Abstract
Citrus fruits enjoy widespread consumption globally, being among the most popular fruits. They are highly regarded for their nutritional composition, offering a range of beneficial nutrients. However, it is important to acknowledge that they can also elicit allergic reactions in sensitized individuals, which presents a contrasting aspect. The Bet v 1 cross-reacting allergen is a major birch pollen allergen, and it is the most commonly sensitizing allergen in central Europe. Bet v 1 belongs to the group of PR-10 proteins in the plant kingdom that cause various allergic reactions. The Bet v 1 allergen has a number of isoforms and homologs. These homolog genes are inherited from a common ancestor and subsequent amino acid similarity. They can cause the phenomenon of cross-reactivity in food allergies. The aim of the study was to analyze the length of polymorphism variability of the Bet v 1 homolog in orange varieties by using degenerated and nondegenerated primers. A total of eight varieties of Citrus sinensis L. Osbeck were used in the analysis. The BBAP technique (Bet v 1 based amplified polymorphism) was used to detect the length variability of fingerprints of allergen encoding genes of Bet v 1 homologs. Degenerated primer combinations and only one of the nondegenerated variants of primers provided fingerprints that were unique for every individual variety of analyzed oranges. By using other primer variants, from two up to the four varieties generated by the same BBAP profile, indicate a higher degree of Bet v 1 homolog sequential conservativity when compared to other fruit species.
1. Introduction
Oranges are one of the most grown species and account for more than half of the world’s citrus production [1]. Citrus sinensis L. Osbeck contains various flavonoids, flavonols, polymethoxy flavonoids, flavanones, and coumarins [2]. In addition to its excellent nutritional value, Citrus sinensis L. Osbeck is also known for its use in medicine because it possesses antiproliferative activity, antibacterial activity, antifungal activity, antiparasitic activity, insecticidal activity, and many other health benefits [3]. Oranges are divided into sweet oranges, which are divided into navel oranges, white oranges, and blood oranges. And then, we have a group of sour oranges [4].
Plant species have a large number of proteins in common, including allergenic proteins. An increasing number of proteins potentially expressed in all plant species have been identified from decades of molecular biology studies and genome sequencing. Many genes encoding homologous proteins have been found in the genomes. Bet v 1 (major allergen of birch pollen) and its homologs belong to the PR-10 (pathogenesis-related class 10) protein family [5]. Research about the Bet v 1 allergen dates back to 1989 [6]. Many plants contain food allergens that are Bet v 1 homologs, suggesting that people allergic to birch pollen often suffer from PFS (pollen food syndrome) syndrome. When such a phenomenon occurs, we talk about cross-reactivity [7]. Cross-reactivity can be described as the similarity between two allergens, and the more similar they are, the more likely it is that cross-reactivity will occur [8]. Franzese and co-authors reported that the cross-reaction is a consequence of a similar epitope structure of the allergen to which the same antibodies bind [9]. Reactions to plant foods associated with birch pollen (Bet v 1) are considered to be the most common form of food allergy in adults in Central and Northern Europe [10]. Studies show that cross-reactions occur between oranges and other foods, such as peanuts [11]. The database AllerBase contains up to 27 isoallergens of Bet v 1 [12]. When we refer to isoallergens, we are talking about allergens that are homologous, and exhibit shared biochemical characteristics. These shared properties include a similar molecular size, comparable or identical known biological functions, and an amino acid sequence identity of at least 67%. It is important to note that each isoallergen can have multiple highly similar forms (>90% identity), which are commonly referred to as variants or isoforms [13]. Structurally homologous Bet v1 isoforms may have different properties in terms of allergic sensitization and Th2 polarization. This is probably due to differential susceptibility to proteolytic cleavage [14]. The AllerBase database also contains cross-reactive allergens that include the allergen Mal d 1, which is the main allergen of Malus domestica [15]; Api g 1 is a major allergen of Apium graveolens [16] and many others. Knowledge of the Bet v 1 homologs is increasing, but the knowledge of the ypr10 gene and its potential applicability in different genomic techniques is still limited. However, a certain percentage of homology has been found [17].
