Bivalent System of Deoxyribozymes for Efficient RNA Cleavage †
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA and RNA Fragments Used
2.2. Buffers and Reagents Used
2.3. Experimental Procedure
2.3.1. Cleaving RNA and Cleavage Agents
2.3.2. PAGE Assay and Data Analysis
3. Results and Discussion
3.1. Choosing Target RNA STR-58
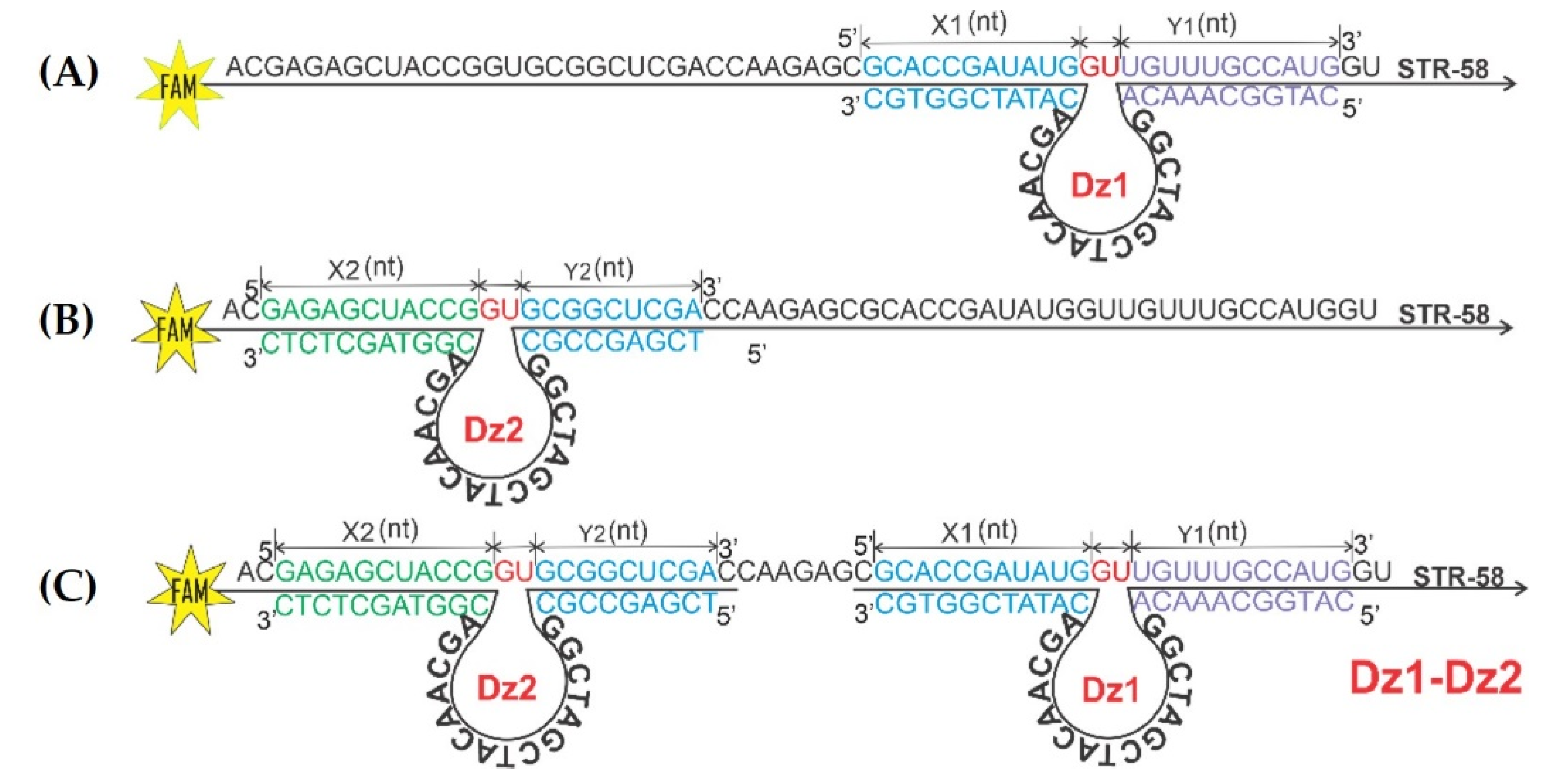
3.2. Design of Catalytic DNAzymes Used in the Study
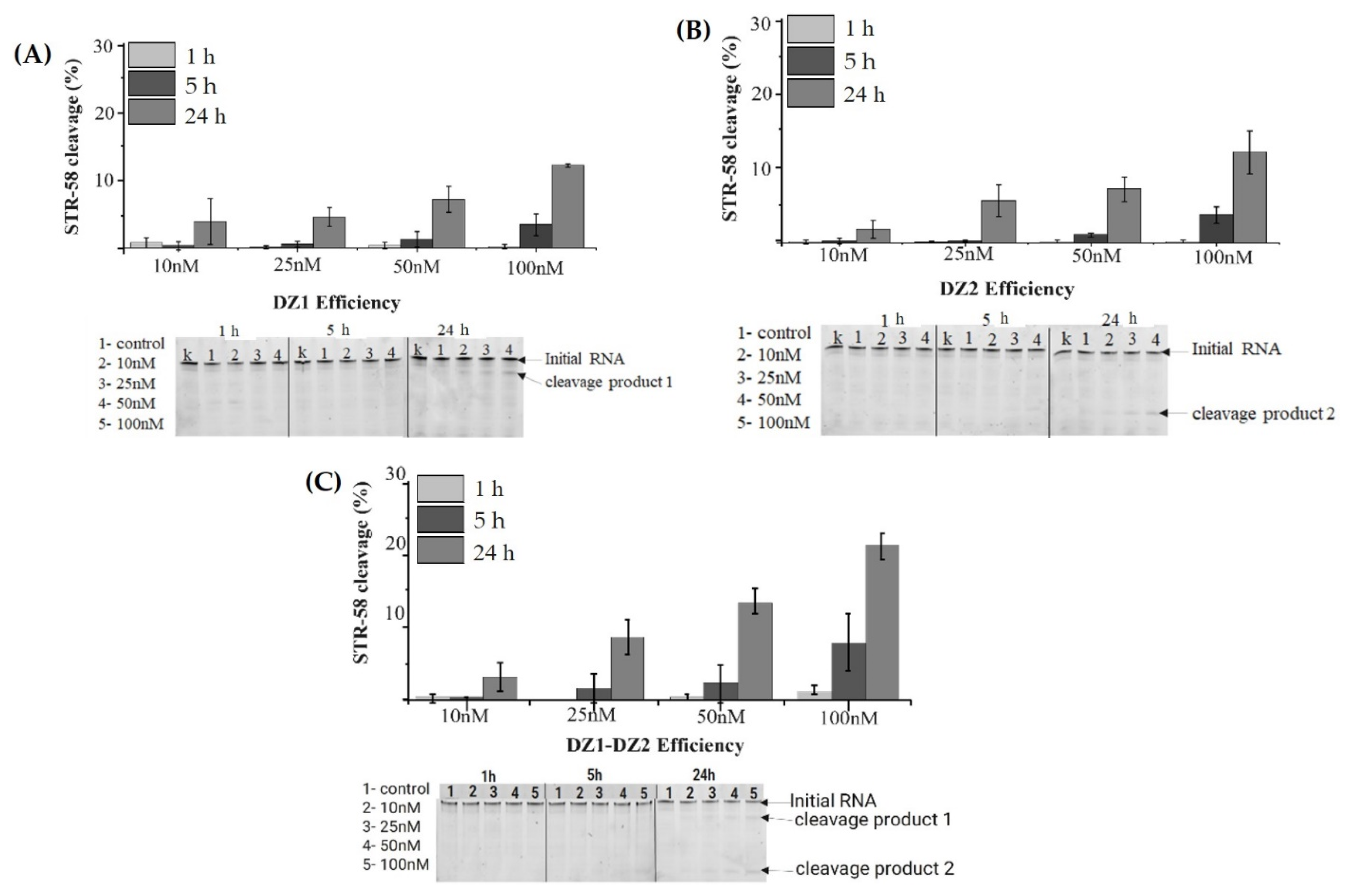
3.3. Cleavage Efficiency of 10-23 DNAzymes (Dz1, Dz2 and Dz1-Dz2 Association)
