Abstract
Lactic acid bacteria (LAB) have a vast genetic potential and are used in food production. The aims of this work were to identify forty-six 16S ribosomal gene sequences of LAB isolated from goat’s raw milk cheeses and to evaluate the phylogenetic relationship between the sequences. Using Sanger sequencing and the BLAST algorithm, the species were identified. Phylogenetic analysis was carried out using the R 4.3.0 software, and the genetic distance was calculated using the MEGA 11 software. The phylogenetic tree revealed variability between sequences of the same species, which may have implications for the technological capabilities of LAB sought for the production of cheeses of standardized quality.
1. Introduction
Cheese is a generic name for a group of fermented food products based on milk [1]. Goat’s milk is generally consumed as a product derived from raw milk and used to make cheese and yogurt [2]. Raw milk cheeses differ from pasteurized milk cheeses in their intense and strong taste [3].
Cheese derived from raw milk has a rich microbiota, mainly from the genera Lactococcus, Lactobacillus, Streptococcus, Leuconostoc, and Enterococcus [4]. Nonetheless, it may also include unwanted microorganisms such as Staphylococcus aureus, Shiga-toxin-producing Escherichia coli, Listeria monocytogenes, and Salmonella spp. [5].
Lactic acid bacteria (LAB) are part of a group of bacteria that have been widely studied and applied in the dairy industry, as they play various roles in the manufacturing and ripening of cheese, from the fermentation of sugars and hydrolysis of proteins to synergism and control of adventitious flora, as well as having an influence on the organoleptic characteristics of fermented products [6]. These bacteria can be selected among the varieties based on their metabolic characteristics and antimicrobial activity.
Phylogenetic studies assess the evolutionary relationships of a group of organisms and have various applications in basic biology, such as studying the evolution of a gene and identifying species in samples [7]. The aim of this work was to molecularly identify the LAB present in samples of goat’s raw milk cheese and to use taxonomy and phylogenetics to analyze the diversity and evolutionary proximity between the samples obtained.
2. Materials and Methods
2.1. Reactivation of Cryopreserved Lactic Acid Bacteria Samples
Lactic acid bacteria were previously isolated and cryopreserved from 20 semi-hard Transmontano goat cheeses (DOP) [8], produced with raw goat’s milk in four batches from a company located in Mirandela, Portugal. Of these, forty-six were reactivated by inoculation on MRS or M17 agar, depending upon the type of agar they were originally isolated from. Incubation was carried out at 37 °C for 48 h in a low O2 atmosphere.
2.2. Amplification of the 16S Ribosomal Gene
Colony PCR was performed after bacterial lysis at 95 °C using primers 15f 5′-GCTCAGGAYGAACGCYGG-3′ and 687r 5′-CACCGCTACACATGRADTTC-3′ [9]. The PCR consisted of an initial denaturation phase at 94 °C for 2 min, followed by 35 cycles with three phases: (1) denaturation at 94 °C for 10 s, (2) hybridization at 61 °C for 30 s, and (3) extension at 72 °C for 45 s. At the end of the cycles, a final extension step was carried out at 72 °C for 1 min. The DNA polymerase used was DFS-Taq (BIORON Life Science, Portugal). The PCR product was evaluated and visualized on a 1% (w/w) agarose gel in Tris-acetate-EDTA 1Xbuffer and stained with ethidium bromide, and then purified using the ExoSAP-IT™ reagent (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA) with two incubation periods: (1) digestion, exposure to 37 °C for 15 min, and (2) heat deactivation, exposure to 80 °C for 15 min.
2.3. Sequencing by the Sanger Method
The first sequencing reaction carried out was PCR using the BigDye™ Terminator v3.1 reagent (Applied Biosystems, Thermo Fisher Scientific) using primers 15f and 687r. For capillary electrophoresis, purification was carried out using the SAM/BigDyeXTerminator™ bead solution (Applied Biosystems, Thermo Fisher Scientific), and its product was applied to the SeqStudio Genetic Analyzer (Applied Biosystems, Thermo Fisher Scientific) using the MediumSeq-BDX module. The sequences and chromatograms were visualized using Sequencing Analysis Software v7.0 (Applied Biosystems, Thermo Fisher Scientific).
2.4. Taxonomic Identification of the Biological Sequence
The sequences obtained during sequencing were aligned using the Basic Local Alignment Search Tool (BLAST) of the National Center for Biotechnology Information (NCBI, Bethesda, MD, USA) in the rRNA/ITS database, specifying a search for 16S ribosomal RNA sequences. Matches with an identity percentage greater than or equal to 97% were considered the best identification at the species level.
2.5. Phylogenetic Analysis
The phylogenetic analysis was carried out using the software R 4.3.0, following the steps: (1) performing the multiple alignment of the sequences; (2) determining the distance matrix; and (3) creating the phylogenetic tree using an agglomerative method, Neighbor-Joining. The multiple alignment was carried out using the “MSA” package with the ClustalOmega algorithm, while the “seqinr” package determined the distance matrix, returning an identity matrix with the square root of the pairwise alignment distances. The phylogenetic trees were obtained with the “nj()” function of the “APE” package. With the “ggtree” package and iTol v6.8 software, it was possible to visualize the phylogenetic tree and add annotations.
