Assessing the Flowering Genetic Regulatory Network in Neotropical Orchids †
Abstract
:1. Introduction
2. Experiments
2.1. Phylogenetic Analyses of Flowering Candidate Genes
2.2. Morpho-Anatomical Characterization of the Flowering Transition in Orchidaceae
2.3. RT-PCR Expression Analysis of GRN Candidate Genes
3. Results
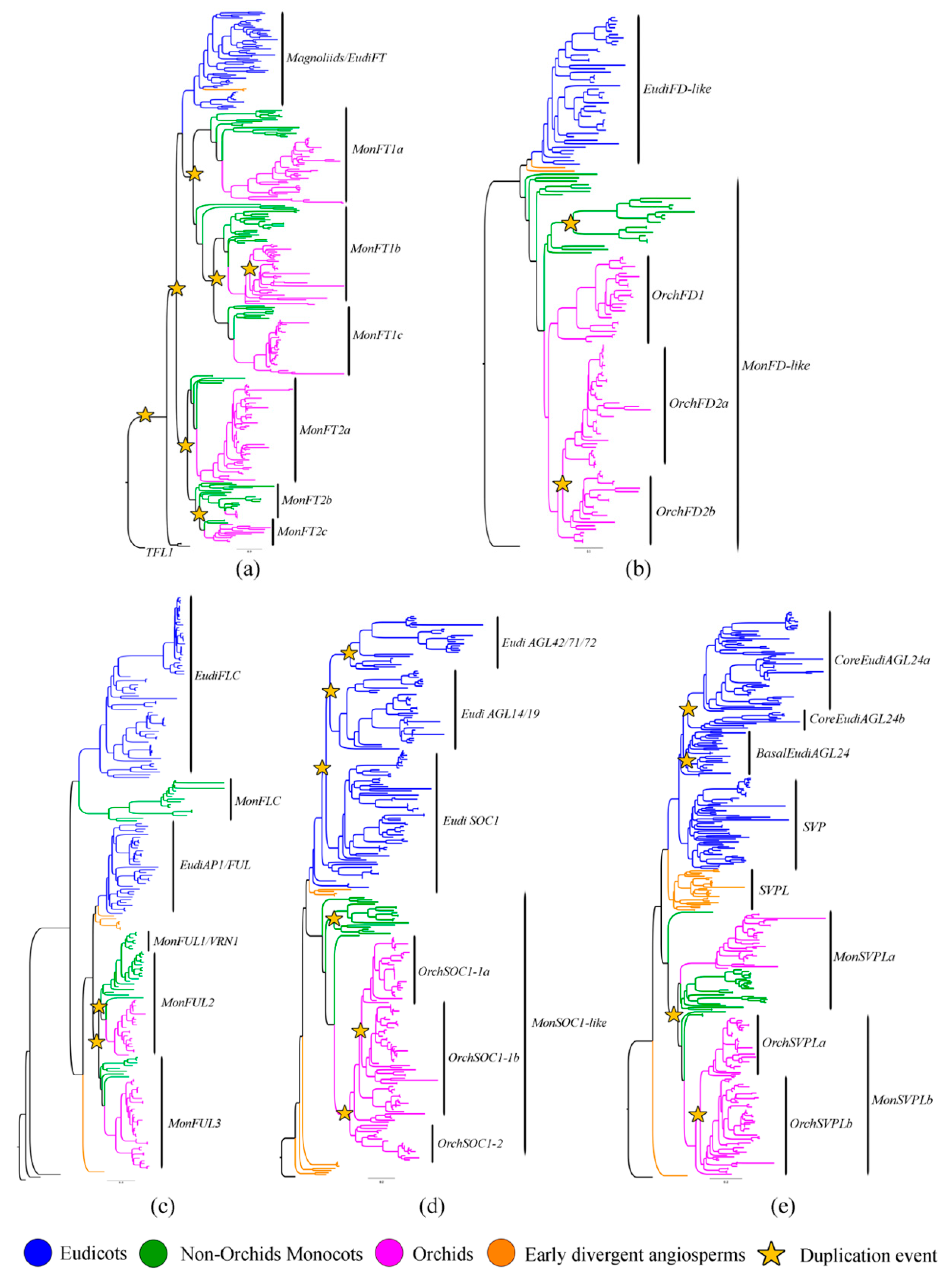
3.1. Flowering GRN Genes Have Undergone Multiple Duplication Events
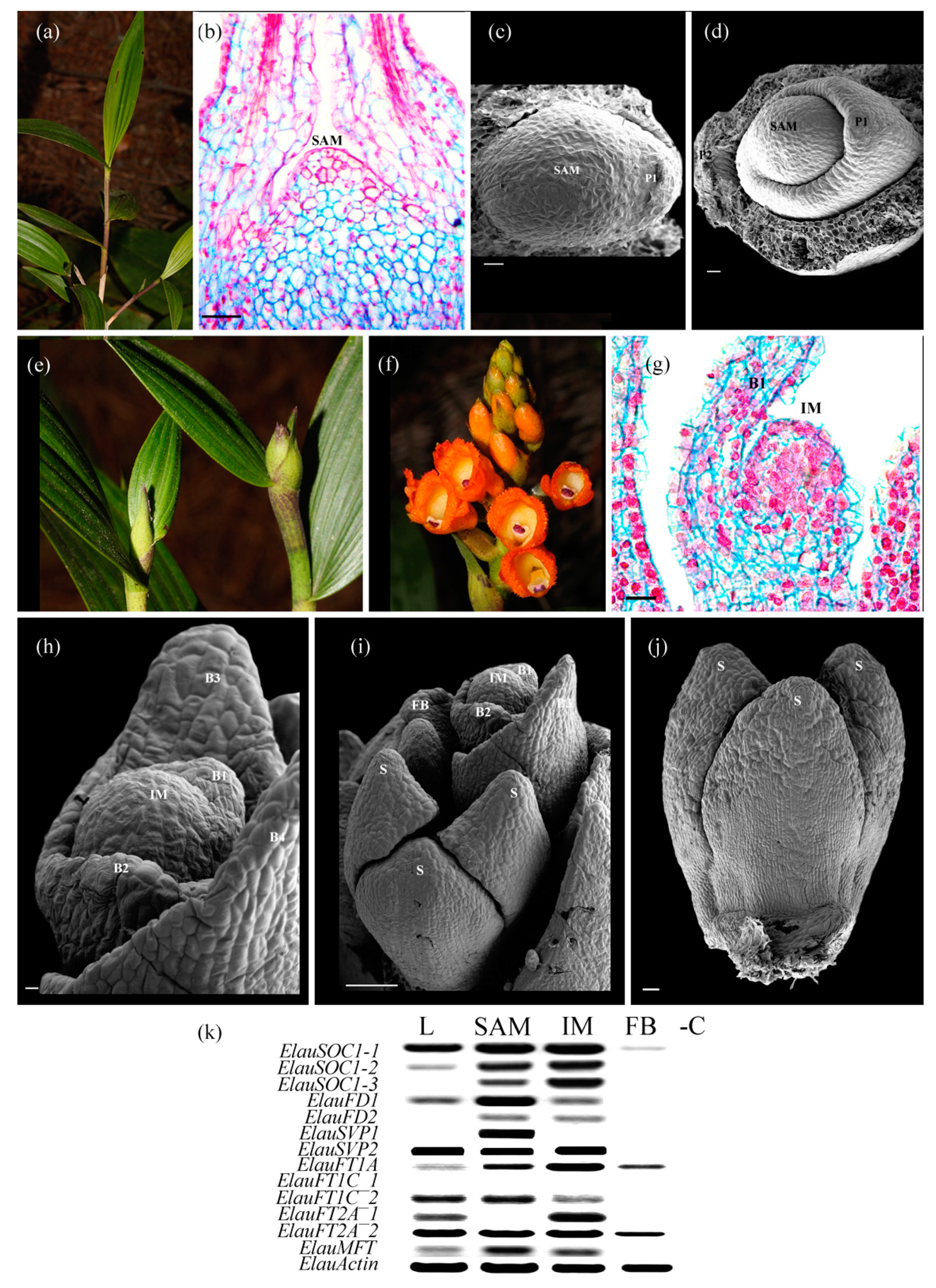
3.2. The Flowering Transition in Orchidaceae Recruits Several Flowering GRN Genes, Actively Expressed in the SAM and the IM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AGL24/SVP | AGAMOUS LIKE 24/SHORT VEGETATIVE PHASE |
| FD | FLOWERING LOCUS D |
| FLC | FLOWERING LOCUS C |
| FT | Flowering Locus T |
| FUL | FRUITFULL |
| IM | Inflorescence Meristem |
| GRN | Genetic Regulatory Network |
| ML | Maximum Likelihood |
| SAM | Shoot Apical Meristem |
| SOC1 | SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 |
| WGD | Whole Genome Duplication |
Appendix A
| Primer Name | Sequence | Amplicon Size (bp) |
|---|---|---|
| ACTIN7a_fwd | GCATTGTGCTTGATTCCGGTGATGGTGT | 450 |
| ACTIN7a_rev | CCACCTTAATCTTCATGCTGC | |
| ElauSOC1-3_fwd | GGAAAGACGGAGATGAGAC | 534 |
| ElauSOC1-3_rev | CTTATGCTGATGATTGTCATC | |
| ElauSOC1-1_fwd | GAAGGACGGAGATGAGACG | 555 |
| ElauSOC1-1_rev | CAGTTCGGTCTCTACATCCT | |
| ElauSOC1-2_fwd | CGGAGATGAAGCGTATAGAA | 457 |
| ElauSOC1-2_rev | CATCCTTATAGTGGCTATCA | |
| ElauFD2_Rev | AGCGGATGAGGTTCTTTGAA | 425 |
| ElauFD2_Fwd | CCACCGTGCTTAGCCTTAGT | |
| ElauFD1_Rev | ATAGTGGTGATCGCCTCCTG | 357 |
| ElauFD1_Fwd | CCCCAAACACCTAAGCGTAA |
References
- Levy, Y.Y.; Dean, C. The Transition to Flowering. Plant Cell 1998, 10, 1973–1989. [Google Scholar] [CrossRef] [PubMed]
- Parcy, F. Flowering: A Time for Integration. Int. J. Dev. Biol. 2005, 49, 585–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komiya, R.; Ikegami, A.; Tamaki, S.; Yokoi, S.; Shimamoto, K. Hd3a and RFT1 Are Essential for Flowering in Rice. Development 2008, 135, 767–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamaki, S.; Matsuo, S.; Wong, H.L.; Yokoi, S.; Shimamoto, K. Hd3a