Four primer pairs were designed for the development of the BBAP technique for the comparison of triplets for different amino acids (histidine/asparagine/glutamine/lysine). In developing the primers, the authors focused specifically on two amino acid segments. These segments were subjected to a BLAST (Basic Local Alignment Search Tool) analysis with fruit species with established genomic sequences. Forward primer was designed for a region of high homology in Malus domestica [18]. A degenerate primer has degeneracy situated at positions 12 (S) and 14 (K), meaning that position 12 can be occupied by either guanine or cytosine, and position 14 can be filled with either thymine or guanine [19,20].
2. Methods
2.1. Plant Material and DNA Isolation
In the study, we used 8 varieties of Citrus sinesis L. Osbeck (Salustiana, Navelina, Navel Late, Mid Knights, Odmiana, Lane Late, Valencia, Spain). Total genomic DNA was isolated by using Thermo scientific GeneJET purification mini kit, according to the manufacturer’s protocol. The samples were subjected to a PCR analysis to confirm their functionality. In this analysis, the presence of ITS (Internal Transcribed Spacer) sequences, which are universally present in organisms belonging to the Eukarya domain, was examined. The presence of ITS was verified on 1.5% agarose gel.
2.2. BBAP Analysis
In our analysis, the BBAP technique was used to detect the homologs of Bet v 1 allergen in Citrus sinensis L. Osbeck. Five different reverse primers were used in the PCR analysis according to the methodology of Žiarovská and Urbanová [18]: R1 (5′- aaccacaccatcaccgac-3′), R2 (5′-aaccacaccatcaacgac-3′), R3 (5′-aaccacaccatgaccgac-3′), R4 (5′-aaccacaccatgaacgac-3′) and one degenerated primer (5′-ttggtgtggtastkgctg-3′). One forward primer was used in analysis (5′-cctggaaccatcaagaag-3′). The premix itself consisted of 5 μL of Mastermix (2 × Elizyme HS Robust Mix), forward primer, and reverse primer in 400 nM concentrations, H2O, and 4 μL of DNA. All components were pipetted to a final volume of 10 μL. The following temperature and time regimes were used for thermal cycling on the PCR cyclers: initial denaturation 95 °C for 5 min, denaturation 95 °C for 45 s, annealing 54 °C for 45 s, elongation 72 °C for 35 s, and the last step was final elongation of 72° for 10 min. Results from electroforeogram were processed by using GelAnalyzer. Binary matrix was created from fragments distribution in gel and followed dendrogram by using DendroUPGMA [21]. Jaccard coefficient has been used to compare between sets of variables.
3. Results and Discussion
Due to a phenomenon called cross-reaction, allergy to oranges is often associated with pollinosis and sensitization to other plants [22]. Using reverse primer R1, a total of 56 fragments were amplified in all Citrus sinensis varieties. These fragments were visualized and evaluated on agarose gels, indicating the presence of approximately 200 bp (base pair) fragments in each variety. When the reverse primer R2 was used, slightly more fragments (58) were produced. Conversely, when the reverse primer R3 was used, the smallest number of fragments, only 38 were detected. The reverse primer R4 resulted in the production of 57 fragments. Additionally, the use of the degenerate primer led to the amplification of 48 fragments. By using primers R2, R3, and R4, we detected Bet v 1 homologs with a size of around 388 bp. The main allergen of birch Bet v 1 gained its notoriety thanks to the phenomenon of cross-reaction. It is likely that homologs of Bet v 1 found in several plants cause the so-called cross-reaction phenomenon in humans [10]. The variability of homologs of Bet v 1 with the BBAP technique used was monitored in Malus domestica [20] and in cereals, specifically in Avena Sativa [23]. In a study conducted by Žiarovská and co-authors, they performed an analysis where allergen Bet v 1 homologs were identified in a range of 30 plant species. These species included Ficus carica, Carica papaya, Pyrus communis, Punica granatum, Vaccinium myrtillus, Ananas comosus, Citrus reticulata, Annona cherimola, Castanea sativa, and Citrus × limon [17]. Notably, the study also investigated Citrus sinensis, the focus of our own investigation. The presence of Bet v 1 homologs in these diverse plant species suggests a potential role in allergenicity and highlights the relevance of understanding these homologs across various plant taxa. Urbanová and Žiarovská applied the BBAP technique to different vegetable species to see what profiles and how much variability there is between species. The vegetables included Allium cepa, Beta vulgaris, Spinacia oleracea, Daucus carota, and others. Apium graveolens was also analyzed in the same study [24]. Bohle, in their study, already reported that in these vegetable species, the Bet v 1 homologs are present [25]. Several techniques have already been used to detect Bet v 1 homologs in plants such as Cannabis sativa [26]. Comparing and searching for conserved stretches of PR genes has also been addressed by Juskyté and her colleagues. In their study, they compared the sequences of a putative PR genes among different crops, including Citrus sinensis [27].
