Abstract
Hyperspectral images (HSI) offer detailed spectral reflectance information about sensed objects through provision of information on hundreds of narrow spectral bands. HSI have a leading role in a broad range of applications, such as in forestry, agriculture, geology, and environmental sciences. The monitoring and management of agricultural lands is of great importance for meeting the nutritional and other needs of a rapidly and continuously increasing world population. In relation to this, classification of HSI is an effective way for creating land use and land cover maps quickly and accurately. In recent years, classification of HSI using convolutional neural networks (CNN), which is a sub-field of deep learning, has become a very popular research topic and several CNN architectures have been developed by researchers. The aim of this study was to investigate the classification performance of CNN model on agricultural HSI scenes. For this purpose, a 3D-2D CNN framework and a well-known support vector machine (SVM) model were compared using the Indian Pines and Salinas Scene datasets that contain crop and mixed vegetation classes. As a result of this study, it was confirmed that use of 3D-2D CNN offers superior performance for classifying agricultural HSI datasets.
1. Introduction
Remote sensing data obtained from airborne and spaceborne sensors has come to provide more detailed spatial and spectral resolution with developments in recent years. Through these improvements, remotely sensed data is now a time-saving and low-cost alternative for precision agriculture and forestry applications. Applications, such as the detection and separation of various vegetation species, and determination of the conditions of crops, require rich spectral and spatial resolution. Hyperspectral images (HSI) are the most suitable data for these types of analysis, by providing high spectral resolution with hundreds of spectral bands. Many studies in the literature have been performed to produce highly accurate classification maps and identify land cover types by classifying HSIs. However, high spectral information produces a huge volume of data and high dimensionality. This causes the Hughes phenomenon which is one of main problems in HSI classification [1]. Traditional classification methods, such as maximum likelihood and the Spectral Angle Mapper, cannot handle HSI data with high classification accuracy due to these problems.
In the last three decades, various studies have been conducted to apply the high classification success of machine learning (ML) methods to HSI classification problems. Gualtieri et al. [2] applied the SVM model with an ad-hoc kernel to Indian Pines (IP) and Salinas Scene (SS) datasets and achieved 87.3% and 98.6% overall accuracy (OA), respectively. Chan & Paelinckx [3] compared tree-based random forest and Adaboost algorithms on HSI obtained with a HyMap sensor. The results of this study were that Adaboost showed slightly better OA while random forest required less processing time. In recent years, convolutional neural network (CNN) algorithms, that have become more widespread in various fields of application, have been used in HSI classification. Luo et al. [4] applied HSI-CNN and HSI-CNN + XGBoost architectures to HSI datasets. Experimental results with common HSI benchmark datasets showed that the proposed methods provided more than 99% OA. Roy et al. [5] proposed the HybridSN architecture providing both spatial-spectral and spatial feature extraction capability from HSIs. The architecture demonstrated over 99% OA on various benchmark datasets. However, the HyRANK dataset has hardly ever been used as a benchmark in studies in the literature despite the widespread use of IP and SS datasets in studies.
In this study, the classification performance of SVM and CNN algorithms was evaluated. For this purpose, two publicly available HSI datasets, namely SS and HyRANK Loukia (HL), were used. The datasets contain various land classes related to agriculture and forestry. In the data preparation stage, principal components analysis (PCA) was applied to both datasets to reduce band numbers and avoid high dimensionality of HSI. A total of 150 training samples were selected of almost every class from both datasets to simulate a limited training sample scenario. After applying the classification models to HSIs, the classification performances of the algorithms were evaluated by examining OA, producer accuracy (PA), user accuracy (UA), f scores, and the kappa coefficient (κ).