2.6. Genetic Distance Determination
The calculation of the genetic distance between and within species was carried out using MEGA 11 software with the Maximum Likelihood Composite model, returning results that show the average number of base substitutions per site.
3. Results and Discussion
Table 1 shows the absolute frequency of the genus and species identified by sequencing the 16S ribosomal sequence of the LAB isolated. The two most frequent species among the 46 samples were Lacticaseibacillus paracasei and Leuconostoc mesenteroides. Research has revealed the presence of the genera and species of LABs identified in fermented products, such as foods from Brazil and raw graviera cheese [10,11]. Penland et al. identified the dominance, at different stages of goat cheese production, of the species L. paracesei and L. mesenteroides [12]. Lactobacilli are shown to be more associated with cheese ripening [11], and the L. paracasei species is closely related to goat cheese aroma [12].

Table 1.
Absolute frequency of genus and species identified by sequencing 46 16S ribosomal sequences of lactic acid bacteria isolated from goat cheese.
Table 2 and Table 3 compile the results obtained from the analysis of the genetic distances of the 46 sequences analyzed using MEGA 11 software. Table 2 shows the genetic distance between sequences from the same species, and Table 3 shows the genetic distance between species. L. plantarum was the species that registered only one representative and was excluded from the analysis of genetic distance within the same species.

Table 2.
Genetic distance within the same species, obtained by MEGA 11 software, which returns the average number of substitutions per site. L. plantarum was excluded from the analysis as it has only one representative of the species.

Table 3.
Triangular matrix of genetic distance between species obtained using MEGA 11 software. The results presented are the average number of substitutions per site.
The species with the most uniform sequences was Lactococcus cremoris, and the two species with the closest sequences were L. plantarum and Lactococcus lactis. Genetic distance analysis can shed light on the formation of phylogenetic trees, which are based on evolutionary relationships and the proximity of biological sequences [13]. Figure 1 illustrates the phylogenetic tree built using the Neighbor-Joining method with the 16S ribosomal sequences of lactic acid bacteria isolated from goat cheese. It was possible to observe that the species that showed the lowest average substitution values per site in the analysis of genetic distance within the same species are the ones that form clusters, such as L. paracasei, L. mesenteroides, L. cremoris, and L. lactis.
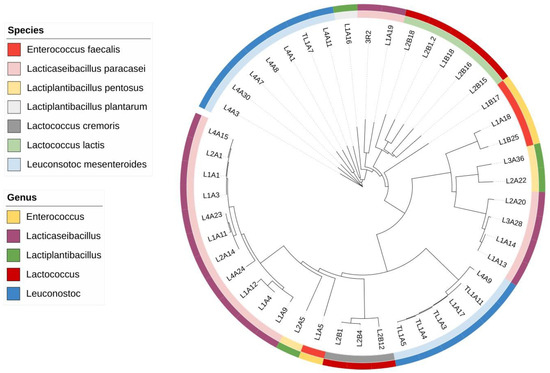
Figure 1.
Phylogenetic tree of the forty-six 16S ribosomal sequences of lactic acid bacteria isolated from raw goat’s cheese. The tree shows the species identified for each sample (inner circle) and the genus of each sample (outer circle).
The construction of the phylogenetic tree highlighted the fact that samples of the same species can have different positions in the tree’s conformation. This dynamic can be explained by the limitation of the 16S ribosomal region in not allowing the identification of subspecies [14] and by the fact that the primers used are only efficient for identification down to the species level [9]. In addition, sequences of the same species that differ in position can mean mutation, which generates genetic variation, and these substitutions reflect a mutation that can either increase or decrease the metabolic functionalities [15] of a group or taxon of LAB, such as antimicrobial capacity and capacities for the development of the sensory characteristics of a product.
4. Conclusions
It can be concluded that Transmontano goat’s raw milk cheese has a diversity of lactic acid bacteria species, a group of bacteria that play different roles in cheese production. The Lacticaseibacillus paracasei species and its representative genus proved to be the most frequent in the 46 samples identified.
Lactococcus cremoris was the species with the most uniform sequences, and the species closest to each other were Lactiplantibacillus plantarum and Lactococcus lactis. Lacticaseibacillus paracasei, Leuconostoc mesenteroides, Lactococcus cremoris, and Lactococcus lactis were the species that formed clusters.
The observation that sequences from the same species can have different evolutionary positions is an important conclusion for future studies that analyze mutations and their implications for the technological capabilities of lactic acid bacteria. It can therefore be concluded that phylogenetics is a powerful tool for the fermented food industry and that the identification of LAB is an investment in increasing food quality and safety.