The Cophenetic correlation coefficient when using primer R1 is 0.83; for R2, it is 0.91; for R3, it is 0.94; for R4, it is 0.79; and for D, it is 0.86. The genetic distance was from 0.000 (Odmiana and Lane Late) to 0.545 (Odmiana and Navel Late, Lane Late, and Navel Late) by using primer R1, from 0.000 (Odmiana and Mid Knights, Valencia and Mid Knights, Valencia, and Odmiana) to 0.556 (Navel and Salustiana) by using primer R2, from 0.000 (Navel and Mid Knights, Navelina, and Navel Late) to 0.667 (Salustiana and Valencia, Mid Knights, and Valencia) by using primer R3, from 0.222 (Navel and Lane Late) to 0.636 (Navel and Valencia) by using primer R4, and from 0.167 (Navel and Navelina) to 0.889 (Mid Knights and Navel Late) by using primer D. From the results of the distance matrix and dendrograms (Figure 1), we can conclude that when we use primer R1 and R2, the same profile was degenerated between some varieties, but on the contrary, by using primers R3, R4, and D, there were different profiles between each variety.
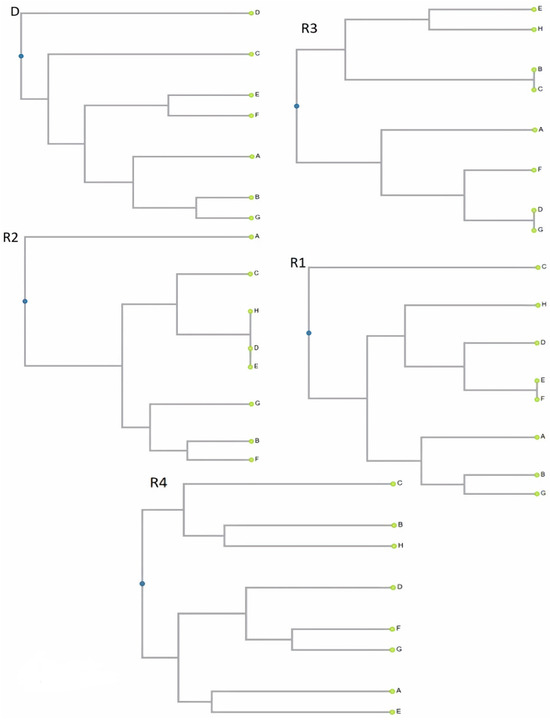
Figure 1.
Dissimilarity dendrograms for length polymorphism of Bet v 1 homologs in analysis of different varieties in Citrus sinenes L. Osbeck. The letters represent the following varieties of Citrus sinensis: A = Salustiana, B = Navelina, C = Navel Late, D = Mid Knights, E = Odmiana, F = Lane Late, G = Navel, and H = Valencia.
4. Conclusions
It is likely that in Central Europe, where birch is abundant, homologs of the Bet v1 allergen in plants such as Citrus sinensis L. Osbeck may play a role in allergy to Citrus sinensis. This analysis provides valuable information about the variability between each variety of Citrus sinensis L. Osbeck. The successful application of this BBAP technique to Citrus sinensis L. Osbeck varieties suggests that they have a wide range of practical uses, and as we have found out from other studies, this technique can be applied to different types of vegetables and fruits with consistent results. This universal applicability indicates that the BBAP technique can be utilized across multiple vegetable and fruit species, allowing for an efficient and reliable analysis of genetic variability.
Author Contributions
D.M. and J.Ž. contributed to the proceeding paper equally. All authors have read and agreed to the published version of the manuscript.