Protein Is a Mobile Flowering Signal in Rice. Science 2007, 316, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a Rice Ortholog of the Arabidopsis FT Gene, Promotes Transition to Flowering Downstream of Hd1 under Short-Day Conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef] [Green Version]
- Taoka, K.; Ohki, I.; Tsuji, H.; Furuita, K.; Hayashi, K.; Yanase, T.; Yamaguchi, M.; Nakashima, C.; Purwestri, Y.A.; Tamaki, S.; et al. 14-3-3 Proteins Act as Intracellular Receptors for Rice Hd3a Florigen. Nature 2011, 476, 332–335. [Google Scholar] [CrossRef]
- Tsuji, H.; Nakamura, H.; Taoka, K.; Shimamoto, K. Functional Diversification of FD Transcription Factors in Rice, Components of Florigen Activation Complexes. Plant Cell Physiol. 2013, 54, 385–397. [Google Scholar] [CrossRef] [Green Version]
- Taoka, K.; Ohki, I.; Tsuji, H.; Kojima, C.; Shimamoto, K. Structure and Function of Florigen and the Receptor Complex. Trends Plant Sci. 2013, 18, 287–294. [Google Scholar] [CrossRef]
- Fornara, F.; Gregis, V.; Pelucchi, N.; Colombo, L.; Kater, M. The Rice StMADS11-like Genes OsMADS22 and OsMADS47 Cause Floral Reversions in Arabidopsis without Complementing the Svp and Agl24 Mutants. J. Exp. Bot. 2008, 59, 2181–2190. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Park, S.H.; Ahn, J.H. Functional Conservation and Diversification between Rice OsMADS22/OsMADS55 and Arabidopsis SVP Proteins. Plant Sci. 2012, 185–186, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, J.; Han, J.J.; Han, M.J.; An, G. Functional Analyses of the Flowering Time Gene OsMADS50, the Putative Suppressor of Overexpression of CO 1/Agamous-Like 20 (SOC1/AGL20) Ortholog in Rice. Plant J. 2004, 38, 754–764. [Google Scholar] [CrossRef]
- Qin, Z.; Wu, J.; Geng, S.; Feng, N.; Chen, F.; Kong, X.; Song, G.; Chen, K.; Li, A.; Mao, L.; et al. Regulation of FT Splicing by an Endogenous Cue in Temperate Grasses. Nat. Commun. 2017, 8, 14320. [Google Scholar] [CrossRef] [Green Version]