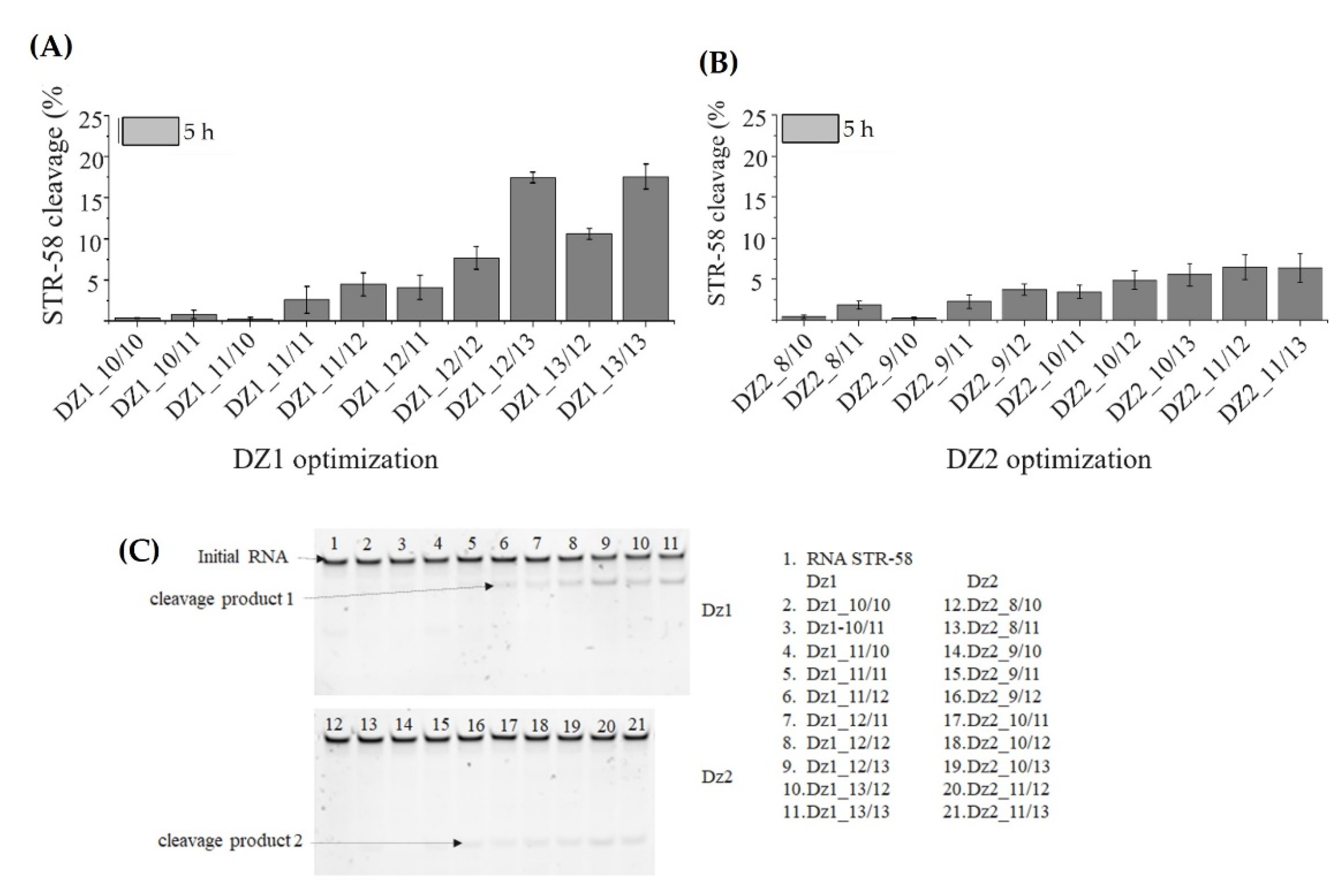
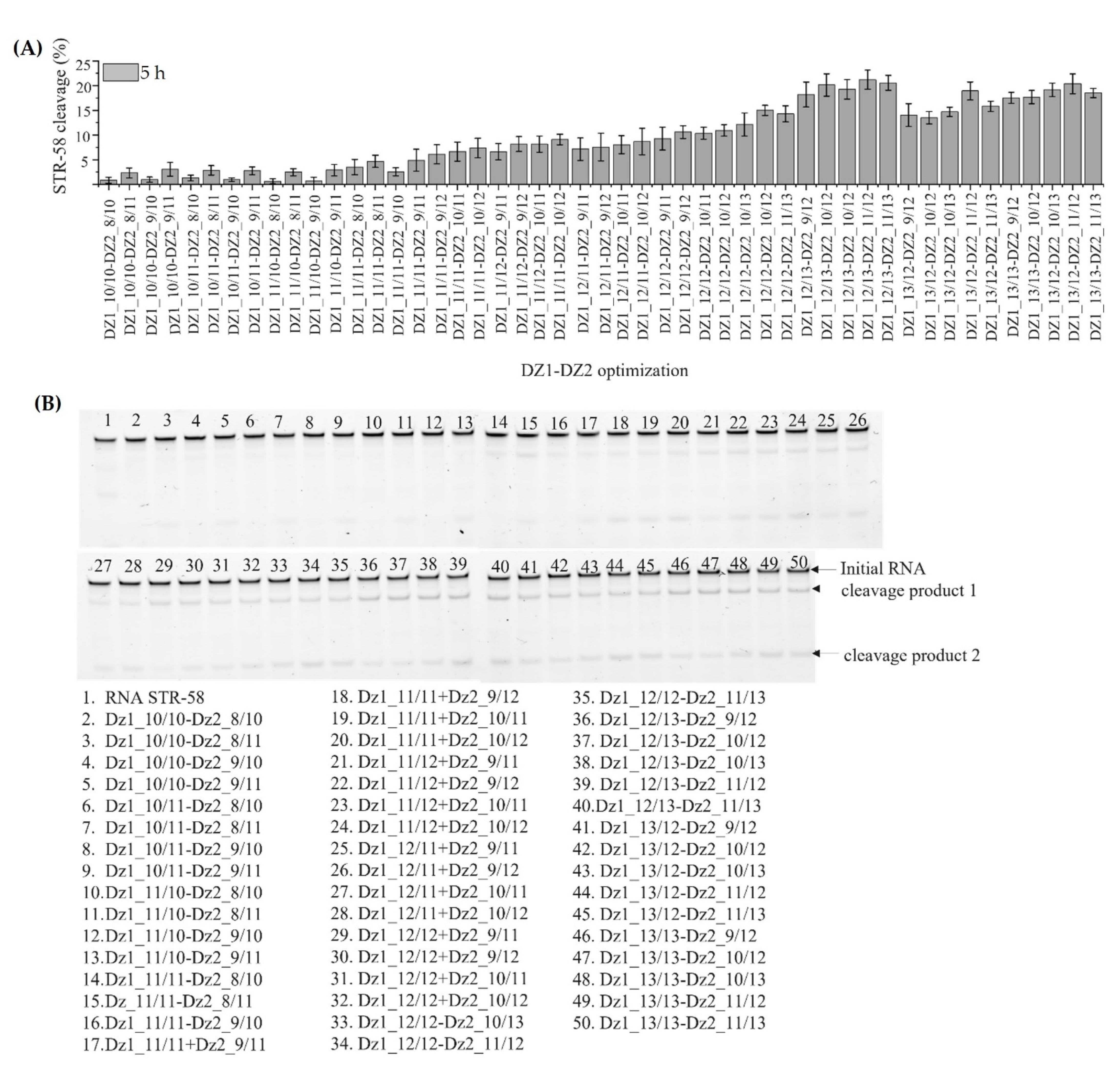
3.4. Optimization of Dz1, Dz2 and Dz1-Dz2 Association
3.5. Discussion of Results
4. Conclusions
- (1)
- Multivalently associated DNAzymes are higher in catalytic cleavage activity in cleaving STR-58 than monovalent constructions.
- (2)
- Multivalent association of DNAzymes improves DNAzyme hybridization and affinity to targeted substrates, hence increasing their catalytic activity as compared to monovalent DNAzymes.