Funding
This publication was supported by the Operational program Integrated Infrastructure within the project: Demand-driven research for the sustainable and innovative food, Drive4SIFood 313011V336, co-financed by the European Regional Development Fund.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- FAO. Citrus©; FAO: Rome, Italy, 2021. [Google Scholar]
- Luo, J.; Yuan, H.; Mao, L.; Wu, J.; Jiang, S.; Yang, Y.; Fu, Y.; Liu, L.; Chen, S.; Wang, W. The Young Fruit of Citrus Aurantium L. or Citrus Sinensis Osbeck as a Natural Health Food: A Deep Insight into the Scientific Evidence of Its Health Benefits. Arab. J. Chem. 2023, 16, 104681. [Google Scholar] [CrossRef]
- Dongre, P.; Doifode, C.; Choudhary, S.; Sharma, N. Botanical Description, Chemical Composition, Traditional Uses and Pharmacology of Citrus Sinensis: An Updated Review. Pharmacol. Res. -Mod. Chin. Med. 2023, 8, 100272. [Google Scholar] [CrossRef]
- Calabrese, F. Origin and history. In Citrus; Dugo, G., Giacomo, A.D., Eds.; The Taylor & Francis: Abingdon, UK, 2002; p. 642. ISBN 0-203-21661-X. [Google Scholar]
- Alessandri, C.; Ferrara, R.; Bernardi, M.L.; Zennaro, D.; Tuppo, L.; Giangrieco, I.; Tamburrini, M.; Mari, A.; Ciardiello, M.A. Diagnosing Allergic Sensitizations in the Third Millennium: Why Clinicians Should Know Allergen Molecule Structures. Clin. Transl. Allergy 2017, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Kraft, D. The History and Science of the Major Birch Pollen Allergen Bet v 1. Biomolecules 2023, 13, 1151. [Google Scholar] [CrossRef] [PubMed]
- Biedermann, T.; Winther, L.; Till, S.J.; Panzner, P.; Knulst, A.; Valovirta, E. Birch Pollen Allergy in Europe. Allergy 2019, 74, 1237–1248. [Google Scholar] [CrossRef] [PubMed]
- Aalberse, R.C. Assessment of allergen cross-reactivity. Clin. Mol. Allergy 2007, 5, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Franzese, C.B.; Damask, C.C.; Wise, S.K.; Ryan, M.W. Handbook of Otolaryngic Allergy; Thieme Medical Publisher: New York, NY, USA, 2019; p. 306. ISBN 9781626239074. [Google Scholar]
- Kleine-Tebbe, J.; Ballmer-Weber, B.K.; Breitneder, H.; Vieths, S. Bet v 1 and its Homologs: Triggers of Tree-Pollen Allergy and Birch Pollen-Associated Cross-Reactions. In Molecular Allergy Diagnostics; Klene-Tebbe, J., Jacob, T., Eds.; Springer: Cham, Switzerland, 2017; pp. 21–42. ISBN 978-3-319-42499-6. [Google Scholar]
- Glaspole, I.N.; de Leon, M.P.; Rolland, J.M.; O’Hehir, R.E. Anaphylaxis to Lemon Soap: Citrus Seed and Peanut Allergen Cross-Reactivity. Ann. Allergy Asthma Immunol. 2007, 98, 286–289. [Google Scholar] [CrossRef] [PubMed]
- Kadam, K.; Karbhal, R.; Jayaraman, V.K.; Sawant, S.; Kulkarni-Kale, U. AllerBase: A Comprehensive Allergen Knowledgebase. Database 2017, 2017, bax066. [Google Scholar] [CrossRef] [PubMed]
- Pomés, A.; Davies, J.M.; Gadermaier, G.; Hilger, C.; Holzhauser, T.; Lidholm, J.; Lopata, A.L.; Mueller, G.A.; Nandy, A.; Radauer, C.; et al. WHO/IUIS Allergen Nomenclature: Providing a Common Language. Mol. Immunol. 2018, 100, 3–13. [Google Scholar] [CrossRef]
- Grutsch, S.; Fuchs, J.E.; Ahammer, L.; Kamenik, A.S.; Liedl, K.R.; Tollinger, M. Conformational Flexibility Differentiates Naturally Occurring Bet v 1 Isoforms. Int. J. Mol. 2017, 18, 1192. [Google Scholar] [CrossRef]
