Variability of Allergen-Based Length Polymorphism of Glycine max L. Varieties †
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Material
2.2. DNA Isolation
2.3. PBAP and VBAP Analysis
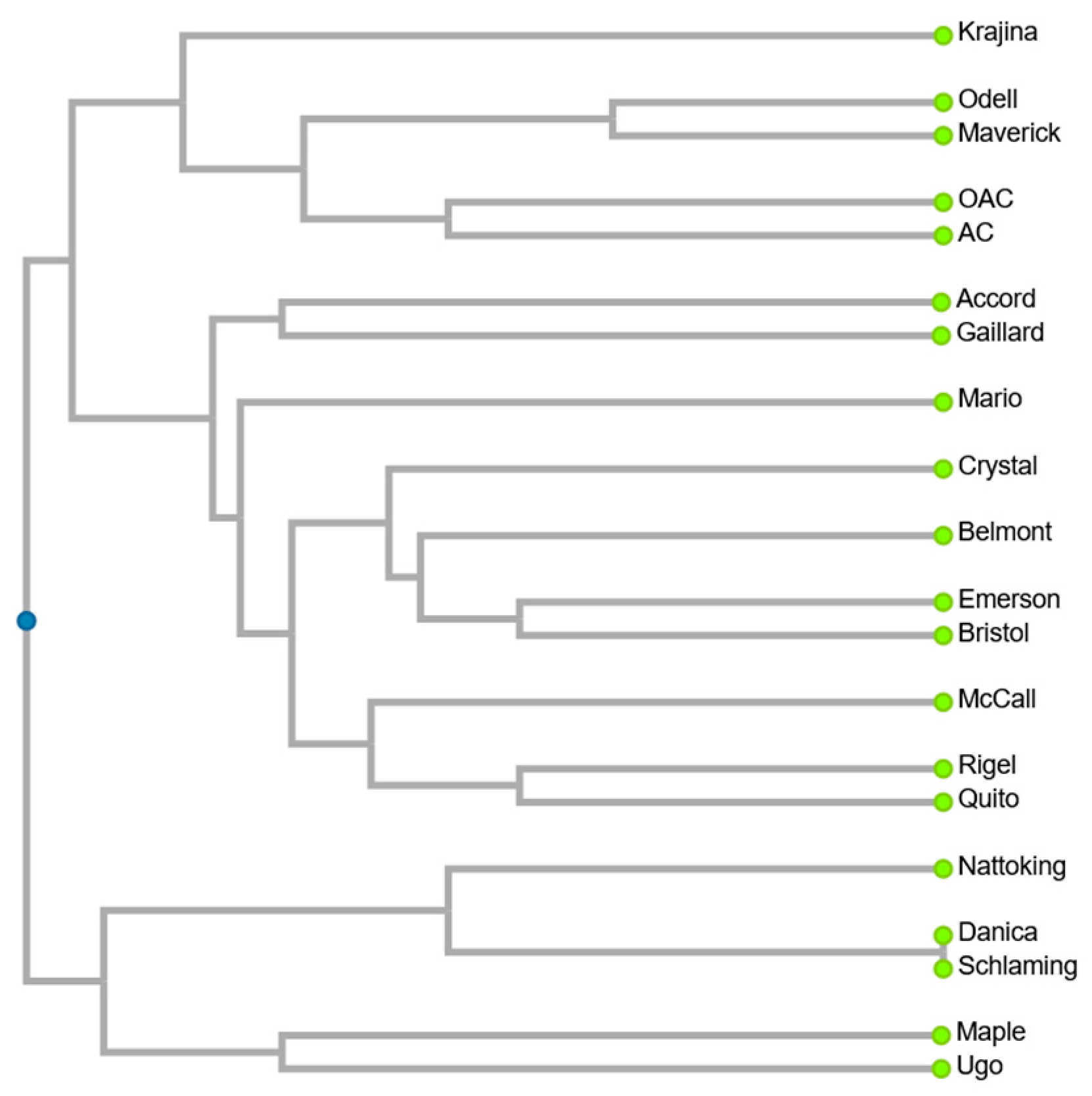
3. Results and Discussion
3.1. PBAP of Analysed G. max Varieties
3.2. VBAP of Analysed G. max Varieties
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smýkal, P.; Coyne, C.J.; Ambrose, M.J.; Maxted, N.; Schaefer, H.; Blair, M.W.; Berger, J.; Greene, S.L.; Nelson, M.N.; Besharat, N.; et al. Legume crops phylogeny and genetic diversity for science and breeding. Crit. Rev. Plant Sci. 2015, 34, 43–104. [Google Scholar]
- Bennetau-Pelissero, C. Plant proteins from Legumes. In Bioactive Molecules in Food; Mérillon, J.M., Ramawat, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 223–265. [Google Scholar]
- Colletti, A.; Attrovio, A.; Boffa, L.; Mantegna, S.; Cravotto, G. Valorisation of by-products from soybean (Glycine max (L.) Merr.) processing. Molecules 2020, 25, 2129. [Google Scholar] [CrossRef] [PubMed]
- Modgil, R.; Tanwar, B.; Goyal, A.; Kumar, V. Soybean (Glycine max). In Oilseeds: Health Attributes and Food Applications; Springer: Singapore, 2021; pp. 1–46. [Google Scholar]
- Verma, A.K.; Kumar, S.; Das, M.; Dwivedi, P.D. A comprehensive review of legume allergy. Clin. Rev. Allergy Immunol. 2013, 45, 30–46. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Radauer, C. A classification of plant food allergens. J. Allergy Clin. Immunol. 2004, 113, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Jardim-Botelho, A.; de Oliveira, L.C.L.; Motta-Franco, J.; Solé, D. Nutritional management of immediate hypersensitivity to legumes in vegetarians. Allergol. Immunopathol. 2022, 50, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Burney, J.; Ramanathan, V. Recent climate and air pollution impacts on Indian agriculture. Proc. Natl. Acad. Sci. USA 2014, 111, 16319–16324. [Google Scholar] [CrossRef] [PubMed]
- Nwaru, B.I.; Hickstein, L.; Panesar, S.S.; Roberts, G.; Muraro, A.; Sheikh, A.; EAACI Food Allergy and Anaphylaxis Guidelines Group. Prevalence of common food allergies in Europe: A systematic review and meta-analysis. Allergy 2014, 69, 992–1007. [Google Scholar] [CrossRef] [PubMed]
- Klemans, R.J.; Knol, E.F.; Michelsen-Huisman, A.; Pasmans, S.G.; de Kruijf-Broekman, W.; Bruijnzeel-Koomen, C.A.; van Hoffen, E.; Knulst, A.C. Components in soy allergy diagnostics: Gly m 2S albumin has the best diagnostic value in adults. Allergy 2013, 68, 1396–1402. [Google Scholar] [CrossRef] [PubMed]
- Klongová, L.; Kováčik, A.; Urbanová, L.; Kyseľ, M.; Ivanišová, E.; Žiarovská, J. Utilization of specific primers in legume allergens based polymorphism screening. Sci. Technol. Innov. 2021, 13, 12–21. [Google Scholar] [CrossRef]