- Leiboff, S.; Hake, S. Reconstructing the Transcriptional Ontogeny of Maize and Sorghum Supports an Inverse Hourglass Model of Inflorescence Development. Curr. Biol. 2019, 29, 3410–3419. [Google Scholar] [CrossRef] [Green Version]
- Higgins, J.A.; Bailey, P.C.; Laurie, D.A. Comparative Genomics of Flowering Time Pathways Using Brachypodium Distachyon as a Model for the Temperate Grasses. PLoS ONE 2010, 5, e10065. [Google Scholar] [CrossRef]
- Trevaskis, B.; Bagnall, D.J.; Ellis, M.H.; Peacock, W.J.; Dennis, E.S. MADS Box Genes Control Vernalization-Induced Flowering in Cereals. Proc. Natl. Acad. Sci. USA 2003, 100, 13099–13104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trevaskis, B.; Tadege, M.; Hemming, M.N.; Peacock, W.J.; Dennis, E.S.; Sheldon, C. Short Vegetative Phase-like MADS-BOX Genes Inhibit Floral Meristem Identity in Barley. Plant Physiol. 2007, 143, 225–235. [Google Scholar] [CrossRef] [Green Version]
- Preston, J.C.; Kellogg, E.A. Discrete Developmental Roles for Temperate Cereal Grass Vernalization1/Fruitfull-like Genes in Flowering Competency and the Transition to Flowering. Plant Physiol. 2008, 146, 265–276. [Google Scholar] [CrossRef] [Green Version]
- Trevaskis, B.; Hemming, M.N.; Dennis, E.S.; Peacock, W.J. The Molecular Basis of Vernalization-Induced Flowering in Cereals. Trends Plant Sci. 2007, 12, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tong, C.; Jang, S. Current Progress in Orchid Flowering / Flower Development Research. Plant Signal. Behav. 2017, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.L.; Viswanath, K.K.; Tong, C.G.; An, H.R.; Jang, S.; Chen, F.C. Floral Induction and Flower Development of Orchids. Front. Plant Sci. 2019, 10, 1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGinnis, S.; Madden, T.L. BLAST: At the Core of a Powerful and Diverse Set of Sequence Analysis Tools. Nucleic Acids Res. 2004, 32, W20–W25. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ospina-Zapata, D.A.; Madrigal, Y.; Alzate, J.F.; Pabón-Mora, N. Evolution and Expression of Reproductive Transition Regulatory Genes FT/TFL1 With Emphasis in Selected Neotropical Orchids. Front. Plant Sci. 2020, 11, 469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, F.; Pabón-Mora, N. Floral Development and Morphoanatomy in the Holoparasitic Pilostyles Boyacensis (Apodanthaceae, Cucurbitales) Reveal Chimeric Half-Staminate and Half-Carpellate Flowers. Int. J. Plant Sci. 2017, 178, 522–536. [Google Scholar] [CrossRef]