- (3)
- Increasing the length of 10-23 Dz binding arms increased the performance of DNAzymes.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kaufmann, S.H.E.; Dorhoi, A.; Hotchkiss, R.S.; Bartenschlager, R. Host-directed therapies for bacterial and viral infections. Nat. Rev. Drug Discov. 2018, 17, 35–56. [Google Scholar] [CrossRef] [PubMed]
- Lapteva, L.; Purohit-Sheth, T.; Serabian, M.; Puri, R.K. Clinical Development of Gene Therapies: The First Three Decades and Counting. Mol. Ther. Methods Clin. Dev. 2020, 19, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Deverman, B.E.; Ravina, B.M.; Bankiewicz, K.S.; Paul, S.M.; Sah, D.W.Y. Gene therapy for neurological disorders: Progress and prospects. Nat. Rev. Drug Discov. 2018, 17, 641–659. [Google Scholar] [CrossRef] [PubMed]
- Moore, N.A.; Morral, N.; Ciulla, T.A.; Bracha, P. Gene therapy for inherited retinal and optic nerve degenerations. Expert Opin. Biol. Ther. 2018, 18, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Veroniaina, H.; Su, N.; Sha, K.; Jiang, F.; Wu, Z.; Qi, X. Applications and developments of gene therapy drug delivery systems for genetic diseases. Asian J. Pharm. Sci. 2021, 16, 687–703. [Google Scholar] [CrossRef]
- Vakulskas, C.A.; Behlke, M.A. Evaluation and Reduction of CRISPR Off-Target Cleavage Events. Nucleic. Acid. Ther. 2019, 29, 167–174. [Google Scholar] [CrossRef]
- Gonçalves, G.A.R.; Paiva, R.D.M.A. Gene therapy: Advances, challenges and perspectives. Einstein 2017, 15, 369–375. [Google Scholar] [CrossRef]
- He, M.; He, M.; Nie, C.; Yi, J.; Zhang, J.; Chen, T.; Chu, X. mRNA-activated multifunctional DNAzyme nanotweezer for intracellular mRNA sensing and gene therapy. ACS Appl. Mater. Interfaces 2021, 13, 8015–8025. [Google Scholar] [CrossRef]
- Xue, T.; Sheng, A.; Mao, D.; Zhang, Y.; Liu, Z.; Zhang, J. DNAzyme-based colorimetric assay and its application for lipopolysaccharide analysis assisted by oxime chemistry. Biosens. Bioelectron. 2021, 189, 113379. [Google Scholar] [CrossRef]
- Watts, J.K.; Corey, D.R. Silencing disease genes in the laboratory and the clinic. J. Pathol. 2012, 226, 365–379. [Google Scholar] [CrossRef]
- Nedorezova, D.D.; Dubovichenko, M.V.; Belyaeva, E.P.; Grigorieva, E.D.; Peresadina, A.V.; Kolpashchikov, D.M. Specificity of oligonucleotide gene therapy (OGT) agents. Theranostics 2022, 12, 7132–7157. [Google Scholar] [CrossRef] [PubMed]
- Warashina, M.; Kuwabara, T.; Kato, Y.; Sano, M.; Taira, K. RNA–protein hybrid ribozymes that efficiently cleave any mRNA independently of the structure of the target RNA. Proc. Natl. Acad. Sci. USA 2001, 98, 5572–5577. [Google Scholar] [CrossRef] [PubMed]