- Garcia-Vallve, S.; Palau, J.; Romeu, A. Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol. Biol. Evololution 1999, 16, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Marzano, V.; Tilocca, B.; Fiocchi, A.G.; Vernocchi, P.; Mortera, S.L.; Urbani, A.; Roncada, P.; Putignani, L. Perusal of food allergens analysis by mass spectrometry-based proteomics. J. Proteom. 2020, 215, 103636. [Google Scholar] [CrossRef]
- Nakamura, R.; Teshima, R. Proteomics-based allergen analysis in plants. J. Proteom. 2013, 93, 40–49. [Google Scholar] [CrossRef] [PubMed]
- National Centre of Biotechnology Information. Available online: www.ncbi.nlm.nih.gov (accessed on 12 September 2023).
- UniProt: The Universal Protein Knowledgebase in 2023. Available online: www.uniprot.org (accessed on 13 September 2023).

| Sample Number | Variety | Sample Number | Variety | Sample Number | Variety |
|---|---|---|---|---|---|
| 1 | Maria | 11 | Nattoking—K88 | 21 | Gaillard |
| 2 | Mivak | 12 | Nattoking—K87 | 22 | Mario |
| 3 | Arkadija | 13 | Danica | 23 | Rigel |
| 4 | Odesskaja | 14 | Balkan | 24 | Ugo |
| 5 | Maple Ridge | 15 | Schladming | 25 | Quito |
| 6 | McCall | 16 | Krajina | 26 | Dorota |
| 7 | OAC Scorpio | 17 | Odell | 27 | Emerson |
| 8 | Sibley | 18 | Maverick | 28 | Bristol |
| 9 | Sturdy | 19 | Accord | 29 | Belmont |
| 10 | Simpson | 20 | AC Glengarry | 30 | Crystal |
| Specific primers: | |
| Forward: | 5′-ACCGGCCAAGATCTGGTTTT-3′ |
| Reverse: | 5′-AGGTAGTCTCCCAACCTCTCC-3′ |
| Degenerated primers: | |
| Forward: | 5′-AGAGAATTCCATATGTCGTGCCARRCGTACGT-3′ |
| Reverse: | 5′-AGAAAGCTTYTACAKGCCYTGTTCABVAGGTA-3′ |
| Specific primers: | |
| Forward: | 5′-AGGGATCTTTATTGTTGCCA-3′ |
| Reverse: | 5′-TCATTTCTTTGACCCACAAG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kováčik, A.; Žiarovská, J.; Urbanová, L. Variability of Allergen-Based Length Polymorphism of Glycine max L. Varieties. Biol. Life Sci. Forum 2024, 30, 20. https://doi.org/10.3390/IOCAG2023-16879
Kováčik A, Žiarovská J, Urbanová L. Variability of Allergen-Based Length Polymorphism of Glycine max L. Varieties. Biology and Life Sciences Forum. 2024; 30(1):20. https://doi.org/10.3390/IOCAG2023-16879
Chicago/Turabian StyleKováčik, Adam, Jana Žiarovská, and Lucia Urbanová. 2024. "Variability of Allergen-Based Length Polymorphism of Glycine max L. Varieties" Biology and Life Sciences Forum 30, no. 1: 20. https://doi.org/10.3390/IOCAG2023-16879
APA StyleKováčik, A., Žiarovská, J., & Urbanová, L. (2024). Variability of Allergen-Based Length Polymorphism of Glycine max L. Varieties. Biology and Life Sciences Forum, 30(1), 20. https://doi.org/10.3390/IOCAG2023-16879