- Madrigal, Y.; Alzate, J.F.; González, F.; Pabón-mora, N. Evolution of RADIALIS and DIVARICATA Gene Lineages in Flowering Plants with an Expanded Sampling in Non-Core Eudicots. Am. J. Bot. 2019, 106, 334–351. [Google Scholar] [CrossRef]
- Ramirez-Ramirez, J.A. Evolución y Expersion de Genes MADS-BOX AGL24 (AGAMOUS LIKE 24) y SVP (SHORT VEGETATIVE PHASE) en Orquídeas Selectas Neotropicales. Bachelor’s Thesis, Universidad de Antioquia, Colombia, 2020. [Google Scholar]
- Teo, Z.W.N.; Zhou, W.; Shen, L. Dissecting the Function of MADS-Box Transcription Factors in Orchid Reproductive Development. Front. Plant Sci. 2019, 10, 1474. [Google Scholar] [CrossRef]
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 Gene Family: Functional Evolution and Molecular Mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef] [Green Version]
- Fudge, J.B.; Lee, R.H.; Laurie, R.E.; Mysore, K.S.; Wen, J.; Weller, J.L.; Macknight, R.C. Medicago Truncatula SOC1 Genes Are Up-Regulated by Environmental Cues That Promote Flowering. Front. Plant Sci. 2018, 9, 496. [Google Scholar] [CrossRef]
- Preston, J.C.; Zhong, J.; McKeown, M.; den Bakker, M.; Friedman, J. Comparative Transcriptomics Indicates a Role for SHORT VEGETATIVE PHASE (SVP) Genes in Mimulus Guttatus Vernalization Response. G3 Genes Genomes Genet. 2016, 6, 1239–1249. [Google Scholar] [CrossRef] [Green Version]
- Smaczniak, C.; Immink, R.G.H.; Angenent, G.C.; Kaufmann, K. Developmental and Evolutionary Diversity of Plant MADS-Domain Factors: Insights from Recent Studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef] [Green Version]
- Chardon, F.; Damerval, C. Phylogenomic Analysis of the PEBP Gene Family in Cereals. J. Mol. Evol. 2005, 61, 579–590. [Google Scholar] [CrossRef]