- Böhmer, V.I.; Szymanski, W.; Feringa, B.L.; Elsinga, P.H. Multivalent Probes in Molecular Imaging: Reality or Future? Trends Mol. Med. 2021, 27, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Choi, H.; Heo, Y.; Kim, C.; Kim, M.; Kim, K.T. Biosensors Based on Bivalent and Multivalent Recognition by Nucleic Acid Scaffolds. Appl. Sci. 2022, 12, 1717. [Google Scholar] [CrossRef]
- Bernardi, A.; Jiménez-Barbero, J.; Casnati, A.; de Castro, C.; Darbre, T.; Fieschi, F.; Finne, J.; Funken, H.; Jaeger, K.; Lahmann, M.; et al. Multivalent glycoconjugates as anti-pathogenic agents. Chem. Soc. Rev. 2013, 42, 4709–4727. [Google Scholar] [CrossRef]
- Yang, D.K.; Kuo, C.J.; Chen, L.C. Synthetic multivalent DNAzymes for enhanced hydrogen peroxide catalysis and sensitive colorimetric glucose detection. Anal. Chim. Acta 2015, 856, 96–102. [Google Scholar] [CrossRef]
- Chen, F.; Li, Z.; Wang, R.; Liu, B.; Zeng, Z.; Zhang, H.; Zhang, J. Inhibition of ampicillin-resistant bacteria by novel mono-DNAzymes and di-DNAzyme targeted to beta-lactamase mRNA. Oligonucleotides 2004, 14, 80–89. [Google Scholar] [CrossRef]
- Sunde, M.; Norström, M. The genetic background for streptomycin resistance in Escherichia coli influences the distribution of MICs. J. Antimicrob. Chemother. 2005, 56, 87–90. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, G.H.; Koh, Y.J.; Jung, J.S. Identification of strA-strB Genes in Streptomycin-Resistant Pseudomonas syringae pv. actinidiae Biovar 2 Strains Isolated in Korea. Plant. Pathol. J. 2021, 37, 489–493. [Google Scholar] [CrossRef]
- Nedorezova, D.D.; Fakhardo, A.F.; Nemirich, D.V.; Bryushkova, E.A.; Kolpashchikov, D.M. Towards DNA Nanomachines for Cancer Treatment: Achieving Selective and Efficient Cleavage of Folded RNA. Angew. Chem. Int. Ed. 2019, 58, 4654–4658. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.L.; Ai, S.M.; Liang, J.; Sang, P.; Ji, X.L.; Liu, S.Q. Insights into Protein-Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef] [PubMed]
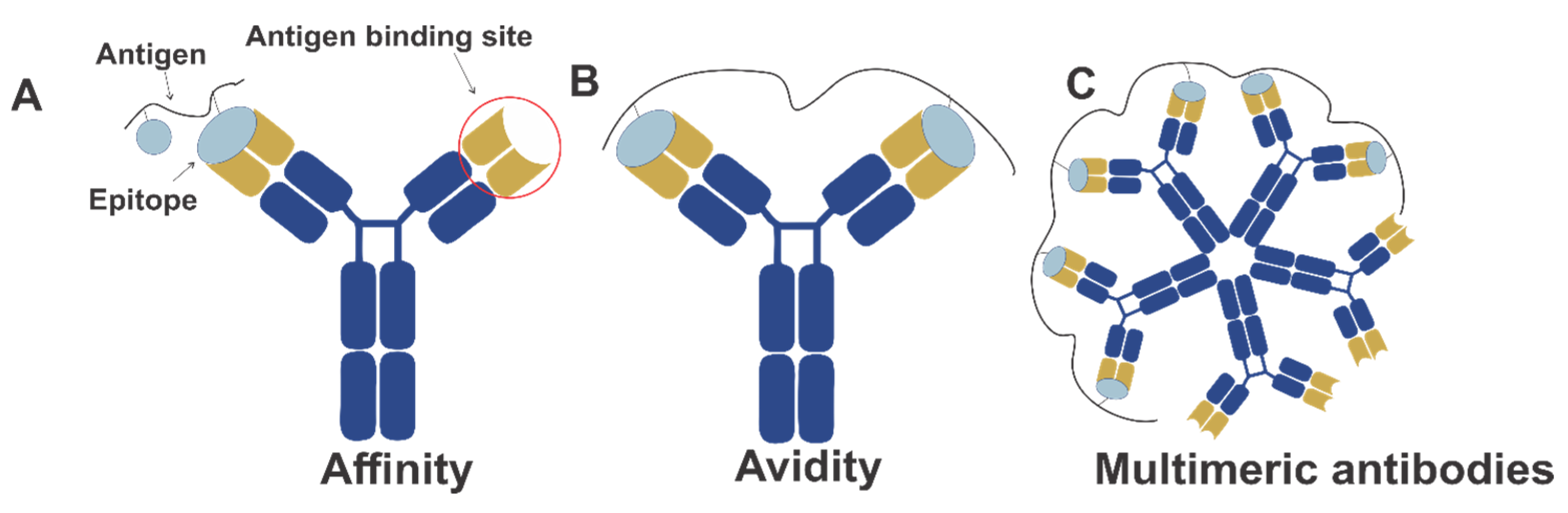
- Rudnick, S.I.; Adams, G.P. Affinity and avidity in antibody-based tumor targeting. Cancer Biother. Radiopharm. 2009, 24, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Schubert, S.; Gül, D.C.; Grunert, H.-P.; Zeichhardt, H.; Erdmann, V.A.; Kurreck, J. RNA cleaving ‘10-23′ DNAzymes with enhanced stability and activity. Nucleic. Acids. Res. 2003, 31, 982–5992. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Zhang, D. Linker Design Impacts Antibody-Drug Conjugate Pharmacokinetics and Efficacy via Modulating the Stability and Payload Release Efficiency. Front. Pharmacol. 2021, 12, 687926. [Google Scholar] [CrossRef]


| Name | Arms | Sequence | Melting Point (°C) |
|---|---|---|---|
| Dz1 | X1 | CATATCGGTGC | 32.4 |
| Y1 | CATGGCAAACA | 34.5 | |
| Dz2 | X2 | CGGTAGCTCTC | 34.1 |
| Y2 | TCGAGCCGC | 34.9 | |
| Catalytic core | A GGC TAG CTA CAA |
| Reagents | Usage | Company |
|---|---|---|
| Oligonucleotides | For building DNA constructs | DNAsynthesis |