2. Classification Methods
2.1. Support Vector Machines (SVM)
SVM is a supervised and non-parametric ML algorithm based on statistical learning theory, developed by Vapnik [6]. There are no assumptions about data distribution. In binary classification problems in which classes can be separated linearly, classes can be separated with an infinite number of linear decision boundaries. The main approach of SVM is to identify the best decision boundary that minimizes generalization error, called an optimum hyperplane [7,8]. Data samples that were closest to the hyperplane were used to measure the margin, referred to as support vectors (SV) [7]. Since it considers only SVs, SVM can be useful with limited training sets, where collecting training data is costly in terms of both time and resources [9,10]. In most classification problems, such as remotely sensed images, classes cannot be separated by linear hyperplanes. To overcome this situation, kernel functions are used to transform to a larger feature space. Commonly used kernels are linear, sigmoid, polynomial, and radial basis function (RBF) [11]. However, the RBF kernel outperforms on most classification problems [11,12]. Therefore, the RBF was used in this study when implementing the SVM model, by determining C and γ parameters with the grid search algorithm.
2.2. Convolutional Neural Networks (CNNs)
CNN is a form of deep learning that processes data in the form of multiple arrays, such as 1-dimensional data including sequences and signals, 2-dimensional data including images and audio spectrograms, 3-dimensional data including volumetric images, and videos [13]. CNNs are generally formed of three fundamental components which serve different purposes, referred to as the convolution layer, the pooling layer, and the fully connected layer. In the convolution layer, various kernels are utilized such that the entire input data (tensors) and feature maps are created [14]. Subsequently, feature maps are put into an activation function to generate output feature maps, such as ReLU. A pooling layer is applied to the convolution layer in most CNN models in order to reduce the dimensionality of output feature maps. The third and last type of layer is the fully connected layer, where neurons are fully connected to all neurons in the previous layer, as in a regular artificial neural network [15]. Following this, the layer is connected to a classifier, such as Softmax, and classification is performed.
In this study, a hybrid CNN model was used to classify HSIs which was able to extract spectral and spatial features along bands using 3D and 2D convolution operations. To avoid spectral redundancy of HSI, PCA transformation was applied along bands and the first 15 principal components were used. The model contained two 3D convolution layers and one 2D convolution layer. The 2D convolution operation can extract only spatial features of input data. In the 3D convolution operation, spectral and spatial learning is performed simultaneously. The PReLU activation function was selected for its advantages over ReLU [16]. All weights were randomly initialized and trained using a back-propagation algorithm with the Adam optimizer using the Softmax classifier. Epoch and batch size parameters were detected as 256 and 500, respectively. A summary of the CNN model is given at Table 1.

Table 1.
Summary of the CNN model.
3. Experiments
3.1. Dataset Description
SS data was acquired on 9th October using an AVIRIS sensor with a 3.7-m spatial resolution and 224 bands. To train the algorithms, 150 samples were selected randomly from each class. The numbers of training and testing samples are given in Table 2. The final size of the hypercube was 512 × 217 × 204. The image composition and ground truth of SS are given in Figure 1.

Table 2.
Number of samples of the SS and HL datasets.
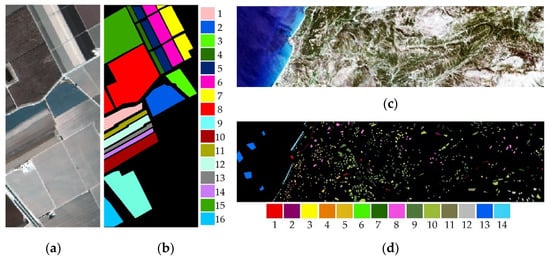
Figure 1.
True color and labeled views of HSIs: (a) RGB composite and (b) ground truth of the SS with legend, and (c) RGB composite, and (d) ground truth of the HL.
The HyRANK dataset was developed through a scientific initiative of the ISPRS, WG III/4 [17]. The data was obtained using an EO-1 Hyperion sensor with 30 m spatial resolution and 220 bands. Only the Loukia (HL) data was considered in this study, while the HyRANK dataset contained 5 HSI data. The ground truth data of the HL contains 14 of land use/land cover classes. The size of HL was 250 × 1376 × 176. The image composition and ground truth of HL is given in Figure 1. Since the 2nd and 4th classes had limited samples on the HL, 150 samples were chosen randomly from all classes, except the aforementioned classes, equally to train the algorithms. There were 30 samples chosen from the 2nd and 4th classes. The numbers of the training and testing samples for HL are given in Table 2.