- Karlgren, A.; Gyllenstrand, N.; Kallmam, T.; Sundstrom, J.F.; Moore, D.; Lascoux, M.; Lagercrantz, U. Evolution of the PEBP Gene Family in Plants: Functional Diversification in Seed Plant Evolution. Plant Physiol. 2011, 156, 1967–1977. [Google Scholar] [CrossRef] [Green Version]
- Preston, J.C.; Kellogg, E.A. Reconstructing the Evolutionary History of Paralogous APETALA1/FRUITFULL-like Genes in Grasses (Poaceae). Genetics 2006, 174, 421–437. [Google Scholar] [CrossRef] [Green Version]
- Alter, P.; Bircheneder, S.; Zhou, L.Z.; Schlüter, U.; Gahrtz, M.; Sonnewald, U.; Dresselhaus, T. Flowering Time-Regulated Genes in Maize Include the Transcription Factor ZmMADS1. Plant Physiol. 2016, 172, 389–404. [Google Scholar] [CrossRef] [Green Version]
- Ruelens, P.; De Maagd, R.A.; Proost, S.; Geuten, K.; Kaufmann, K. FLOWERING LOCUS C in Monocots and the Tandem Origin of Angiosperm-Specific MADS-Box Genes. Nat. Commun. 2013, 4, 2280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, F.; Zhang, X.; Liu, X.; Zhang, L. Evolutionary Analysis of MIKCc-Type MADS-Box Genes in Gymnosperms and Angiosperms. Front. Plant Sci. 2017, 8, 895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, A.C.K.; Rozano, L.; Bakar, U.K.A.; Svp, P. Isolation and Phylogenetic Characterisation of LdSVP, SHORT VEGETATIVE PHASE (SVP) Homologous Gene from Lansium Domesticum. J. Trop. Agric. Food Sci. 2018, 46, 75–89. [Google Scholar]
- Jiao, F.; Pahwa, K.; Manning, M.; Dochy, N.; Geuten, K. Cold Induced Antisense Transcription of FLOWERING LOCUS C in Distant Grasses. Front. Plant Sci. 2019, 10, 72. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Wang, Y.; Yu, H. Overexpression of DOSOC1, an Ortholog of Arabidopsis SOC1, Promotes Flowering in the Orchid Dendrobium Chao Parya Smile. Plant Cell Physiol. 2013, 54, 595–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Liu, L.; Song, S.; Li, Y.; Shen, L.; Yu, H. DOFT and DOFTIP1 Affect Reproductive Development in the Orchid Dendrobium Chao Praya Smile. J. Exp. Bot. 2017, 68, 5759–5772. [Google Scholar] [CrossRef] [Green Version]
| Species | FT | FD | FUL | SOC1 | AGL24/SVP | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MonFT1 | MonFT2 | OrchFD1 | OrchFD2 | VRN1 | MonFUL2 | MonFUL3 | OrchSOC1 | OrchSOC2 | MonSVPa | MonSVPb | |