Author Contributions
Conceptualization, L.C., N.F., U.G.-B., and V.C.; methodology, L.C., N.F., B.N.S., U.G.-B. and V.C.; software, L.C., N.F. and V.C.; validation, V.C.; formal analysis, L.C., N.F., U.G.-B. and V.C.; investigation, L.C., N.F. and V.C.; resources, B.N.S.,U.G.-B. and V.C.; data curation, L.C., N.F.,U.G.-B. and V.C.; writing—original draft preparation, L.C.; writing—review and editing, L.C., N.F.,U.G.-B. and V.C.; visualization, L.C., N.F.,U.G.-B. and V.C.; supervision, U.G.-B. and V.C.; project administration, U.G.-B. and V.C.; funding acquisition, U.G.-B. and V.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Foundation for Science and Technology (FCT, Portugal) through FCT/MCTES (PIDDAC), CIMO (UIDB/00690/2020 and UIDP/00690/2020), and SusTEC (LA/P/0007/2021). EU PRIMA program, FCT for funding the ArtiSaneFood project (PRIMA/0001/2018). B.N.S. acknowledges the financial support provided by the Foundation for Science and Technology (FCT, Portugal) through the PhD grant SFRH/BD/137801/2018.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Summary data are available upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fox, P.F.; McSweeney, P.L.H.; Cogan, T.M.; Guinee, T.P. Cheese: Chemistry, Physics and Microbiology, Volume 1: General Aspects; Elsevier: Cork, Ireland, 2004; ISBN 978-0-08-050093-5. [Google Scholar]
- Hernández-Saldaña, O.F.; Valencia-Posadas, M.; de la Fuente-Salcido, N.M.; Bideshi, D.K.; Barboza-Corona, J.E. Bacteriocinogenic Bacteria Isolated from Raw Goat Milk and Goat Cheese Produced in the Center of México. Indian J. Microbiol. 2016, 56, 301–308. [Google Scholar] [CrossRef]
- Yoon, Y.; Lee, S.; Choi, K.-H. Microbial Benefits and Risks of Raw Milk Cheese. Food Control. 2016, 63, 201–215. [Google Scholar] [CrossRef]
- Scientific Opinion on the Public Health Risks Related to the Consumption of Raw Drinking Milk|EFSA. Available online: https://www.efsa.europa.eu/en/efsajournal/pub/3940 (accessed on 26 June 2023).
- Gonzales-Barron, U.; Gonçalves-Tenório, A.; Rodrigues, V.; Cadavez, V. Foodborne Pathogens in Raw Milk and Cheese of Sheep and Goat Origin: A Meta-Analysis Approach. Curr. Opin. Food Sci. 2017, 18, 7–13. [Google Scholar] [CrossRef]
- Olson, N.F. The Impact of Lactic Acid Bacteria on Cheese Flavor. FEMS Microbiol. Rev. 1990, 7, 131–147. [Google Scholar] [CrossRef]
- Horiike, T. An Introduction to Molecular Phylogenetic Analysis. Rev. Agric. Sci. 2016, 4, 36–45. [Google Scholar] [CrossRef]
- Silva, B.; Faria, A.S.; Cadavez, V.; Teixeira, J.A.; Gonzales-Barron, U. Technological Potential of Lactic Acid Bacteria Isolated from Portuguese Goat’s Raw Milk Cheeses. Biol. Life Sci. Forum 2021, 6, 9. [Google Scholar] [CrossRef]
- Hou, Q.; Bai, X.; Li, W.; Gao, X.; Zhang, F.; Sun, Z.; Zhang, H. Design of Primers for Evaluation of Lactic Acid Bacteria Populations in Complex Biological Samples. Front. Microbiol. 2018, 9, 2045. [Google Scholar] [CrossRef]
- Lima, T.T.M.; de Oliveira Hosken, B.; Venturim, B.C.; Lopes, I.L.; Martin, J.G.P. Traditional Brazilian Fermented Foods: Cultural and Technological Aspects. J. Ethn. Foods 2022, 9, 35. [Google Scholar] [CrossRef]
- Psomas, E.; Sakaridis, I.; Boukouvala, E.; Karatzia, M.-A.; Ekateriniadou, L.V.; Samouris, G. Indigenous Lactic Acid Bacteria Isolated from Raw Graviera Cheese and Evaluation of Their Most Important Technological Properties. Foods 2023, 12, 370. [Google Scholar] [CrossRef]
- Penland, M.; Falentin, H.; Parayre, S.; Pawtowski, A.; Maillard, M.-B.; Thierry, A.; Mounier, J.; Coton, M.; Deutsch, S.-M. Linking Pélardon Artisanal Goat Cheese Microbial Communities to Aroma Compounds during Cheese-Making and Ripening. Int. J. Food Microbiol. 2021, 345, 109130. [Google Scholar] [CrossRef]
- Woese, C.R. Interpreting the Universal Phylogenetic Tree. Proc. Natl. Acad. Sci. USA 2000, 97, 8392–8396. [Google Scholar] [CrossRef]
- Forouhandeh, H.; Vahed, S.Z.; Ahangari, H.; Tarhriz, V.; Hejazi, M.S. Phenotypic and Phylogenetic Characterization of Lactobacillus Species Isolated from Traditional Lighvan Cheese. Food Prod. Process. Nutr. 2021, 3, 20. [Google Scholar] [CrossRef]
- Hershberg, R. Mutation—The Engine of Evolution: Studying Mutation and Its Role in the Evolution of Bacteria. Cold Spring Harb. Perspect. Biol. 2015, 7, a018077. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).