| Fluorescent Substrate | Produce a fluorescent signal after detection of the nucleic acids | DNAsynthesis |
| H2O RNAse Free | Dilution of oligos and buffer preparation | QIAGEN |
| KCl, | Cleavage buffer preparation | ROTH |
| MgCl2, | Cleavage buffer preparation | ROTH |
| HEPES | Cleavage buffer preparation | ROTH |
| NaCl | Cleavage buffer preparation | ROTH |
| Hepes | Cleavage buffer preparation | ROTH |
| Tris | TBE buffer preparation | Helicon |
| Boric acid | TBE buffer preparation | Helicon |
| EDTA | TBE buffer preparation | Helicon |
| Acrylamide (AA) | PAGE preparation | Helicon |
| Bis-acrylamide (BA) | PAGE preparation | Helicon |
| APS | PAGE preparation | |
| Urea | Stop buffer and PAGE buffer preparation | ROTH |
| TEMED | Stop buffer and PAGE buffer preparation | |
| Bromophenol blue (optional) | Stop buffer preparation | |
| Ethidium Bromide | Staining nucleic acid | |
| Molecular rulers | DNA ladders | Everogene |
| Loading buffer 4×, 6× | Sample loading | Everogene |
| Name of Oligonucleotides | Length of ‘X’ Arm (nt) | Y arm Tm (°C) | Length of ‘Y’ Arm (nt) | X arm Tm (°C) |
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 |
| Dz1_10/10 | 10 | 26.2 | 10 | 23.4 °C |
| Dz1-10/11 | 10 | 26.2 | 11 | 32.4 °C |
| Dz1_11/10 | 11 | 32.3 | 10 | 23.4 °C |
| Dz1_11/11 | 11 | 32.3 | 11 | 32.4 |
| Dz1_11/12 | 11 | 32.3 | 12 | 39.6 |
| Dz1_12/11 | 12 | 38.3 | 11 | 32.4 |
| 1 | 2 | 3 | 4 | 5 |
| Dz1_12/12 | 12 | 38.3 | 12 | 39.6 |
| Dz1_12/13 | 12 | 38.3 | 13 | 45.8 |
| Dz1_13/12 | 13 | 40.7 | 12 | 39.6 |
| Dz1_13/13 | 13 | 40.7 | 13 | 45.8 |
| Dz2_8/10 | 8 | 28.3 | 10 | 28.7 |
| Dz2_8/11 | 8 | 28.3 | 11 | 34.1 |
| Dz2_9/10 | 9 | 34.9 | 10 | 28.7 |
| Dz2_9/11 | 9 | 34.9 | 11 | 34.1 |
| Dz2_9/12 | 9 | 34.9 | 12 | 41.2 |
| Dz2_10/11 | 10 | 39.4 | 11 | 39.4 |
| Dz2_10/12 | 10 | 39.4 | 12 | 41.2 |
| Dz2_10/13 | 10 | 39.4 | 13 | 49.6 |
| Dz2_11/12 | 11 | 45.3 | 12 | 41.2 |
| Dz2_11/13 | 11 | 45.3 | 13 | 49.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batsa, M.; Dubovichenko, M.V.; Kolpashchikov, D.M. Bivalent System of Deoxyribozymes for Efficient RNA Cleavage. Biol. Life Sci. Forum 2022, 20, 6. https://doi.org/10.3390/IECBM2022-13510
Batsa M, Dubovichenko MV, Kolpashchikov DM. Bivalent System of Deoxyribozymes for Efficient RNA Cleavage. Biology and Life Sciences Forum. 2022; 20(1):6. https://doi.org/10.3390/IECBM2022-13510
Chicago/Turabian StyleBatsa, Michael, Mikhail V. Dubovichenko, and Dmitry M. Kolpashchikov. 2022. "Bivalent System of Deoxyribozymes for Efficient RNA Cleavage" Biology and Life Sciences Forum 20, no. 1: 6. https://doi.org/10.3390/IECBM2022-13510
APA StyleBatsa, M., Dubovichenko, M. V., & Kolpashchikov, D. M. (2022). Bivalent System of Deoxyribozymes for Efficient RNA Cleavage. Biology and Life Sciences Forum, 20(1), 6. https://doi.org/10.3390/IECBM2022-13510






