32fg |
| PKDP4505 | 171.80bcd | 17.64cd | 10.23bcdef |
| PKDP4529 | 176.42abc | 16.30de | 11.03abc |
| PKDP4535 | 108.44j | 11.84ij | 8.62gh |
| PKDP4539 | 152.02efgh | 17.69cd | 8.22hi |
| PKDP4540 | 168.31bcde | 18.57bc | 9.09fgh |
| PKDP4548 | 175.01abc | 18.28bc | 9.63defg |
| PKDP4550 | 158.99cdefg | 16.41de | 9.95cdef |
| PKDP4570 | 110.86j | 14.09fgh | 7.61ij |
| PKDP4591 | 180.07ab | 16.25de | 11.25ab |
| PKDP4621 | 94.99jk | 10.86j | 6.99jk |
| PKDP4648 | 141.25ghi | 14.58efg | 9.72defg |
| PKDP4651 | 138.35hi | 19.78ab | 6.71jk |
| PKDP4674 | 158.72cdefg | 15.24ef | 10.48abcde |
| PKDP4679 | 160.48cdef | 14.20fg | 10.72abcd |
| PKDP4841 | 129.91i | 12.90ghi | 10.09cdef |
| Mean | 145.41 | 15.41 | 9.34 |
| SE | 2.08 | 2.26 | 0.12 |
| CV | 23.16 | 27.16 | 22.50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swaray, S.; Rafii, M.Y.; Amiruddin, M.D.; Ismail, M.F.; Jamian, S.; Marjuni, M.; Jalloh, M.; Yusuff, O.; Mohamad, M.M. Study on Yield Variability in Oil Palm Progenies and Their Genetic Origins. Biol. Life Sci. Forum 2021, 4, 68. https://doi.org/10.3390/IECPS2020-08760
Swaray S, Rafii MY, Amiruddin MD, Ismail MF, Jamian S, Marjuni M, Jalloh M, Yusuff O, Mohamad MM. Study on Yield Variability in Oil Palm Progenies and Their Genetic Origins. Biology and Life Sciences Forum. 2021; 4(1):68. https://doi.org/10.3390/IECPS2020-08760
Chicago/Turabian StyleSwaray, Senesie, Mohd Y. Rafii, Mohd Din Amiruddin, Mohd Firdaus Ismail, Syari Jamian, Marhalil Marjuni, Momodu Jalloh, Oladosu Yusuff, and Mohd Mustakim Mohamad. 2021. "Study on Yield Variability in Oil Palm Progenies and Their Genetic Origins" Biology and Life Sciences Forum 4, no. 1: 68. https://doi.org/10.3390/IECPS2020-08760
APA StyleSwaray, S., Rafii, M. Y., Amiruddin, M. D., Ismail, M. F., Jamian, S., Marjuni, M., Jalloh, M., Yusuff, O., & Mohamad, M. M. (2021). Study on Yield Variability in Oil Palm Progenies and Their Genetic Origins. Biology and Life Sciences Forum, 4(1), 68. https://doi.org/10.3390/IECPS2020-08760