3.2. Experimental Results
The classification models were built using Python’s Tensorflow [18] and Scikit-learn [19] libraries. The best C and γ parameters were determined for both datasets as 100 and 0.1 respectively with grid search. To compare the classification performances of the algorithms, OA, PA, UA, f scores, and κ were calculated. Accuracy metrics for datasets are given in Table 3. Note that bold characters indicate higher value on the rows in the same category for each dataset. As can be seen from the OA and κ accuracy metrics in the table, CNN outperformed SVM for both datasets. For the SS dataset, CNN showed slightly better performance compared to SVM on the 3rd, 4th, 5th, and 10th classes, while CNN’s PA, UA and f score values for the 8th and 15th classes were significantly higher than those for SVM. SVM showed a slightly better performance for PA on the 16th class, 0.01 higher than CNN’s. When comparing processing times, it was clear that CNN was faster because the SVM parameter tuning process was included in the training time.

Table 3.
The performance analysis of classification algorithms.
Figure 2a,b shows the classification maps for the SS dataset. Some pixels of the parcel on the upper-left of the image labeled as Fallow_rough_plow class on the ground truth data were misclassified by both classification algorithms. Other significant misclassifications included the upper-right parcel, which was classified as Vinyard_vertical_trellis by CNN, and classified by SVM as Lettuce_romaine_6wk and Vinyard_vertical_trellis, where there was no available ground truth data.
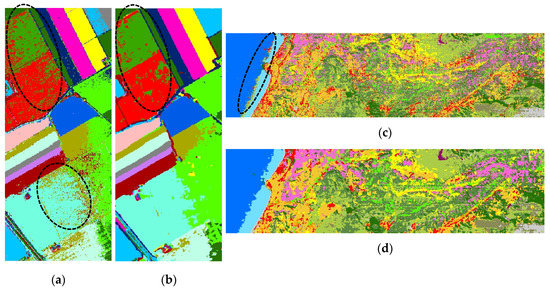
Figure 2.
The classification maps for the SS dataset using (a) SVM and (b) CNN, and for the HL dataset using (c) SVM and (d) CNN, respectively.
For the HL dataset, CNN showed better classification performance in almost every class in terms of OA and κ metrics. In the 2nd, 10th, and 15th classes, SVM’s PA values were slightly higher than CNN’s PA values. The 12th class showed better classification performance on the SVM. The PA of the 1st, 6th, and 7th classes obtained a low value in a range from 0.16 to 0.62 for both classification algorithms, indicating that classification performance was considerably worse than for other classes. The reason for misclassification could be that the boundary limits for these classes on feature space could not be defined properly. Moreover, low spatial resolution of the spaceborne HSI causes a mixed pixel problem. Considering the training time of the algorithms, the SVM was outperformed by CNN.
Figure 2c,d shows the classification maps for the HL dataset. It can be seen from Figure 2d that some pixels were misclassified on the sea along the line where the coastal water and water classes joined. On the other hand, visual analysis of the result was harder than for the other dataset since ground data was not collected for the larger region of interest from wider areas.
4. Conclusions
In this study, the classification of HSI datasets was evaluated with SVM and CNN algorithms. For this purpose, two publicly available datasets that included agricultural and forestry classes were evaluated. The experimental results given in Table 3 show that the CNN algorithm outperformed for both HSI datasets. For the SS dataset, CNN showed a better performance on PA, UA and f scores compared to SVM. For the HL dataset, CNN again achieved better f scores in 10 of 14 land-cover classes. Despite the high accuracy of the measured UA values, the calculated PA metrics were very low in the broad-leaved forest and coniferous forest classes. The reason for the low accuracy of HL could be the unbalanced ground truth data that was labelled without field studies and the low spatial resolution of the data provided by the spaceborne sensor. However, the classification performance was sufficient despite the limited training data. The results showed that CNN models are useable for HSI classification problems that include agricultural and forestry areas.
Supplementary Materials
The poster presentation is available online at https://www.mdpi.com/article/10.3390/IECAG2021-09739/s1.