- Fahlbusch, B.; Rudeschko, O.; Müller, W.D.; Schlenvoigt, G.; Vettermann, S.; Jäger, L. Purification and Characterization of the Major Allergen from Apple and Its Allergenic Cross-Reactivity with Bet v 1. Int. Arch. Allergy Immunol. 1995, 108, 119–126. [Google Scholar] [CrossRef]
- Gepp, B.; Lengger, N.; Bublin, M.; Hemmer, W.; Breiteneder, H.; Radauer, C. Chimeras of Bet v 1 and Api g 1 Reveal Het ogeneous IgE Responses in Patients with Birch Pollen Allergy. J. Allergy Clinnical Immunol. 2014, 134, 188–194. [Google Scholar] [CrossRef]
- Žiarovská, J.; Zeleňáková, L. Application of genomic data for PCR Screening of BET v 1 conserved sequence in clinically relevant plant species. In Systems Biology; IntechOpen: London, UK, 2018. [Google Scholar]
- Žiarovská, J.; Urbanová, L. Utilization of Bet v 1 homologs based amplified profile (BBAP) variability in allergenic plants fingerprinting. Biologia 2022, 77, 517–523. [Google Scholar] [CrossRef]
- Fu, L.; Cherayil, B.; Shi, H.; Wang, Y.; Zhu, Y. Species and structure of food allergens: Epitopes and cross-reactivity. In Food Allergy: From Molecular Mechanisms to Control Strategies; Fu, L., Cherayil, B., Shi, H., Wang, Y., Zhu, Y., Eds.; Springer: Singapore, 2019; pp. 13–39. ISBN 978-981-13-6927-8. [Google Scholar]
- Speváková, I.; Urbanová, L.; Kyseľ, M.; Bilcikova, J.; Farkasová, S.; Ziarovska, J. BBAP Amplification Profiles of Apple Varieties. Sci. Technol. Innov. 2021, 13, 1–6. [Google Scholar] [CrossRef]
- Garcia-Vallvé, S.; Palau, J.; Romeu, A. Horizontal Gene Transfer in Glycosyl Hydrolases Inferred from Codon Usage in Escherichia Coli and Bacillus Subtilis. Mol. Biol. Evol. 1999, 16, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Iorio, R.A.; Del Duca, S.; Calamelli, E.; Pula, C.; Lodolini, M.; Scamardella, F.; Pession, A.; Ricci, G. Citrus Allergy from Pollen to Clinical Symptoms. PLoS ONE 2013, 8, e53680. [Google Scholar] [CrossRef][Green Version]
- Farkasová, S.; Droppa, M.; Žiarovská, J. variability of amplified profiles generated by BBAP in avena sativa l. J. Microbiol. Biotechnol. Food Sci. 2023, 12, e9545. [Google Scholar] [CrossRef]
- Urbanová, L.; Žiarovská, J. Variability of DNA based amplicon profiles generated by Bet v 1 homologous among different vegetable species. Acta Fytotech. Et Zootech. 2021, 24, 1–6. [Google Scholar] [CrossRef]
- Bohle, B.; Radakovics, A.; Jahn-Schmid, B.; Hoffmann-Sommergruber, K.; Fischer, G.F.; Ebner, C. Bet v 1, the Major Birch Pollen Allergen, Initiates Sensitization to Api g 1, the Major Allergen in Celery: Evidence at the T Cell Level. Eur. J. Immunol. 2003, 33, 3303–3310. [Google Scholar] [CrossRef]
- Ebo, D.G.; Decuyper, I.I.; Rihs, H.P.; Mertens, C.; Van Gasse, A.L.; Van der Poorten, M.L.M.; De Puysseleyr, L.; Faber, M.A.; Hagendorens, M.M.; Bridts, C.H.; et al. IgE-Binding and Mast Cell–Activating Capacity of the Homologue of the Major Birch Pollen Allergen and Profilin from Cannabis Sativa. J. Allergy Clin. Immunol. 2021, 9, 2509–2512.e3. [Google Scholar] [CrossRef]
- Juškytė, A.D.; Mažeikienė, I.; Stanys, V. Putative Genes of Pathogenesis-Related Proteins and Coronatine-Insensitive Protein 1 in Ribes Spp. Plants 2022, 11, 355. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).