| Cattleya trianae | 2 | 1 | 1 | 1 | 0 | 3 | 2 | 2 | 0 | 1 | 2 |
| Elleanthus aurantiacus2 | 3 | 2 | 1 | 1 | 0 | 2 | 0 | 2 | 1 | 3 | 4 |
| Epidendrum fimbriatum | 3 | 5 | 1 | 4 | 0 | 4 | 5 | 2 | 0 | 2 | 8 |
| Gomphichis scaposa | 2 | 3 | 0 | 2 | 0 | 2 | 3 | 2 | 0 | 3 | 4 |
| Masdevalia coccinea “Alba” | 3 | 2 | 2 | 1 | 0 | 2 | 2 | 3 | 0 | 2 | 5 |
| Masdevalia wendlandiana | 5 | 3 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 1 | 6 |
| Maxilaria aurea | 9 | 3 | 1 | 1 | 0 | 2 | 2 | 4 | 6 | 0 | 3 |
| Miltonia roezli | 6 | 0 | 2 | 1 | 0 | 2 | 2 | 2 | 1 | 1 | 4 |
| Oncidium “Gower Ramsey” | 1 | 1 | 1 | 5 | 0 | 0 | 3 | 5 | 0 | 1 | 3 |
| Oncidium “Twinkle” | 2 | 4 | 6 | 1 | 0 | 1 | 4 | 2 | 0 | 2 | 6 |
| Stelis pusilla | 3 | 4 | 2 | 2 | 0 | 1 | 0 | 2 | 0 | 2 | 3 |
| Tolumnia “Cherry red × Ralph yagh” | 2 | 2 | 3 | 0 | 0 | 1 | 4 | 3 | 2 | 1 | 5 |
| Vanilla aphylla | 12 | 3 | 2 | 2 | 0 | 1 | 0 | 7 | 0 | 5 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madrigal, Y.; Ospina-Zapata, D.; Ramírez-Ramírez, J.A.; Alzate, J.F.; Pabón-Mora, N. Assessing the Flowering Genetic Regulatory Network in Neotropical Orchids. Biol. Life Sci. Forum 2021, 4, 53. https://doi.org/10.3390/IECPS2020-08576
Madrigal Y, Ospina-Zapata D, Ramírez-Ramírez JA, Alzate JF, Pabón-Mora N. Assessing the Flowering Genetic Regulatory Network in Neotropical Orchids. Biology and Life Sciences Forum. 2021; 4(1):53. https://doi.org/10.3390/IECPS2020-08576
Chicago/Turabian StyleMadrigal, Yesenia, Diego Ospina-Zapata, Jessica A. Ramírez-Ramírez, Juan Fernando Alzate, and Natalia Pabón-Mora. 2021. "Assessing the Flowering Genetic Regulatory Network in Neotropical Orchids" Biology and Life Sciences Forum 4, no. 1: 53. https://doi.org/10.3390/IECPS2020-08576
APA StyleMadrigal, Y., Ospina-Zapata, D., Ramírez-Ramírez, J. A., Alzate, J. F., & Pabón-Mora, N. (2021). Assessing the Flowering Genetic Regulatory Network in Neotropical Orchids. Biology and Life Sciences Forum, 4(1), 53. https://doi.org/10.3390/IECPS2020-08576