Acknowledgments
The authors thank the ISPRS Commission III, WG III/4 for providing the HyRANK dataset used in the experiments. This study was supported by Afyon Kocatepe University Scientific Research Projects Coordination Unit within the 20.FEN.BİL.12 project.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dalponte, M.; Bruzzone, L.; Vescovo, L.; Gianelle, D. The Role of Spectral Resolution and Classifier Complexity in the Analysis of Hyperspectral Images of Forest Areas. Remote Sens. Environ. 2009, 113, 2345–2355. [Google Scholar] [CrossRef]
- Gualtieri, J.A.; Chettri, S.R.; Cromp, R.F.; Johnson, L.F. Support Vector Machine Classifiers as Applied to AVIRIS Data. In Proceedings of the Eighth JPL Airborne Geoscience Workshop, Pasadena, CA, USA, 9–11 Feburary 1999. [Google Scholar]
- Chan, J.C.-W.; Paelinckx, D. Evaluation of Random Forest and Adaboost Tree-based Ensemble Classification and Spectral Band Selection for Ecotope Mapping Using Airborne Hyperspectral Imagery. Remote Sens. Environ. 2008, 112, 2999–3011. [Google Scholar] [CrossRef]
- Luo, Y.; Zou, J.; Yao, C.; Zhao, X.; Li, T.; Bai, G. HSI-CNN: A Novel Convolution Neural Network for Hyperspectral Image. In Proceedings of the 2018 International Conference on Audio, Language and Image Processing (ICALIP), IEEE, Beijing, China, 16 July 2018. [Google Scholar]
- Roy, S.K.; Krishna, G.; Dubey, S.R.; Chaudhuri, B.B. HybridSN: Exploring 3-D–2-D CNN Feature Hierarchy for Hyperspectral Image Classification. IEEE Geosci. Remot. Sens. Lett. 2019, 17, 277–281. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Vapnik, V. The Nature of Statistical Learning Theory; Springer: New York, NY, USA, 1995; p. 188. [Google Scholar]
- Pal, M.; Mather, P. Support Vector Machines for Classification in Remote Sensing. Int. J. Remote Sens. 2005, 26, 1007–1011. [Google Scholar] [CrossRef]
- Christovam, L.E.; Pessoa, G.G.; Shimabukuro, M.H.; Galo, M.L. Land Use and Land Cover Classification Using Hyperspectral Imagery: Evaluating the Performance of Spectral Angle Mapper, Support Vector Machine and Random Forest. In Proceedings of the International Archives of the Photogrammetry, Remote Sensing & Spatial Information Sciences, Enschede, The Netherlands, 10–14 June 2019; pp. 1841–1847. [Google Scholar]
- Tuia, D.; Volpi, M.; Copa, L.; Kanevski, M.; Munoz-Mari, J. A survey of active learning algorithms for supervised remote sensing image classification. IEEE J. Sel. Top. Signal Process. 2011, 5, 606–617. [Google Scholar] [CrossRef]
- Kavzoglu, T.; Colkesen, I. A Kernel Functions Analysis for Support Vector Machines for Land Cover Classification. Int. J. Appl. Earth Obs. Geoinf. 2009, 11, 352–359. [Google Scholar] [CrossRef]
- Mountrakis, G.; Im, J.; Ogole, C. Support Vector Machines in Remote Sensing: A Review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio YHinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Liu, Y.; Oerlemans, A.; Lao, S.; Wu, S.; Lew, M.S. Deep Learning for Visual Understanding: A Review. Neurocomputing 2016, 187, 27–48. [Google Scholar] [CrossRef]
- Aloysius, N.; Geetha, M. A Review on Deep Convolutional Neural Networks. In Proceedings of the 2017 International Conference on Communication and Signal Processing (ICCSP), IEEE, Chennai, India, 6–8 April 2017. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015. [Google Scholar]
- Karantzalos, K.; Karakizi, C.; Kandylakis, Z.; Antoniou, G. (Eds.) HyRANK Hyperspectral Satellite Dataset I (Version v001); I.W. III/4; ISPRS: Hannover, Germany, 2018. [Google Scholar] [CrossRef]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th {USENIX} Symposium on Operating Systems Design and Implementation ({OSDI} 16), Savannah, GA, USA, 2–4 November 2016. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).